Abstract
Breast cancer (BC) is a multifactorial condition and it primarily expresses the estrogen receptor α (ERα) that is encoded by the gene estrogen receptor 1 (ESR1), which modulates estrogen signaling. ESR1, by facilitating estrogen overproduction, plays an indispensable role in the progression and survival of the majority of BCs. To obtain molecular insights into these phenomena, we analyzed The Cancer Genome Atlas (TCGA) breast invasive carcinoma (BRCA) RNA-Seq datasets for the expression of ESR1 and its correlation to microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), along with its methylation patterns. Regulation of ESR1 was also assessed with a total of 43 cancerous and non-cancerous breast cell lines. Analyses of both TCGA BRCA and breast cell line RNA-Seq data revealed that specific lncRNAs, i.e., MEG3, BIK, MLL, and FAS are negatively correlated with the ESR1, in which PARP1 demonstrates a positive association. Additionally, both miR-30a and miR-145 showed negative correlations with the ESR1 expression. Of the 54 ESR1 methylation loci analyzed, the majority of them exhibited a negative correlation with the ESR1 expression, highlighting a potentially modifiable regulatory mechanism. These findings underscore the complex regulatory events influencing ESR1 expression and its interaction with diverse signaling pathways, demonstrating novel insights into breast pathogenesis and its potential therapeutics.
1. Introduction
Breast cancer (BC) is one of the most frequently diagnosed malignancies in women worldwide [1]. BC is the second leading cause of cancer-related death among women, with an estimated 316,950 new cases and 42,170 deaths occurring in 2025 in the United States [2]. BC is a multifactorial disease including four molecular subtypes, i.e., luminal A (ER+, PR+, and human epidermal growth factor receptor-negative, Her−), luminal B (ER+, PR+, and Her+), Her2 (ER−, PR−, and Her+), and basal like (ER−, PR−, and Her−, also called triple-negative BC, TNBC) [3,4,5]. Therefore, BC heterogeneity exhibits unique biological and clinical features, which considerably influence prognosis and treatment strategies [6,7]. For instance, hormone receptor-dependent BCs (≥80% of all cases) largely express ER, especially ESR1, progesterone receptor (PR+), and/or HER2+, all of which are responsible for the growth and maintenance of BCs [8,9]. Treatment typically involves endocrine therapy with aromatase inhibitors (AIs), which either block ER and/or reduce estrogen levels [9,10,11]. In contrast, triple-negative BC (TNBC, 10–15%) is hormone-independent and does not express ER, PR, or HER2; thus, it is unresponsive to endocrine therapy [12,13]. Regardless of the diverse factors and pathways involved, the activation of estrogen signaling, involving its overproduction, is a predominant event in breast pathogenesis [14,15,16].
The actions of estrogens are mediated by nuclear ERs, ERα and ERβ (encoded by ESR1 and ESR2 genes, respectively), which play pivotal roles in a plethora of physiological, as well as pathophysiological, processes that involve both genomic and non-genomic signaling [17,18,19]. Whereas both ESR1 and ESR2 display considerable sequence homology, they exert diverse actions in tumor initiation and development [6,19]. Notably, ESR1 is activated by estrogens and it plays a key role in the growth and progression of hormone-sensitive BC. Mutations and/or variations in ESR1 can affect how BC responds to endocrine therapy, making it a critical factor for the prognosis and treatment of this life-threatening disease [20,21,22,23]. Even so, BC heterogeneity, involving epidemiological patterns, differently influences ESR1 regulation, necessitating an in-depth understanding of the mechanisms with diverse signaling [24,25,26].
Long non-coding RNAs (lncRNAs) constitute those that are longer than 200 nucleotides that do not code for proteins and have been involved in various malignant and non-malignant diseases [27,28]. LncRNAs regulate gene expression through various mechanisms, including chromatin remodeling, transcription, and post-transcriptional events [29,30]. Moreover, a number of lncRNAs have been shown to play crucial roles in the regulation of ESR1 expression and activity, impacting breast pathogenesis [31,32,33].
MicroRNAs (miRNAs) are small non-coding RNAs (~22 nucleotides) that regulate gene expression by binding to complementary sequences, leading to either degradation or translational inhibition [34,35]. Concomitantly, both miR-30a and miR-145 have been implicated in the regulation of ESR1 expression, and their interactions contribute to the progression and treatment of BCs. MicroRNAs have been reported to influence estrogen signaling, thus contributing to endocrine therapy for mitigating BCs [36,37].
Methylation of the ESR1 promoter can silence its expression, resulting in a reduction in estrogen levels. Hypomethylation of ESR1 is characteristically associated with ER+/PR+ BC, maintaining hormone responsiveness. Studies have shown that methylation of the ESR1 promoter is considered a predictive marker in BC [38,39,40]. Moreover, environmental factors, such as diet, obesity, and exposure to endocrine-disrupting chemicals, alter ESR1 methylation patterns. This suggests that ESR1 methylation could be a modifiable factor influencing BC risk via modifications of genetic and epigenetic signaling [41].
An overwhelming amount of evidence indicates that TCGA RNA-Seq datasets offer a comprehensive understanding of the molecular basis of BC and other malignant disorders through high-throughput genome sequencing “https://gdc.cancer.gov/about-data/publications/pancanatlas (accessed on 25 January 2025)” [6,9,42,43,44,45]. To gain knowledge of the molecular events involved in breast pathogenesis, we analyzed both TCGA BRCA [42] and breast cell line [46] RNA-Seq data for the expression of ESR1 and its correlation to miRNA, lncRNA, and methylation. Our data advance the field and provide novel insights into ESR1 regulation and its interaction with a variety of signaling, permitting a better understanding of breast carcinogenesis, in addition to prospective therapeutics, especially for the ER+/PR+ BC subtype.
2. Results and Discussion
BC is characterized by aberrant and uncontrolled growth of mammary epithelial cells that involve genetic abnormality in modulating DNA damage and genomic instability [6,45,47]. Disruption of the equilibrium between oncogenes and tumor suppressor genes results in breast tumorigenesis, and this event is primarily impacted by the upregulation of estrogen signaling [48,49]. Utilizing TCGA BRCA datasets, we reported that genetic and epigenetic irregularities, impacting ESR1 function, are common in human primary breast tumors that are mirrored in pertinent breast cell lines [6,9]. Notably, ESR1 plays a crucial role in the pathogenesis of BCs and many relevant disorders. In accordance with this, we reported that amplification of the ESR1 correlates with poor survival of BC patients [6]. Analyzing TCGA BRCA, as well as breast cell line RNA-Seq datasets, our current data extend these observations and elucidate an improved understanding of breast carcinogenesis by demonstrating the molecular insights into the interacting mechanisms between ESR1, lncRNA, and miRNA, along with methylation.
2.1. Expression of the ESR1 in the TCGA BRCA Datasets with Various BC Subtypes
Since the activation of estrogen signaling, modulating estrogen synthesis is a primary event in breast tumorigenesis [16,50], TCGA BRCA (lrgely ER+/PR+ subtypes [45]) RNA-Seq datasets were examined for the relative expression of the ESR1 in non-cancerous (normal) and cancerous (ER+/PR+ and other BCs) breast tissues. As shown by box plot analyses (Figure 1), ER+/PR+ BC samples showed substantially higher ESR1 expression (p < 0.001), compared with both normal and other BC types, in which the latter includes hormone-independent/TNBC. Other BC subtypes displayed significantly lower expression of ESR1 (p < 0.001) than that of non-cancerous normal breast tissue. These data indicate that ESR1 expression, enhancing estrogen levels, plays an important role for in various BC subtypes in comparison to normal breast tissue.
