SIRT7 Is a Lysine Deacylase with a Preference for Depropionylation and Demyristoylation
Abstract
1. Introduction
2. Results
2.1. Catalytic Efficiency of SIRT7 on Different Acyl-Modified Peptides
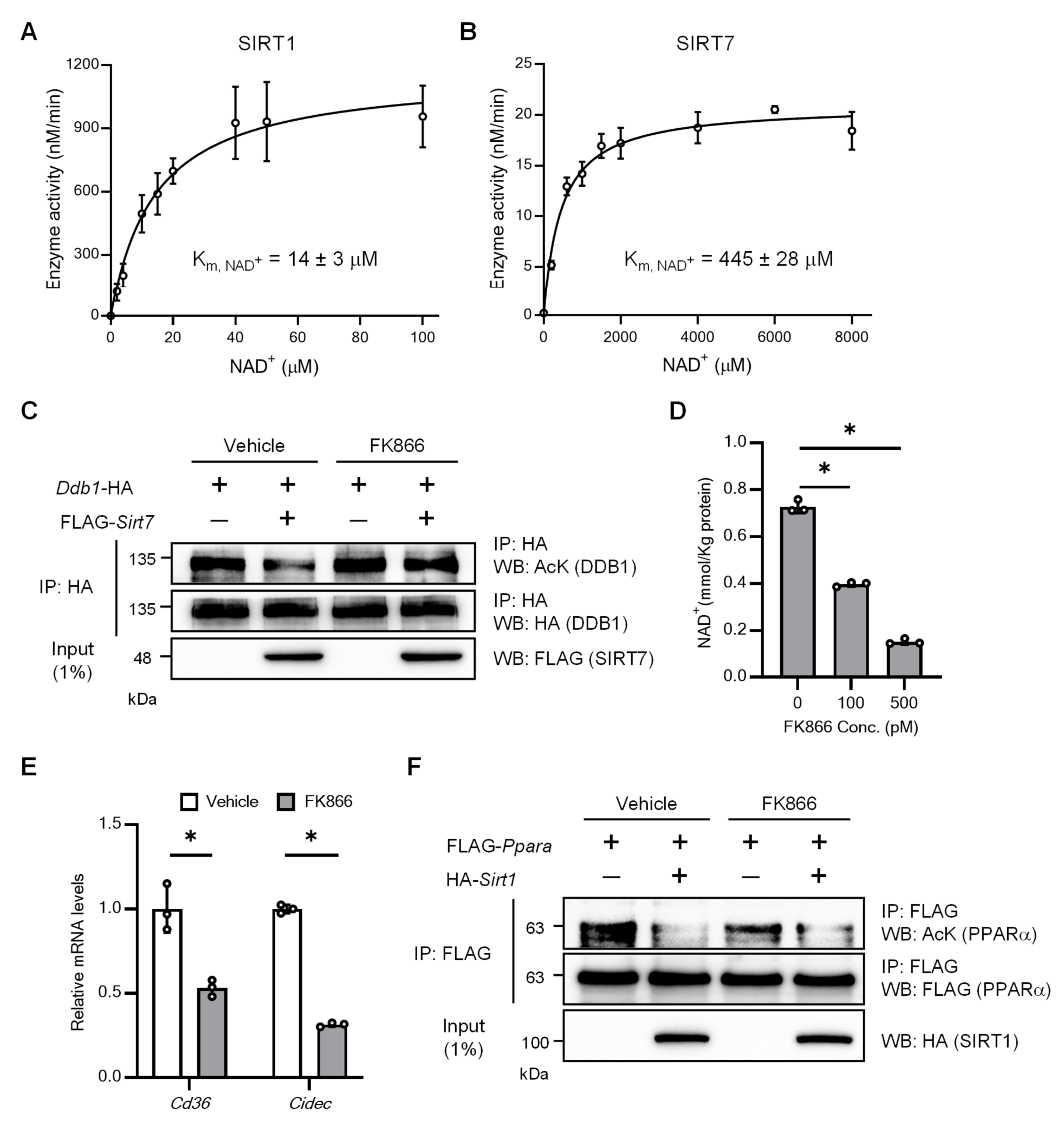
2.2. SIRT7 Has a Low Affinity for NAD+ for Deacetylation
2.3. SIRT7 Has a High Affinity for NAD+ in Depropionylation and Demyristoylation
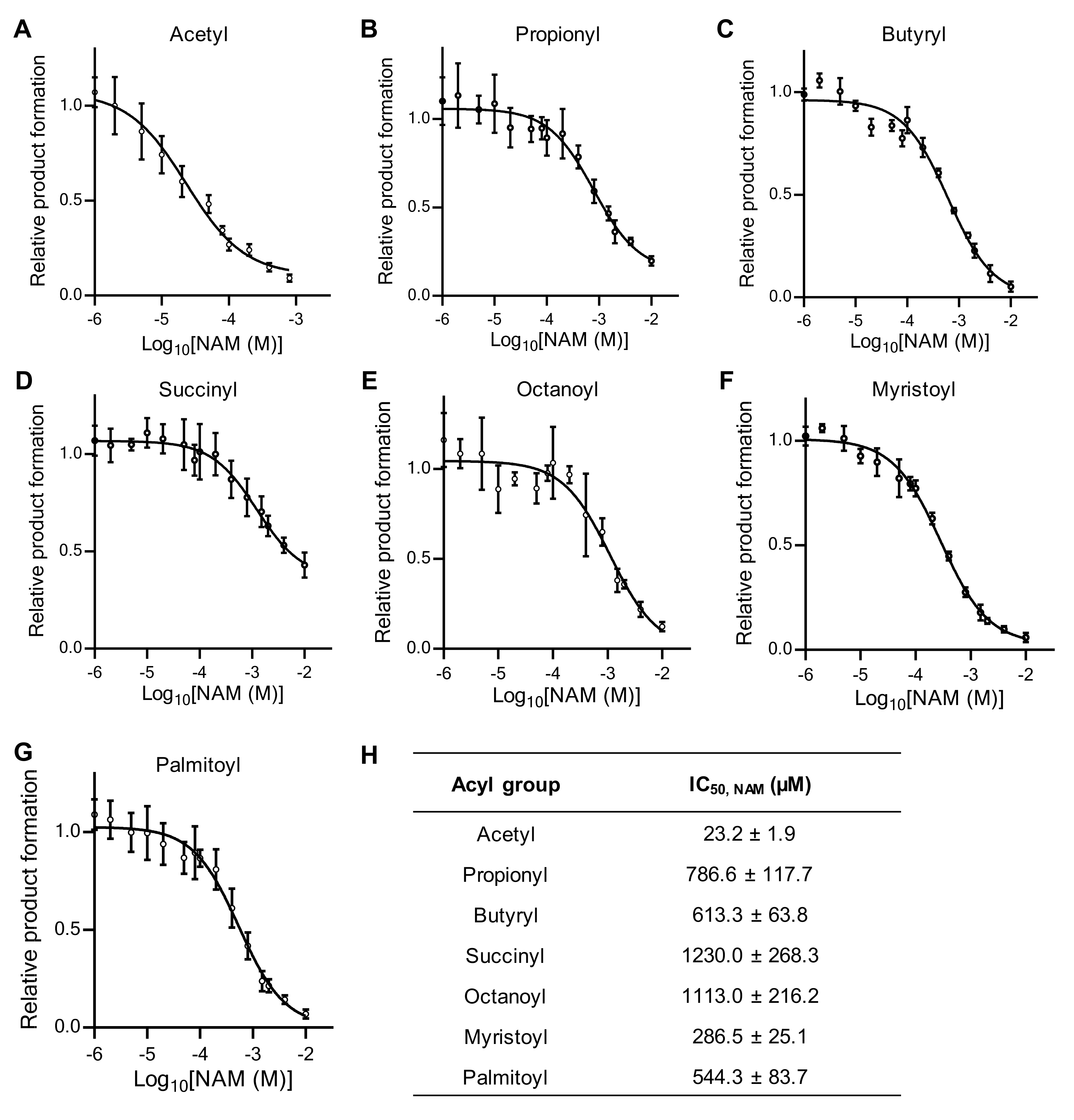
2.4. NAM Is a Poor Inhibitor of SIRT7 with Non-Acetylated Substrates
3. Discussion
4. Materials and Methods
4.1. Peptides
4.2. Preparation of Recombinant SIRT7 and SIRT1
4.3. In Vitro Deacylation Assay
4.4. LC-MS/MS Analysis
4.5. Cell Culture
4.6. Plasmids
4.7. NAD+ Assay
4.8. Detection of Lysine Acetylation and Western Blotting
4.9. Cellular Gene Expression
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SIRT7 | Sirtuin 7 |
| NAD+ | Nicotinamide adenine dinucleotide |
| NAM | Nicotinamide |
| LC-MS/MS | Liquid chromatography–tandem mass spectrometry |
| DDB1 | Damage-specific DNA-binding protein 1 |
| NAMPT | Nicotinamide phosphoribosyltransferase |
| CIDEC | Cell death-inducing DFFA-like effector c |
| IC50 | Half-maximal inhibitory concentration |
References
- Sabari, B.R.; Zhang, D.; Allis, C.D.; Zhao, Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017, 18, 90–101. [Google Scholar] [PubMed]
- Wang, M.; Lin, H. Understanding the Function of Mammalian Sirtuins and Protein Lysine Acylation. Annu. Rev. Biochem. 2021, 90, 245–285. [Google Scholar] [PubMed]
- Houtkooper, R.; Pirinen, E.; Auwerx, J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238. [Google Scholar] [PubMed]
- Wu, Q.-J.; Zhang, T.-N.; Chen, H.-H.; Yu, X.-F.; Lv, J.-L.; Liu, Y.-Y.; Liu, Y.-S.; Zheng, G.; Zhao, J.-Q.; Wei, Y.-F. The sirtuin family in health and disease. Signal Transduct. Target. Ther. 2022, 7, 402. [Google Scholar]
- Tanno, M.; Sakamoto, J.; Miura, T.; Shimamoto, K.; Horio, Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 2007, 282, 6823–6832. [Google Scholar] [CrossRef]
- Kiran, S.; Chatterjee, N.; Singh, S.; Kaul, S.C.; Wadhwa, R.; Ramakrishna, G. Intracellular distribution of human SIRT 7 and mapping of the nuclear/nucleolar localization signal. FEBS J. 2013, 280, 3451–3466. [Google Scholar] [CrossRef]
- Michishita, E.; Park, J.Y.; Burneskis, J.M.; Barrett, J.C.; Horikawa, I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 2005, 16, 4623–4635. [Google Scholar]
- Haigis, M.C.; Mostoslavsky, R.; Haigis, K.M.; Fahie, K.; Christodoulou, D.C.; Murphy, A.J.; Valenzuela, D.M.; Yancopoulos, G.D.; Karow, M.; Blander, G. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell 2006, 126, 941–954. [Google Scholar] [CrossRef]
- Mathias, R.A.; Greco, T.M.; Oberstein, A.; Budayeva, H.G.; Chakrabarti, R.; Rowland, E.A.; Kang, Y.; Shenk, T.; Cristea, I.M. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell 2014, 159, 1615–1625. [Google Scholar] [CrossRef]
- Anderson, K.A.; Huynh, F.K.; Fisher-Wellman, K.; Stuart, J.D.; Peterson, B.S.; Douros, J.D.; Wagner, G.R.; Thompson, J.W.; Madsen, A.S.; Green, M.F. SIRT4 is a lysine deacylase that controls leucine metabolism and insulin secretion. Cell Metab. 2017, 25, 838–855.e815. [Google Scholar]
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Kim, J.H.; Choi, B.H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 2011, 334, 806–809. [Google Scholar] [CrossRef]
- Rardin, M.J.; He, W.; Nishida, Y.; Newman, J.C.; Carrico, C.; Danielson, S.R.; Guo, A.; Gut, P.; Sahu, A.K.; Li, B. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013, 18, 920–933. [Google Scholar] [CrossRef]
- Jiang, H.; Khan, S.; Wang, Y.; Charron, G.; He, B.; Sebastian, C.; Du, J.; Kim, R.; Ge, E.; Mostoslavsky, R. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 2013, 496, 110–113. [Google Scholar] [CrossRef]
- Feldman, J.L.; Baeza, J.; Denu, J.M. Activation of the Protein Deacetylase SIRT6 by Long-chain Fatty Acids and Widespread Deacylation by Mammalian Sirtuins. J. Biol. Chem. 2013, 288, 31350–31356. [Google Scholar] [CrossRef]
- Barber, M.F.; Michishita-Kioi, E.; Xi, Y.; Tasselli, L.; Kioi, M.; Moqtaderi, Z.; Tennen, R.I.; Paredes, S.; Young, N.L.; Chen, K. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 2012, 487, 114–118. [Google Scholar] [CrossRef]
- Yamagata, K.; Mizumoto, T.; Yoshizawa, T. The Emerging Role of SIRT7 in Glucose and Lipid Metabolism. Cells 2023, 13, 48. [Google Scholar] [CrossRef]
- Tong, Z.; Wang, Y.; Zhang, X.; Kim, D.D.; Sadhukhan, S.; Hao, Q.; Lin, H. SIRT7 is activated by DNA and deacetylates histone H3 in the chromatin context. ACS Chem. Biol. 2016, 11, 742–747. [Google Scholar] [CrossRef]