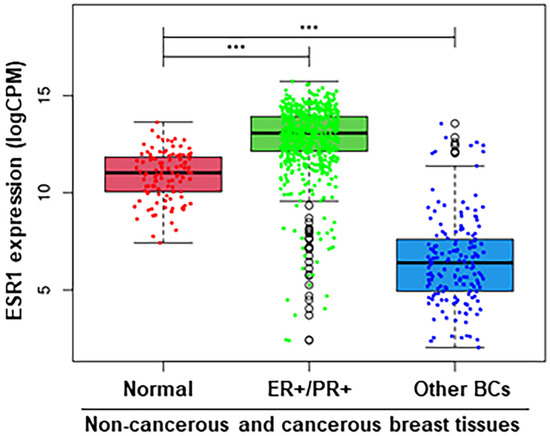
Figure 1.
Boxplot analysis of the ESR1 expression using TCGA BRCA RNA-Seq datasets under three different categories: Normal (114), ER+/PR+ (611), and Other BCs (164), with sample numbers in parentheses. ***, p < 0.001 vs. Normal, as indicated.
2.2. Analyses of TCGA BRCA and Breast Cell Line RNA-Seq Datasets for a Number of lncRNAs and Their Correlation to ESR1
There is increasing evidence that expression of the ESR1 is higher in ER+/PR+ BC, compared with normal breast tissue samples; however, hormone-independent BCs exhibit low to no ESR1 levels [6,8,9]. We observed that a number of lncRNAs such as MEG3, MLL, and BIK show higher expression patterns in both hormone-sensitive and other BC subtypes (Figure 2). To investigate the molecular interaction between ESR1 and target lncRNAs, TCGA BRCA and breast cell line RNA-Seq datasets were analyzed, converging normal tissue and ER+/PR+ BC. The pairwise comparison results revealed that the Spearman’s correlation coefficients between ESR1 and lncRNAs, i.e., H19, MEG3, BIK, FAS, LIN28B, NEAT1, PARP1, and MLL were −0.20, −0.46, 0.28, −0.48, −0.16, 0.12, 0.26, and −0.19, respectively, with TCGA BRCA data. These correlations are significant at p < 0.001 for H19, MEG3, BIK, FAS, LIN28B, NEAT1, PARP1, and MLL. Alternatively, LIN28B is correlated at p = 0.018 (Figure 2A). Additionally, the Spearman’s correlation coefficients for MEG3, BIK, FAS, LIN28B, PARP1, and MLL were −0.06, 0.41, −0.42, −0.11, −0.02, and −0.26, respectively (Figure 2B), using a variety of breast cell line RNA-Seq data [46]. Among these lncRNAs, whereas BIK and FAS were positively correlated at p = 0.002 and p = 0.004, FAS showed a negative association (p = 0.004) with ESR1, respectively. The results obtained in breast cell lines are consistent with human primary breast cancer data associated with TCGA RBCA.
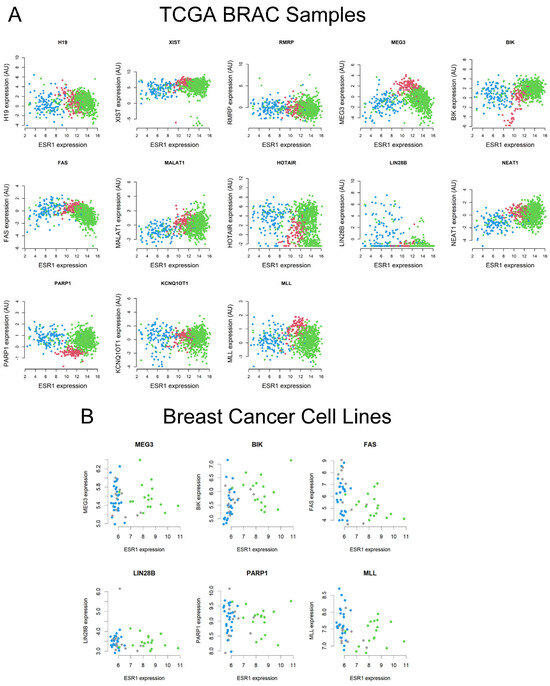
Figure 2.
Analyses of the Spearman’s correlation coefficients between ESR1 and specific lncRNAs (H19, XIST, RMRP, MEG3, BIK, FAS, MALAT1, HOTAIR, LIN28B, NEAT1, PARP1, KCNQ10T1, and MLL) using TCGA BRCA RNA-Seq data (A). The colors used were normal tissue—red; ER+/PR+ samples—green; and other BC subtypes—blue. (B) The Spearman’s correlation coefficients between ESR1 and target lncRNAs (MEG3, BIK, FAS, LIN 28B, PARP1, and MLL) with 43 breast cell line RNA-Seq data. Colors used were ER+ samples—green; ER− samples—blue; and other subtypes—gray.
Breast tissue cell line RNA-Seq datasets were further analyzed for the relative expression of ESR1, BIK, and MLL. Data presented in Figure 3 show that all 43 cancerous and non-cancerous cell lines express ESR1 at varying levels, regardless of ER− and ER+ status. However, the expression of ESR1 mRNA was higher in the ER+ category than in those ER− subtypes. Genomic profiles of BIK and MLL indicated that all breast cell lines analyzed express these lncRNAs to varied levels; however, the expression of BIK and MLL mRNA levels was found to be diverse in ER+ BC cell lines. These data suggest that ESR1, BIK, and MLL play important roles in breast physiology and pathophysiology.
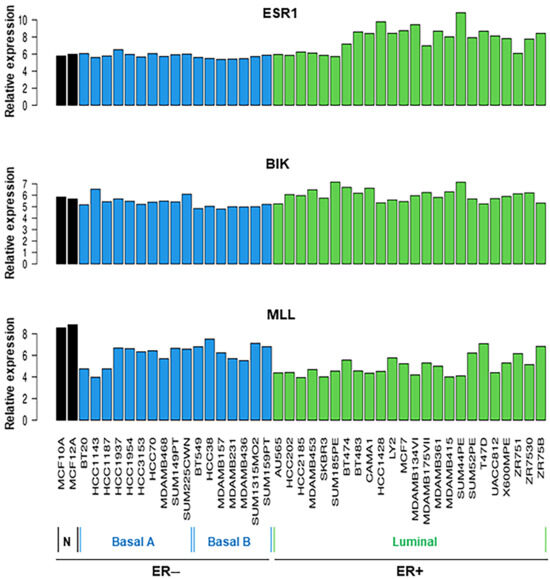
Figure 3.
Analyses of RNA-Seq data for relative expression of ESR1, BIK, and MLL mRNAs in a total of 43 different cancerous and non-cancerous breast cell lines. Shown are the names of various breast cell lines under the ER− and ER+ categories.