- Tong, Z.; Wang, M.; Wang, Y.; Kim, D.D.; Grenier, J.K.; Cao, J.; Sadhukhan, S.; Hao, Q.; Lin, H. SIRT7 is an RNA-activated protein lysine deacylase. ACS Chem. Biol. 2017, 12, 300–310. [Google Scholar] [CrossRef]
- Cantó, C.; Menzies, K.J.; Auwerx, J. NAD+ metabolism and the control of energy homeostasis: A balancing act between mitochondria and the nucleus. Cell Metab. 2015, 22, 31–53. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Cantó, C.; Wanders, R.J.; Auwerx, J. The secret life of NAD+: An old metabolite controlling new metabolic signaling pathways. Endocr. Rev. 2010, 31, 194–223. [Google Scholar] [CrossRef]
- Pacholec, M.; Bleasdale, J.E.; Chrunyk, B.; Cunningham, D.; Flynn, D.; Garofalo, R.S.; Griffith, D.; Griffor, M.; Loulakis, P.; Pabst, B. SRT1720, SRT2183, SRT1460, and Resveratrol Are Not Direct Activators of SIRT1. J. Biol. Chem. 2010, 285, 8340–8351. [Google Scholar] [PubMed]
- Feldman, J.L.; Dittenhafer-Reed, K.E.; Kudo, N.; Thelen, J.N.; Ito, A.; Yoshida, M.; Denu, J.M. Kinetic and structural basis for acyl-group selectivity and NAD+ dependence in sirtuin-catalyzed deacylation. Biochemistry 2015, 54, 3037–3050. [Google Scholar] [PubMed]
- Borra, M.T.; Langer, M.R.; Slama, J.T.; Denu, J.M. Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases. Biochemistry 2004, 43, 9877–9887. [Google Scholar]
- Hirschey, M.D.; Shimazu, T.; Jing, E.; Grueter, C.A.; Collins, A.M.; Aouizerat, B.; Stančáková, A.; Goetzman, E.; Lam, M.M.; Schwer, B. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol. Cell 2011, 44, 177–190. [Google Scholar] [CrossRef]
- Laurent, G.; German, N.J.; Saha, A.K.; de Boer, V.C.; Davies, M.; Koves, T.R.; Dephoure, N.; Fischer, F.; Boanca, G.; Vaitheesvaran, B. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol. Cell 2013, 50, 686–698. [Google Scholar]
- Fischer, F.; Gertz, M.; Suenkel, B.; Lakshminarasimhan, M.; Schutkowski, M.; Steegborn, C. Sirt5 Deacylation Activities Show Differential Sensitivities to Nicotinamide Inhibition. PLoS ONE 2012, 7, e45098. [Google Scholar] [CrossRef]
- Kugel, S.; Feldman, J.L.; Klein, M.A.; Silberman, D.M.; Sebastián, C.; Mermel, C.; Dobersch, S.; Clark, A.R.; Getz, G.; Denu, J.M. Identification of and molecular basis for SIRT6 loss-of-function point mutations in cancer. Cell Rep. 2015, 13, 479–488. [Google Scholar] [CrossRef]
- Zhang, X.; Khan, S.; Jiang, H.; Antonyak, M.A.; Chen, X.; Spiegelman, N.A.; Shrimp, J.H.; Cerione, R.A.; Lin, H. Identifying the functional contribution of the defatty-acylase activity of SIRT6. Nat. Chem. Biol. 2016, 12, 614–620. [Google Scholar]
- Kuznetsov, V.I.; Liu, W.H.; Klein, M.A.; Denu, J.M. Potent activation of NAD+-dependent deacetylase Sirt7 by nucleosome binding. ACS Chem. Biol. 2022, 17, 2248–2261. [Google Scholar]
- Bitterman, K.J.; Anderson, R.M.; Cohen, H.Y.; Latorre-Esteves, M.; Sinclair, D.A. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 2002, 277, 45099–45107. [Google Scholar]
- Lagunas-Rangel, F.A. SIRT7 in the aging process. Cell. Mol. Life Sci. 2022, 79, 297. [Google Scholar] [PubMed]
- Vaquero, A.; Scher, M.; Lee, D.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell 2004, 16, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, T.; Karim, M.F.; Sato, Y.; Senokuchi, T.; Miyata, K.; Fukuda, T.; Go, C.; Tasaki, M.; Uchimura, K.; Kadomatsu, T. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab. 2014, 19, 712–721. [Google Scholar] [PubMed]
- Li, L.; Shi, L.; Yang, S.; Yan, R.; Zhang, D.; Yang, J.; He, L.; Li, W.; Yi, X.; Sun, L.; et al. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat. Commun. 2016, 7, 12235. [Google Scholar]
- Bao, X.; Liu, Z.; Zhang, W.; Gladysz, K.; Fung, Y.M.E.; Tian, G.; Xiong, Y.; Wong, J.W.H.; Yuen, K.W.Y.; Li, X.D. Glutarylation of histone H4 lysine 91 regulates chromatin dynamics. Mol. Cell 2019, 76, 660–675.e669. [Google Scholar] [CrossRef]
- Karim, M.F.; Yoshizawa, T.; Sobuz, S.U.; Sato, Y.; Yamagata, K. Sirtuin 7-dependent deacetylation of DDB1 regulates the expression of nuclear receptor TR4. Biochem. Biophys. Res. Commun. 2017, 490, 423–428. [Google Scholar]
- Wei, Y.; Xiang, H.; Zhang, W. Review of various NAMPT inhibitors for the treatment of cancer. Front. Pharmacol. 2022, 13, 970553. [Google Scholar] [CrossRef]
- Suh, J.H.; Kim, K.H.; Conner, M.E.; Moore, D.D.; Preidis, G.A. Hepatic PPARα Is destabilized by SIRT1 deacetylase in undernourished male mice. Front. Nutr. 2022, 9, 831879. [Google Scholar] [CrossRef]
- Berndsen, C.E.; Denu, J.M. Assays for mechanistic investigations of protein/histone acetyltransferases. Methods 2005, 36, 321–331. [Google Scholar] [CrossRef]