Accumulating evidence indicates that upregulation of MEG3 inhibits the malignant features of cells in vitro [51,52,53]. Moreover, MEG3 acts as a tumor suppressor by epigenetically regulating target genes [54]. Even so, it has been shown that high MEG3 expression impacts the growth of breast cancer in vivo [55]. Studies have demonstrated that MEG3 exerts multifaceted function in various pathological processes, including BCs, by interacting with miRNA, proteins, and epigenetic signaling, and its relevance has been reported in many clinical applications [54,56,57]. Another lncRNA, MLL, modulates H3K4me3 at the ESR1 promoter, maintaining open chromatin for active transcription. Loss or mutation of MLL has been shown to decrease ESR1 expression, potentially contributing to reducing ER signaling in BCs [58,59]. Additionally, PARP1 plays an important role in regulating ESR1 target genes. Enhanced PARP1 activity may promote hormone-driven cancer progression by impacting ER-mediated transcription. Co-inhibition of PARP1 and ER signaling has been implicated as a potential therapeutic strategy for BCs [60,61]. The inverse relationship between MEG3 and PARP1 designates that MEG3 could inhibit DNA repair mechanisms critical for cancer cell survival. This interaction might make MEG3 a valuable modulator in therapies targeting the PARP1 with inhibitors, particularly in tumors with defective apoptotic pathways. Genetic aberrations of ESR1 and lncRNAs associated with human primary breast tumors in TCGA were closely reflected with various breast cell line data, highlighting an improved understanding of the mechanisms involved in breast tumorigenesis and its therapies. Estrogen signaling via ESR1 upregulates the expression of several metabolic enzymes, including FAS, to support lipid metabolism in BC cells. In support of this, studies have shown that cholesterol and its oxygenated derivatives are drivers of BCs [6,62,63]. FAS converts acetyl-CoA and malonyl-CoA into palmitate and plays a critical role in providing lipid synthesis, energy storage, and signaling in rapidly BC and other malignant cells [64]. Moreover, estrogen can directly or indirectly regulate FAS expression, suggesting a link between hormone signaling and lipid metabolism [65,66,67]. It is likely that the interaction of ESR1 with specific lncRNAs, but not their expression levels, plays an important role in breast pathogenesis and can be targeted for improved BC therapeutics.
2.3. Correlation of Specific miRNAs with ESR1 and Their Relevance to Breast Pathogenesis
MiRNAs post-transcriptionally regulate gene expression and, by doing so, they influence diverse cell signaling processes, including proliferation, differentiation, and apoptosis, and promote breast tumor progression [68,69]. Considering their involvement, miRNAs have been targeted in the diagnosis and treatment of BCs [70,71,72,73]. By analyzing TCGA BRCA RNA-Seq data, we found that the expression of miR-29b and miR-155 is lower in ER+/PR+ BC and higher in hormone-independent subtypes, compared with normal breast tissue. In contrast, miR-145 and miR-30a revealed higher expression in non-tumorous mammary tissue. Pairwise comparisons between ESR1 and miRNAs in normal breast tissue and ER+/PR+ BCs revealed that the Spearman’s correlation coefficients between ESR1 and miR-145, miR-29b, miR-155, and miR-30a were −0.30, −0.21, −0.17, and −0.36, respectively. The corresponding p-values for these correlations were significant at p < 0.001 for miR-145, miR-29b, and miR-30a, and p < 0.003 for miR-155 (Figure 4).
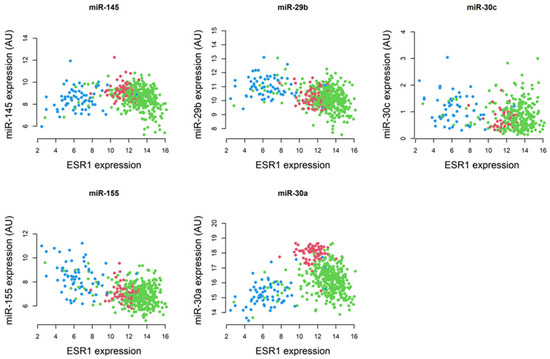
Figure 4.
The Spearman’s correlation coefficients between ESR1 and miRNAs (miR-145, miR-29b, miR-30c, miR-155, and miR-30a) using TCGA BRCA RNA-Seq data. Colors used were normal tissue—red; ER/PR+ samples—green; and other BC subtypes—blue.
MiR-30a directly targets the 3′ UTR of ESR1 mRNA, reducing its expression. This post-transcriptional repression has been reported to affect the ERα protein in BC cells [74]. Also, miR-145 is known to downregulate ESR1 expression by binding to its mRNA, leading to its degradation or translational repression. Studies have demonstrated that miR-145 suppresses cell proliferation and invasion in BC, especially in TNBC [75,76]. The elevated levels of miR-30a in normal breast tissue contribute to cellular homeostasis by disrupting ESR1, which impacts apoptotic pathways. Conversely, its downregulation in ER+/PR+ BCs involves epigenetic silencing and/or transcriptional repression, emphasizing its loss for BC development and progression. Nonetheless, whereas a number of miRNAs, including miR-30a, have been implicated in therapeutic targets for BCs and other relevant diseases [77,78], their clinical applications have yet to be validated. Taken together, regardless of varied correlation patterns of selected lncRNAs and miRNAs with the ESR1, Kaplan–Meier survival curve analyses indicated that none of these lncRNAs and miRNAs were found to affect the overall survival of BC patients (Yang et al., unpublished observations).
2.4. Analyses of TCGA BRCA Data for ESR1 DNA Methylation Patterns
The majority of mammalian gene promoters are comprehended within regions of the genome called CpG islands, which are DNA regions with high CpG frequency and low methylation influencing oncogenic signaling and regulation [79,80,81]. We analyzed TCGA BRCA RNA-Seq data and found that a strong correlation exists between the DNA methylation of various CpG islands and the ESR1 gene expression (Table 1). In particular, a significant negative correlation is observed between ESR1 and the methylation of many CpG sites, such as cg00601836, cg15543523, and cg21265702, with correlation coefficients ranging from −0.26 to −0.68 and unadjusted p-values as low as 6.44 × 10−58. These findings indicate that higher methylation levels in these CpG sites are associated with lower expression of the ESR1. The adjusted p-values for these correlations remain significant even after multiple testing corrections. Interestingly, cg25490334 displayed a positive correlation with the ESR1 expression (correlation = 0.21), with a higher p-value at 5.25 × 10−6. Overall, these data indicate that DNA methylation plays an important regulatory role in the expression of ESR1, with specific CpG sites showing strong inverse relationships with the ESR1 activity. A detailed distribution pattern of DNA methylation and the ESR1 expression is presented in Supplementary Figure S1.

Table 1.
Determination of correlation coefficients between DNA methylation and ESR1 expression using TCGA BRCA RNA-Seq datasets.
2.5. Generation of a Heatmap Depicting a Correlation Matrix with lncRNAs, miRNAs, and ESR1
An interesting aspect of the present findings is strong correlations among various lncRNAs and miRNAs with the ESR1 (Figure 5). A number of lncRNAs and miRNAs, by interacting with ESR1, are known to activate estrogen signaling, triggering breast tumorigenesis [82,83,84,85]. Robust positive associations were observed between MEG3 and FAS (correlation = 0.57; p < 0.001), miR-30a (correlation = 0.59; p < 0.001), and miR-145 (correlation = 0.55; p < 0.001). Alternatively, negative correlations were found with PARP1 (correlation = −0.44; p < 0.001) and ESR1 (correlation = −0.46; p < 0.001). Moreover, miR-30a is negatively correlated with ESR1 and positively correlated with MLL (correlation = 0.41; p < 0.001) and MEG3 and FAS (correlation = 0.49; p < 0.001). In addition, the miR-30a level is much higher in normal breast tissue in comparison to ER+/PR+ BC (correlation = 0.62, p < 0.001). Overall, the relationships observed highlight the complexity of interactions in the regulation of various genes and their implications in normal physiologic, as well as pathophysiologic (hormone-responsive BC), states.
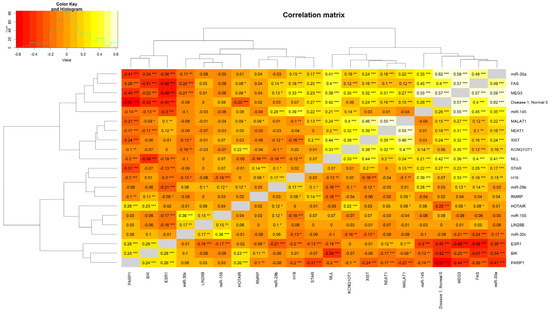
Figure 5.