- Madsen, A.S.; Andersen, C.; Daoud, M.; Anderson, K.A.; Laursen, J.S.; Chakladar, S.; Huynh, F.K.; Colaço, A.R.; Backos, D.S.; Fristrup, P. Investigating the sensitivity of NAD+-dependent sirtuin deacylation activities to NADH. J. Biol. Chem. 2016, 291, 7128–7141. [Google Scholar] [CrossRef]
- Cambronne, X.A.; Stewart, M.L.; Kim, D.; Jones-Brunette, A.M.; Morgan, R.K.; Farrens, D.L.; Cohen, M.S.; Goodman, R.H. Biosensor reveals multiple sources for mitochondrial NAD+. Science 2016, 352, 1474–1477. [Google Scholar] [CrossRef]
- Sallin, O.; Reymond, L.; Gondrand, C.; Raith, F.; Koch, B.; Johnsson, K. Semisynthetic biosensors for mapping cellular concentrations of nicotinamide adenine dinucleotides. eLife 2018, 7, e32638. [Google Scholar] [PubMed]
- Yoshizawa, T.; Sato, Y.; Sobuz, S.U.; Mizumoto, T.; Tsuyama, T.; Karim, M.F.; Miyata, K.; Tasaki, M.; Yamazaki, M.; Kariba, Y.; et al. SIRT7 suppresses energy expenditure and thermogenesis by regulating brown adipose tissue functions in mice. Nat. Commun. 2022, 13, 7439. [Google Scholar] [PubMed]
- Mizumoto, T.; Yoshizawa, T.; Sato, Y.; Ito, T.; Tsuyama, T.; Satoh, A.; Araki, S.; Tsujita, K.; Tamura, M.; Oike, Y.; et al. SIRT7 Deficiency Protects against Aging-Associated Glucose Intolerance and Extends Lifespan in Male Mice. Cells 2022, 11, 3609. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.W.; Liang, Y.L.; Zhang, Q.X.; Wang, D.; Lei, M.Z.; Qu, J.; He, X.H.; Lei, Q.Y.; Wang, Y.P. Arginine methylation of SIRT7 couples glucose sensing with mitochondria biogenesis. EMBO Rep. 2018, 19, e46377. [Google Scholar]
- Tang, X.; Li, G.; Shi, L.; Su, F.; Qian, M.; Liu, Z.; Meng, Y.; Sun, S.; Li, J.; Liu, B. Combined intermittent fasting and ERK inhibition enhance the anti-tumor effects of chemotherapy via the GSK3β-SIRT7 axis. Nat. Commun. 2021, 12, 5058. [Google Scholar]
- Ianni, A.; Kumari, P.; Tarighi, S.; Simonet, N.G.; Popescu, D.; Guenther, S.; Hölper, S.; Schmidt, A.; Smolka, C.; Yue, S.; et al. SIRT7-dependent deacetylation of NPM promotes p53 stabilization following UV-induced genotoxic stress. Proc. Natl. Acad. Sci. USA 2021, 118, e2015339118. [Google Scholar]
- Fukuda, M.; Yoshizawa, T.; Karim, M.F.; Sobuz, S.U.; Korogi, W.; Kobayasi, D.; Okanishi, H.; Tasaki, M.; Ono, K.; Sawa, T.; et al. SIRT7 has a critical role in bone formation by regulating lysine acylation of SP7/Osterix. Nat. Commun. 2018, 9, 2833. [Google Scholar] [CrossRef]