A heatmap illustrating the correlation matrix among lncRNA, miRNA, and ESR1. The genes included on the correlation map that influence diverse signaling pathways were the following: miR-30a, FAS, MEG3, miR-145, MALAT1, NEAT1, XIST, KCNQ10T1, MLL, STAR, H19, miR-29b, RMRP, HOTAIR, miR-155, LIN288, miR-30c, ESR1, BIK, and RARP1. p-values are provided in the correlation map in the format of “*” 0.01 ≤ p < 0.05; “**” 0.001 ≤ p < 0.01; and “***” p < 0.001.
Correlation matrix data revealed the presence of a positive association between miR-30a and MLL, influencing chromatin remodeling and activation of tumor-suppressor genes, which reinforce the anti-tumorigenic effects of miR-30a in BC [86,87]. The tumor-suppressive role of MEG3 appears to be multifaceted, in which it is positively correlated with miR-30a, miR-145, and FAS in promoting apoptosis. These correlations suggest that MEG3 may serve as a platform for orchestrating regulatory networks and enhancing tumor suppression. The negative correlations with PARP1 and ESR1 align with their tumor-suppressive role by diminishing pathways associated with DNA repair and hormone-driven proliferation, respectively. The negative correlations of MEG3 and miR-30a with ESR1 emphasize their potential roles in reducing estrogen-driven proliferation. This inverse relationship supports therapeutic strategies aimed at modulating ESR1 through MEG3 and miR-30a or targeting epigenetic modifications affecting ESR1 expression. There is an interplay between miRNAs and lncRNAs in tumor suppression [88,89]. The positive correlations between MEG3 and miR-30a, as well as miR-145, specify cooperative mechanisms, in which MEG3 may act as a competing endogenous RNA, sequestering oncogenic miRNAs to allow tumor-suppressive function effectively. Co-regulation of these lncRNAs through common upstream pathways, such as the p53 signaling axis, provides further insights into their interdependence and highlights the therapeutic potential in BCs [90].
ESR1 methylation, involving environmental and lifestyle factors, offers perceptions of the epigenetic plasticity of hormone receptor genes. This plasticity represents a promising avenue for preventive strategies and personalized treatments, including dietary interventions and pharmacological modulation of methylation [39,91]. Collectively, the dynamic nature of DNA methylation in genes like ESR1, influenced by diet, chemicals, and/or lifestyle, underscores the potential for reversible epigenetic changes [92]. This opens up new avenues for prevention and intervention through tailored lifestyle modifications and/or drugs that impact the epigenetic landscape. Studying the methylation patterns in breast malignancies could also provide biomarker development for early detection, prognosis, and response to BC therapy.
3. Materials and Methods
3.1. Analyses of the TCGA BRCA and Breast Cell Line RNA-Seq Datasets
RNA-Seq datasets were downloaded from the UCSC Xena “https://xena.ucsc.edu (accessed on 25 January 2025)” browser and evaluated the clinical characteristics of cancerous and non-cancerous breast tissues [9,43,44]. The site provides gene expression profiles along with mean normalization per gene. These values were generated by UCSC Xena in combining gene expression RNA-Seq data from all TCGA BRCA cohorts (n = 1218). The values were mean-centered per gene, and data specific to this cohort were then extracted. Corresponding phenotypic data were also downloaded from the UCSC Xena repository, encompassing information for 1218 patients. Additionally, miRNA mature strand expression by RNA-Seq for 832 patients and Illumina Infinium HumanMethylation450 data for 888 patients were downloaded from “https://xena.ucsc.edu (accessed on 25 January 2025)” [43].
RNA-Seq datasets from a total of 43 cancerous and non-cancerous breast cell lines were downloaded from UCSC Xena “https://ucsc-public-main-xena-hub.s3.us-east-1.amazonaws.com/download/grayBreastCellLines_public%2FgrayBreastCellLineExon_genomicMatrix.gz (accessed on 25 January 2025)” [46,93]. These data contain gene expression profiles for a variety of breast cancer cell lines. In addition, GI50 to 77 therapeutic compounds as well as ER, HER2, and other phenotypic information is available in the TCGA BRCA clinical data.
3.2. Generation of Box and Scatter Plots Utilizing TCGA BRCA Data
The Illumina HTSeq FPKM data were downloaded from the UCSC Xena platform [42,43]. Both box and scatter plots were generated to display the distribution of expression levels [9]. The boxes represent the interquartile range (IQR), with whiskers extending to 1.5 times the IQR, and outliers are shown as individual points. Pairwise comparisons were conducted across groups of normal mammary tissue, ER+/PR+ BC, and other BC subtypes using non-parametric tests, such as the Kruskal–Wallis or Mann–Whitney U tests. Adjusted p-values (e.g., Bonferroni correction) were annotated on the plots when statistical significance was detected at p < 0.05.
3.3. Determination of the Spearman’s Rank Correlation Coefficients with TCGA BRCA Data
The Spearman’s rank correlation coefficients were calculated using the cor.test function in R (R: The R Project for Statistical Computing) to evaluate relationships between two gene expression levels. Correlation matrices were generated and their significance was assessed with p-values adjusted for multiple comparisons. Correlation values closer to either +1 or −1 were considered as strong positive or negative correlations, respectively.
3.4. Development of a Heatmap for Correlation Matrix
Heatmaps were created using the ComplexHeatmap package version 2.16.0 in R to visualize expression patterns across genes, miRNAs, lncRNAs, and ESR1. Hierarchical clustering was applied to samples, with distances calculated using the Spearman correlation coefficients. Log-transformed CPM values were used for data normalization, and color gradients reflected the intensity of expression profiles. A correlation heatmap was generated and analyzed for the enrichment of various genomic factors.
3.5. Statistical Analysis
The R software version 4.3.3 “https://www.r-project.org (accessed on 25 January 2025)” was used to perform descriptive statistical analyses, including mean, standard deviation, and fold changes on relevant gene expression levels across diverse groups. Statistical comparisons in p-values adjusted for multiple testing using the FDR (False Discovery rate) method.
4. Conclusions
It is unequivocal that ESR1 plays a crucial role in promoting the growth and maintenance of BCs, especially the most prevalent hormone-sensitive BC category [6,8]. The interactions between ESR1 and certain lncRNAs and/or miRNAs can contribute to endocrine therapy resistance, making it a critical target for breast pathogenesis and its potential therapeutics [89,94]. These scenarios are exemplified and verified by both publicly available TCGA BRCA and 43 breast cell line RNA-Seq datasets [42,46]. Specifically, MEG3 could play a tumor-suppressive role, resulting in a promising therapeutic target for modulating disease pathways involved in ER+/PR+ BC. MiR-30a likely subsidizes cellular homeostasis by suppressing oncogenic drivers such as ESR1 and ratifying the apoptotic pathway. The positive correlations of miR-30a and MLL with ESR1 indicate their key roles in chromatin remodeling and in the activation of tumor suppression. Additionally, MEG3 can serve as a modulator in therapies that target PARP inhibitors [95]. The negative correlations of MEG3 and miR-30a with ESR1 underscore their potential roles in reducing estrogen-driven proliferation for the management of BCs. These inverse relationships suggest therapeutic opportunities that could modulate ESR1 levels through non-coding RNAs like MEG3 and miR-30a. However, there is a complex interplay between miRNAs and lncRNAs that may result in the suppression of ESR1-regulated genes. Overall, the dynamic nature of DNA methylation in genes like ESR1 provides new insights into BC prevention and/or intervention through tailored lifestyle modifications [92,96]. Based on the above considerations, we conclude that the molecular events involved in ESR1-mediated activation of estrogen signaling are influenced by concerted interactions of a number of miRNAs and/or lncRNAs, which helps enlighten on an improved understanding of breast pathogenesis and its therapeutic interventions. Additional studies involving human primary breast tumor tissues and/or pertinent cell lines provide a detailed understanding of the regulation of ESR1 and its correlation to specific lncRNAs and miRNAs for more targeted therapies for BCs, especially for the prevention and/or treatment of the most prevalent hormone-dependent subtype.
This study has certain limitations, even though it identifies strong correlations between ESR1 and specific lncRNAs, miRNAs, and DNA methylation sites, utilizing both TCGA BRCA and breast cell line RNA-Seq datasets, and supports the conclusions. While these correlations are significant, TCGA BRCA samples involve heterogeneity, including pathological stages, ages, demographics, and numbers in cancerous and non-cancerous tissues; thus, data interpretation should be made carefully. Further experimental studies are necessary to reveal if these molecules analyzed directly impact ESR1 expression and whether they are part of broader regulatory networks. However, the study’s cross-sectional design limits the ability to infer temporal relationships with lncRNAs, miRNAs, and ESR1 expression. Therefore, analyses with more cancerous and non-cancerous breast samples involving demographic information and advanced technological settings would be beneficial for specific conclusions.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms26073101/s1.
Author Contributions
Conceptualization, writing, and original draft preparation, methodology, S.Y. and P.R.M.; interpretation and validation of data, S.Y. and P.R.M.; review, analysis, editing, and finalization of manuscript, S.Y., C.M. and P.R.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by U54 GM104940 from the National Institute of General Medical Sciences of the National Institutes of Health, which funds the Louisiana Clinical and Translational Science Center to S.Y., and the Department of Internal Medicine and The CH Foundation (NFR 21206) to P.R.M.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data reported in this study are included in this manuscript. The datasets analyzed here are available from the corresponding authors upon reasonable request.
Acknowledgments
The authors would like to acknowledge the utilization of TCGA BRCA “https://tcga-data.nci.nih.gov”, breast cell line (https://ucsc-public-main-xena-hub.s3.us-east-1.amazonaws.com/download/grayBreastCellLines_public%2FgrayBreastCellLineExon_genomicMatrix.gz (accessed on 25 January 2025)), and UCSC Xena “https://xenabrowser.net/ (accessed on 25 January 2025)” RNA-Seq research network datasets.
Conflicts of Interest
The authors declare that there are no competing interests that could be perceived as prejudicing the impartiality of this work. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
| BC | Breast cancer |
| ER | Estrogen receptor |
| PR | Progesterone receptor |
| HER2 | Human epidermal growth factor receptor 2 |
| ESR1 | Estrogen receptor 1 |
| lncRNA | long non-coding RNA |
| miRNA | Micro RNA |
| TNBC | Triple-negative breast cancer |
| BRCA | Breast invasive carcinoma |
| RNA-Seq | RNA sequencing |
| FAS | Fatty acid synthetase |
| TCGA | The cancer genomic atlas |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef] [PubMed]
- Lawton, T.J. Update on the Use of Molecular Subtyping in Breast Cancer. Adv. Anat. Pathol. 2023, 30, 368–373. [Google Scholar]
- Hurson, A.N.; Ahearn, T.U.; Koka, H.; Jenkins, B.D.; Harris, A.R.; Roberts, S.; Fan, S.; Franklin, J.; Butera, G.; Keeman, R.; et al. Risk factors for breast cancer subtypes by race and ethnicity: A scoping review. J. Natl. Cancer Inst. 2024, 116, 1992–2002. [Google Scholar]
- Rios-Hoyo, A.; Shan, N.L.; Karn, P.L.; Pusztai, L. Clinical Implications of Breast Cancer Intrinsic Subtypes. Adv. Exp. Med. Biol. 2025, 1464, 435–448. [Google Scholar]
- Manna, P.R.; Ahmed, A.U.; Molehin, D.; Narasimhan, M.; Pruitt, K.; Reddy, P.H. Hormonal and Genetic Regulatory Events in Breast Cancer and Its Therapeutics: Importance of the Steroidogenic Acute Regulatory Protein. Biomedicines 2022, 10, 1313. [Google Scholar] [CrossRef]
- Dikoglu, E.; Pareja, F. Molecular Basis of Breast Tumor Heterogeneity. Adv. Exp. Med. Biol. 2025, 1464, 237–257. [Google Scholar] [PubMed]
- Manna, P.R.; Ramachandran, S.; Pradeepkiran, J.A.; Molehin, D.; Castro-Piedras, I.; Pruitt, K.; Ganapathy, V.; Reddy, P.H. Expression and Function of StAR in Cancerous and Non-Cancerous Human and Mouse Breast Tissues: New Insights into Diagnosis and Treatment of Hormone-Sensitive Breast Cancer. Int. J. Mol. Sci. 2023, 24, 758. [Google Scholar] [CrossRef]
- Manna, P.R.; Yang, S.; Reddy, P.H. Epigenetic Dysregulation and Its Correlation with the Steroidogenic Machinery Impacting Breast Pathogenesis: Data Mining and Molecular Insights into Therapeutics. Int. J. Mol. Sci. 2023, 24, 16488. [Google Scholar] [CrossRef]
- Musgrove, E.A.; Sutherland, R.L. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer 2009, 9, 631–643. [Google Scholar] [CrossRef]
- Manna, P.R.; Molehin, D.; Ahmed, A.U. Dysregulation of Aromatase in Breast, Endometrial, and Ovarian Cancers: An Overview of Therapeutic Strategies. Prog. Mol. Biol. Transl. Sci. 2016, 144, 487–537. [Google Scholar] [PubMed]
- Bianchini, G.; Balko, J.M.; Mayer, I.A.; Sanders, M.E.; Gianni, L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016, 13, 674–690. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Jovanovic, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar]
- Kerdivel, G.; Flouriot, G.; Pakdel, F. Modulation of estrogen receptor alpha activity and expression during breast cancer progression. Vitam. Horm. 2013, 93, 135–160. [Google Scholar] [PubMed]
- Farcas, A.M.; Nagarajan, S.; Cosulich, S.; Carroll, J.S. Genome-Wide Estrogen Receptor Activity in Breast Cancer. Endocrinology 2021, 162, bqaa224. [Google Scholar]
- Clusan, L.; Ferriere, F.; Flouriot, G.; Pakdel, F. A Basic Review on Estrogen Receptor Signaling Pathways in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 6834. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, N.; Silveyra, P. Estrogen receptor signaling mechanisms. Adv. Protein Chem. Struct. Biol. 2019, 116, 135–170. [Google Scholar]