- Avalos, J.L.; Bever, K.M.; Wolberger, C. Mechanism of Sirtuin Inhibition by Nicotinamide: Altering the NAD+ Cosubstrate Specificity of a Sir2 Enzyme. Mol. Cell 2005, 17, 855–868. [Google Scholar]
- Sobuz, S.U.; Sato, Y.; Yoshizawa, T.; Karim, F.; Ono, K.; Sawa, T.; Miyamoto, Y.; Oka, M.; Yamagata, K. SIRT7 regulates the nuclear export of NF-κB p65 by deacetylating Ran. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2019, 1866, 1355–1367. [Google Scholar]
- Karim, M.F.; Yoshizawa, T.; Sato, Y.; Sawa, T.; Tomizawa, K.; Akaike, T.; Yamagata, K. Inhibition of H3K18 deacetylation of Sirt7 by Myb-binding protein 1a (Mybbp1a). Biochem. Biophys. Res. Commun. 2013, 441, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, K.; Liu, J.; Kato, D.; Kurumizaka, H.; Yamatsugu, K.; Kanai, M.; Kawashima, S.A. LC–MS/MS-based quantitative study of the acyl group-and site-selectivity of human sirtuins to acylated nucleosomes. Sci. Rep. 2018, 8, 2656. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-J.; Ogryzko, V.V.; Nishikawa, J.-I.; Howard, B.H.; Nakatani, Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 1996, 382, 319–324. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kibria, M.G.; Yoshizawa, T.; Zhang, T.; Ono, K.; Mizumoto, T.; Sato, Y.; Sawa, T.; Yamagata, K. SIRT7 Is a Lysine Deacylase with a Preference for Depropionylation and Demyristoylation. Int. J. Mol. Sci. 2025, 26, 3153. https://doi.org/10.3390/ijms26073153
Kibria MG, Yoshizawa T, Zhang T, Ono K, Mizumoto T, Sato Y, Sawa T, Yamagata K. SIRT7 Is a Lysine Deacylase with a Preference for Depropionylation and Demyristoylation. International Journal of Molecular Sciences. 2025; 26(7):3153. https://doi.org/10.3390/ijms26073153
Chicago/Turabian StyleKibria, Mohammad Golam, Tatsuya Yoshizawa, Tianli Zhang, Katsuhiko Ono, Tomoya Mizumoto, Yoshifumi Sato, Tomohiro Sawa, and Kazuya Yamagata. 2025. "SIRT7 Is a Lysine Deacylase with a Preference for Depropionylation and Demyristoylation" International Journal of Molecular Sciences 26, no. 7: 3153. https://doi.org/10.3390/ijms26073153
APA StyleKibria, M. G., Yoshizawa, T., Zhang, T., Ono, K., Mizumoto, T., Sato, Y., Sawa, T., & Yamagata, K. (2025). SIRT7 Is a Lysine Deacylase with a Preference for Depropionylation and Demyristoylation. International Journal of Molecular Sciences, 26(7), 3153. https://doi.org/10.3390/ijms26073153