- Mangani, S.; Piperigkou, Z.; Koletsis, N.E.; Ioannou, P.; Karamanos, N.K. Estrogen receptors and extracellular matrix: The critical interplay in cancer development and progression. FEBS J. 2024. Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Gui, Z.; Shi, W.; Zhou, F.; Yan, Y.; Li, Y.; Xu, Y. The role of estrogen receptors in intracellular estrogen signaling pathways, an overview. J. Steroid Biochem. Mol. Biol. 2025, 245, 106632. [Google Scholar]
- Dustin, D.; Gu, G.; Fuqua, S.A.W. ESR1 mutations in breast cancer. Cancer 2019, 125, 3714–3728. [Google Scholar]
- Herzog, S.K.; Fuqua, S.A.W. ESR1 mutations and therapeutic resistance in metastatic breast cancer: Progress and remaining challenges. Br. J. Cancer 2022, 126, 174–186. [Google Scholar] [PubMed]
- Hancock, G.R.; Gertz, J.; Jeselsohn, R.; Fanning, S.W. Estrogen Receptor Alpha Mutations, Truncations, Heterodimers, and Therapies. Endocrinology 2024, 165, bqae051. [Google Scholar]
- Xiong, X.; Zheng, L.W.; Ding, Y.; Chen, Y.F.; Cai, Y.W.; Wang, L.P.; Huang, L.; Liu, C.C.; Shao, Z.M.; Yu, K.D. Breast cancer: Pathogenesis and treatments. Signal Transduct. Target. Ther. 2025, 10, 49. [Google Scholar] [PubMed]
- Sun, T.; Lian, R.; Liang, X.; Sun, D. Association Between ESR1 XBAI and Breast Cancer Susceptibility: A Systematic Review and Meta-Analysis. Clin. Investig. Med. 2022, 45, E21–E34. [Google Scholar]
- Xie, J.; Gan, L.; Xue, B.; Wang, X.; Pei, X. Emerging roles of interactions between ncRNAs and other epigenetic modifications in breast cancer. Front. Oncol. 2023, 13, 1264090. [Google Scholar]
- Noyan, S.; Gur Dedeoglu, B. Upregulation of miR-99b-5p Modulates ESR1 Expression as an Adaptive Mechanism to Circumvent Drug Response via Facilitating ER/HER2 Crosstalk. Balkan Med. J. 2025, 42, 150–156. [Google Scholar]
- Fan, N.; Fu, H.; Feng, X.; Chen, Y.; Wang, J.; Wu, Y.; Bian, Y.; Li, Y. Long non-coding RNAs play an important regulatory role in tumorigenesis and tumor progression through aerobic glycolysis. Front. Mol. Biosci. 2022, 9, 941653. [Google Scholar]
- Huang, Y.; Mo, W.; Ding, X.; Ding, Y. Long non-coding RNAs in breast cancer stem cells. Med. Oncol. 2023, 40, 177. [Google Scholar]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar]
- Garcia-Padilla, C.; Duenas, A.; Garcia-Lopez, V.; Aranega, A.; Franco, D.; Garcia-Martinez, V.; Lopez-Sanchez, C. Molecular Mechanisms of lncRNAs in the Dependent Regulation of Cancer and Their Potential Therapeutic Use. Int. J. Mol. Sci. 2022, 23, 764. [Google Scholar] [CrossRef]
- Hassani, B.; Taheri, M.; Asgari, Y.; Zekri, A.; Sattari, A.; Ghafouri-Fard, S.; Pouresmaeili, F. Expression Analysis of Long Non-Coding RNAs Related with FOXM1, GATA3, FOXA1 and ESR1 in Breast Tissues. Front. Oncol. 2021, 11, 671418. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hamid, N.R.; Mohammed, E.A.; Toraih, E.A.; Kamel, M.M.; Abdelhafiz, A.S.; Badr, F.M. Circulating ESR1, long non-coding RNA HOTAIR and microRNA-130a gene expression as biomarkers for breast cancer stage and metastasis. Sci. Rep. 2023, 13, 22654. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Z.; Jeselsohn, R. ESR1 fusions and therapeutic resistance in metastatic breast cancer. Front. Oncol. 2022, 12, 1037531. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef]
- Treeck, O.; Haerteis, S.; Ortmann, O. Non-Coding RNAs Modulating Estrogen Signaling and Response to Endocrine Therapy in Breast Cancer. Cancers 2023, 15, 1632. [Google Scholar] [CrossRef]
- Drula, R.; Pardini, B.; Fu, X.; De Los Santos, M.C.; Jurj, A.; Pang, L.; El-Daly, S.M.; Fabris, L.; Knutsen, E.; Dragomir, M.P.; et al. 17beta-estradiol promotes extracellular vesicle release and selective miRNA loading in ERalpha-positive breast cancer. Proc. Natl. Acad. Sci. USA 2023, 120, e2122053120. [Google Scholar] [CrossRef]
- Martinez-Galan, J.; Torres-Torres, B.; Nunez, M.I.; Lopez-Penalver, J.; Del Moral, R.; Ruiz De Almodovar, J.M.; Menjon, S.; Concha, A.; Chamorro, C.; Rios, S.; et al. ESR1 gene promoter region methylation in free circulating DNA and its correlation with estrogen receptor protein expression in tumor tissue in breast cancer patients. BMC Cancer 2014, 14, 59. [Google Scholar] [CrossRef]
- Kirn, V.; Strake, L.; Thangarajah, F.; Richters, L.; Eischeid, H.; Koitzsch, U.; Odenthal, M.; Fries, J. ESR1-promoter-methylation status in primary breast cancer and its corresponding metastases. Clin. Exp. Metastasis 2018, 35, 707–712. [Google Scholar] [CrossRef]
- Quintas-Granados, L.I.; Cortes, H.; Carmen, M.G.; Leyva-Gomez, G.; Bustamante-Montes, L.P.; Rodriguez-Morales, M.; Villegas-Vazquez, E.Y.; Lopez-Reyes, I.; Alcaraz-Estrada, S.L.; Sandoval-Basilio, J.; et al. The high methylation level of a novel 151-bp CpG island in the ESR1 gene promoter is associated with a poor breast cancer prognosis. Cancer Cell Int. 2021, 21, 649. [Google Scholar]
- Pogribny, I.P.; Rusyn, I. Environmental toxicants, epigenetics, and cancer. Adv. Exp. Med. Biol. 2013, 754, 215–232. [Google Scholar] [PubMed]
- Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar]
- Casper, J.; Zweig, A.S.; Villarreal, C.; Tyner, C.; Speir, M.L.; Rosenbloom, K.R.; Raney, B.J.; Lee, C.M.; Lee, B.T.; Karolchik, D.; et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 2018, 46, D762–D769. [Google Scholar]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e310. [Google Scholar] [PubMed]
- Manna, P.R.; Ahmed, A.U.; Yang, S.; Narasimhan, M.; Cohen-Tannoudji, J.; Slominski, A.T.; Pruitt, K. Genomic Profiling of the Steroidogenic Acute Regulatory Protein in Breast Cancer: In Silico Assessments and a Mechanistic Perspective. Cancers 2019, 11, 623. [Google Scholar] [CrossRef]
- Heiser, L.M.; Sadanandam, A.; Kuo, W.L.; Benz, S.C.; Goldstein, T.C.; Ng, S.; Gibb, W.J.; Wang, N.J.; Ziyad, S.; Tong, F.; et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 2724–2729. [Google Scholar] [PubMed]
- Garcia-Chico, C.; Lopez-Ortiz, S.; Penin-Grandes, S.; Pinto-Fraga, J.; Valenzuela, P.L.; Emanuele, E.; Ceci, C.; Graziani, G.; Fiuza-Luces, C.; Lista, S.; et al. Physical Exercise and the Hallmarks of Breast Cancer: A Narrative Review. Cancers 2023, 15, 324. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, Y.; Murase, K.; Saito, M.; Imamura, M.; Oh, K. Mechanisms of estrogen receptor-alpha upregulation in breast cancers. Med. Mol. Morphol. 2010, 43, 193–196. [Google Scholar]
- Miziak, P.; Baran, M.; Blaszczak, E.; Przybyszewska-Podstawka, A.; Kalafut, J.; Smok-Kalwat, J.; Dmoszynska-Graniczka, M.; Kielbus, M.; Stepulak, A. Estrogen Receptor Signaling in Breast Cancer. Cancers 2023, 15, 4689. [Google Scholar] [CrossRef]
- Toumba, M.; Kythreotis, A.; Panayiotou, K.; Skordis, N. Estrogen receptor signaling and targets: Bones, breasts and brain (Review). Mol. Med. Rep. 2024, 30, 144. [Google Scholar]
- Dong, S.; Ma, M.; Li, M.; Guo, Y.; Zuo, X.; Gu, X.; Zhang, M.; Shi, Y. LncRNA MEG3 regulates breast cancer proliferation and apoptosis through miR-141-3p/RBMS3 axis. Genomics 2021, 113, 1689–1704. [Google Scholar] [PubMed]
- Wu, J.; Pang, R.; Li, M.; Chen, B.; Huang, J.; Zhu, Y. m6A-Induced LncRNA MEG3 Suppresses the Proliferation, Migration and Invasion of Hepatocellular Carcinoma Cell Through miR-544b/BTG2 Signaling. OncoTargets Ther. 2021, 14, 3745–3755. [Google Scholar]
- Wei, Q.; Liu, G.; Huang, Z.; Huang, Y.; Huang, L.; Huang, Z.; Wu, X.; Wei, H.; Pu, J. LncRNA MEG3 Inhibits Tumor Progression by Modulating Macrophage Phenotypic Polarization via miR-145-5p/DAB2 Axis in Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2023, 10, 1019–1035. [Google Scholar]
- Hussain, M.S.; Majami, A.A.; Ali, H.; Gupta, G.; Almalki, W.H.; Alzarea, S.I.; Kazmi, I.; Syed, R.U.; Khalifa, N.E.; Bin Break, M.K.; et al. The complex role of MEG3: An emerging long non-coding RNA in breast cancer. Pathol. Res. Pract. 2023, 251, 154850. [Google Scholar]
- Zhu, X.; Lv, L.; Wang, M.; Fan, C.; Lu, X.; Jin, M.; Li, S.; Wang, F. DNMT1 facilitates growth of breast cancer by inducing MEG3 hyper-methylation. Cancer Cell Int. 2022, 22, 56. [Google Scholar]
- Shi, Y. MEG3 regulates apoptosis of adipose-derived stem cells. Mol. Med. Rep. 2020, 21, 2435–2442. [Google Scholar]
- Li, Z.; Gao, J.; Sun, D.; Jiao, Q.; Ma, J.; Cui, W.; Lou, Y.; Xu, F.; Li, S.; Li, H. LncRNA MEG3: Potential stock for precision treatment of cardiovascular diseases. Front. Pharmacol. 2022, 13, 1045501. [Google Scholar] [CrossRef]
- Teinturier, R.; Abou Ziki, R.; Kassem, L.; Luo, Y.; Malbeteau, L.; Gherardi, S.; Corbo, L.; Bertolino, P.; Bachelot, T.; Treilleux, I.; et al. Reduced menin expression leads to decreased ERalpha expression and is correlated with the occurrence of human luminal B-like and ER-negative breast cancer subtypes. Breast Cancer Res. Treat. 2021, 190, 389–401. [Google Scholar]
- Jones, R.B.; Farhi, J.; Adams, M.; Parwani, K.K.; Cooper, G.W.; Zecevic, M.; Lee, R.S.; Hong, A.L.; Spangle JM: Targeting, M.L.L. Methyltransferases Enhances the Antitumor Effects of PI3K Inhibition in Hormone Receptor-positive Breast Cancer. Cancer Res. Commun. 2022, 2, 1569–1578. [Google Scholar]
- Gadad, S.S.; Camacho, C.V.; Malladi, V.; Hutti, C.R.; Nagari, A.; Kraus, W.L. PARP-1 Regulates Estrogen-Dependent Gene Expression in Estrogen Receptor alpha-Positive Breast Cancer Cells. Mol. Cancer Res. 2021, 19, 1688–1698. [Google Scholar]
- Bohi, F.; Hottiger, M.O. Expanding the Perspective on PARP1 and Its Inhibitors in Cancer Therapy: From DNA Damage Repair to Immunomodulation. Biomedicines 2024, 12, 1617. [Google Scholar] [CrossRef] [PubMed]
- Nelson, E.R.; Wardell, S.E.; Jasper, J.S.; Park, S.; Suchindran, S.; Howe, M.K.; Carver, N.J.; Pillai, R.V.; Sullivan, P.M.; Sondhi, V.; et al. 27-Hydroxycholesterol links hypercholesterolemia and breast cancer pathophysiology. Science 2013, 342, 1094–1098. [Google Scholar]
- Silvente-Poirot, S.; Poirot, M. Cancer. Cholesterol and cancer, in the balance. Science 2014, 343, 1445–1446. [Google Scholar] [PubMed]
- Menendez, J.A.; Lupu, R. Fatty acid synthase: A druggable driver of breast cancer brain metastasis. Expert. Opin. Ther. Targets 2022, 26, 427–444. [Google Scholar]
- Song, H.J.; Sneddon, A.A.; Heys, S.D.; Wahle, K.W. Regulation of fatty acid synthase (FAS) and apoptosis in estrogen-receptor positive and negative breast cancer cells by conjugated linoleic acids. Prostaglandins Leukot. Essent. Fatty Acids 2012, 87, 197–203. [Google Scholar]
- Menendez, J.A.; Lupu, R. Fatty acid synthase regulates estrogen receptor-alpha signaling in breast cancer cells. Oncogenesis 2017, 6, e299. [Google Scholar]
- Cairns, J.; Ingle, J.N.; Kalari, K.R.; Goetz, M.P.; Weinshilboum, R.M.; Gao, H.; Li, H.; Bari, M.G.; Wang, L. Anastrozole Regulates Fatty Acid Synthase in Breast Cancer. Mol. Cancer Ther. 2022, 21, 206–216. [Google Scholar]
- Kumar, S.; Morton, H.; Sawant, N.; Orlov, E.; Bunquin, L.E.; Pradeepkiran, J.A.; Alvir, R.; Reddy, P.H. MicroRNA-455-3p improves synaptic, cognitive functions and extends lifespan: Relevance to Alzheimer’s disease. Redox Biol. 2021, 48, 102182. [Google Scholar]
- Akshaya, R.L.; Saranya, I.; Selvamurugan, N. MicroRNAs mediated interaction of tumor microenvironment cells with breast cancer cells during bone metastasis. Breast Cancer 2023, 30, 910–925. [Google Scholar] [PubMed]
- Valle-Garcia, D.; Perez de la Cruz, V.; Flores, I.; Salazar, A.; Pineda, B.; Meza-Sosa, K.F. Use of microRNAs as Diagnostic, Prognostic, and Therapeutic Tools for Glioblastoma. Int. J. Mol. Sci. 2024, 25, 2464. [Google Scholar] [CrossRef]
- Mohan Lal, P.; Hamza Siddiqui, M.; Soulat, A.; Mohan, A.; Tanush, D.; Tirath, K.; Raja, S.; Khuzzaim Khan, M.; Raja, A.; Chaulagain, A.; et al. MicroRNAs as promising biomarkers and potential therapeutic agents in breast cancer management: A comprehensive review. Ann. Med. Surg. 2024, 86, 3543–3550. [Google Scholar] [CrossRef] [PubMed]
- Abdul Manap, A.S.; Wisham, A.A.; Wong, F.W.; Ahmad Najmi, H.R.; Ng, Z.F.; Diba, R.S. Mapping the function of MicroRNAs as a critical regulator of tumor-immune cell communication in breast cancer and potential treatment strategies. Front. Cell Dev. Biol. 2024, 12, 1390704. [Google Scholar] [CrossRef]
- Mirzaei, Z.; Barati, T.; Ebrahimi, A.; Derakhshan, S.M.; Khaniani, M.S. The role of mir-7-5p in cancer: Function, prognosis, diagnosis, and therapeutic implications. Mol. Biol. Rep. 2024, 52, 12. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Ding, B.; Lou, W. microRNA-Dependent Modulation of Genes Contributes to ESR1’s Effect on ERalpha Positive Breast Cancer. Front. Oncol. 2020, 10, 753. [Google Scholar] [CrossRef] [PubMed]
- Spizzo, R.; Nicoloso, M.S.; Lupini, L.; Lu, Y.; Fogarty, J.; Rossi, S.; Zagatti, B.; Fabbri, M.; Veronese, A.; Liu, X.; et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ. 2010, 17, 246–254. [Google Scholar]
- Piasecka, D.; Braun, M.; Kordek, R.; Sadej, R.; Romanska, H. MicroRNAs in regulation of triple-negative breast cancer progression. J. Cancer Res. Clin. Oncol. 2018, 144, 1401–1411. [Google Scholar] [CrossRef]
- Wang, H.Y.; Li, Y.Y.; Fu, S.; Wang, X.P.; Huang, M.Y.; Zhang, X.; Shao, Q.; Deng, L.; Zeng, M.S.; Zeng, Y.X.; et al. MicroRNA-30a promotes invasiveness and metastasis in vitro and in vivo through epithelial-mesenchymal transition and results in poor survival of nasopharyngeal carcinoma patients. Exp. Biol. Med. 2014, 239, 891–898. [Google Scholar]
- He, Q.; Chen, Z.; Deng, Y.; Mao, C. Plasma microRNA-30a expression in patients with chronic hepatitis B and its application value in the assessment of the severity of liver fibrosis. Eur. J. Gastroenterol. Hepatol. 2025. Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, N.P.; Klose, R. CpG island chromatin: A platform for gene regulation. Epigenetics 2011, 6, 147–152. [Google Scholar] [CrossRef]
- Sergeeva, A.; Davydova, K.; Perenkov, A.; Vedunova, M. Mechanisms of human DNA methylation, alteration of methylation patterns in physiological processes and oncology. Gene 2023, 875, 147487. [Google Scholar] [CrossRef]
- Bhootra, S.; Jill, N.; Shanmugam, G.; Rakshit, S.; Sarkar, K. DNA methylation and cancer: Transcriptional regulation, prognostic, and therapeutic perspective. Med. Oncol. 2023, 40, 71. [Google Scholar] [PubMed]
- Men, Y.; Fan, Y.; Shen, Y.; Lu, L.; Kallen, A.N. The Steroidogenic Acute Regulatory Protein (StAR) Is Regulated by the H19/let-7 Axis. Endocrinology 2017, 158, 402–409. [Google Scholar] [PubMed]
- Otsuka, K.; Matsubara, S.; Shiraishi, A.; Takei, N.; Satoh, Y.; Terao, M.; Takada, S.; Kotani, T.; Satake, H.; Kimura, A.P. A Testis-Specific Long Noncoding RNA, Start, Is a Regulator of Steroidogenesis in Mouse Leydig Cells. Front. Endocrinol. 2021, 12, 665874. [Google Scholar]
- Kumar, S.; Orlov, E.; Gowda, P.; Bose, C.; Swerdlow, R.H.; Lahiri, D.K.; Reddy, P.H. Synaptosome microRNAs regulate synapse functions in Alzheimer’s disease. NPJ Genom. Med. 2022, 7, 47. [Google Scholar] [PubMed]
- Islam, M.A.; Sultana, O.F.; Bandari, M.; Kshirsagar, S.; Manna, P.R.; Reddy, P.H. MicroRNA-455-3P as a peripheral biomarker and therapeutic target for mild cognitive impairment and Alzheimer’s disease. Ageing Res. Rev. 2024, 100, 102459. [Google Scholar]
- Guo, F.; Chen, H.; Chang, J.; Zhang, L. Mutation R273H confers p53 a stimulating effect on the IGF-1R-AKT pathway via miR-30a suppression in breast cancer. Biomed. Pharmacother. 2016, 78, 335–341. [Google Scholar] [CrossRef]
- Yeap, S.K.; Mohd Ali, N.; Akhtar, M.N.; Razak, N.A.; Chong, Z.X.; Ho, W.Y.; Boo, L.; Zareen, S.; Kurniawan, T.A.; Avtar, R.; et al. Induction of Apoptosis and Regulation of MicroRNA Expression by (2E,6E)-2,6-bis-(4-hydroxy-3-methoxybenzylidene)-cyclohexanone (BHMC) Treatment on MCF-7 Breast Cancer Cells. Molecules 2021, 26, 1277. [Google Scholar] [CrossRef]
- Volovat, S.R.; Volovat, C.; Hordila, I.; Hordila, D.A.; Mirestean, C.C.; Miron, O.T.; Lungulescu, C.; Scripcariu, D.V.; Stolniceanu, C.R.; Konsoulova-Kirova, A.A.; et al. MiRNA and LncRNA as Potential Biomarkers in Triple-Negative Breast Cancer: A Review. Front. Oncol. 2020, 10, 526850. [Google Scholar]
- Schwarzenbach, H.; Gahan, P.B. Interplay between LncRNAs and microRNAs in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 8095. [Google Scholar] [CrossRef]
- Song, J.; Cui, Q.; Gao, J. Roles of lncRNAs related to the p53 network in breast cancer progression. Front. Oncol. 2024, 14, 1453807. [Google Scholar]
- Albrecht, J.; Muller, M.; Hafstaeth, V.; Kaminska, K.; Vallon-Christersson, J.; Honeth, G.; Persson, H. Dynamic methylation and expression of alternative promoters for oestrogen receptor alpha in cell line models of fulvestrant resistance. Mol. Oncol. 2025, 19, 204–224. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Shukla, S.; Khan, S.; Tollefsbol, T.O.; Meeran, S.M. Epigenetic reactivation of p21CIP1/WAF1 and KLOTHO by a combination of bioactive dietary supplements is partially ERalpha-dependent in ERalpha-negative human breast cancer cells. Mol. Cell Endocrinol. 2015, 406, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Castro-Piedras, I.; Sharma, M.; den Bakker, M.; Molehin, D.; Martinez, E.G.; Vartak, D.; Pruitt, W.M.; Deitrick, J.; Almodovar, S.; Pruitt, K. DVL1 and DVL3 differentially localize to CYP19A1 promoters and regulate aromatase mRNA in breast cancer cells. Oncotarget 2018, 9, 35639–35654. [Google Scholar] [CrossRef] [PubMed]
- Giordo, R.; Ahmadi, F.A.M.; Husaini, N.A.; Al-Nuaimi, N.; Ahmad, S.M.S.; Pintus, G.; Zayed, H. microRNA 21 and long non-coding RNAs interplays underlie cancer pathophysiology: A narrative review. Noncoding RNA Res. 2024, 9, 831–852. [Google Scholar] [CrossRef]
- Cheng, J.; Sun, Y.; Zhao, H.; Ren, W.; Gao, D.; Wang, Z.; Lv, W.; Dong, Q. Niraparib restrains prostate cancer cell proliferation and metastasis and tumor growth in mice by regulating the lncRNA MEG3/miR-181-5p/GATA6 pathway. PeerJ 2023, 11, e16314. [Google Scholar] [CrossRef]
- Goncalves, R.; Warner, W.A.; Luo, J.; Ellis, M.J. New concepts in breast cancer genomics and genetics. Breast Cancer Res. 2014, 16, 460. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).