Immunomodulatory Potential of Kaempferol Isolated from Peronema canescens Jack. Leaves Through Inhibition of IL-6 Expression
Abstract
1. Introduction
2. Results
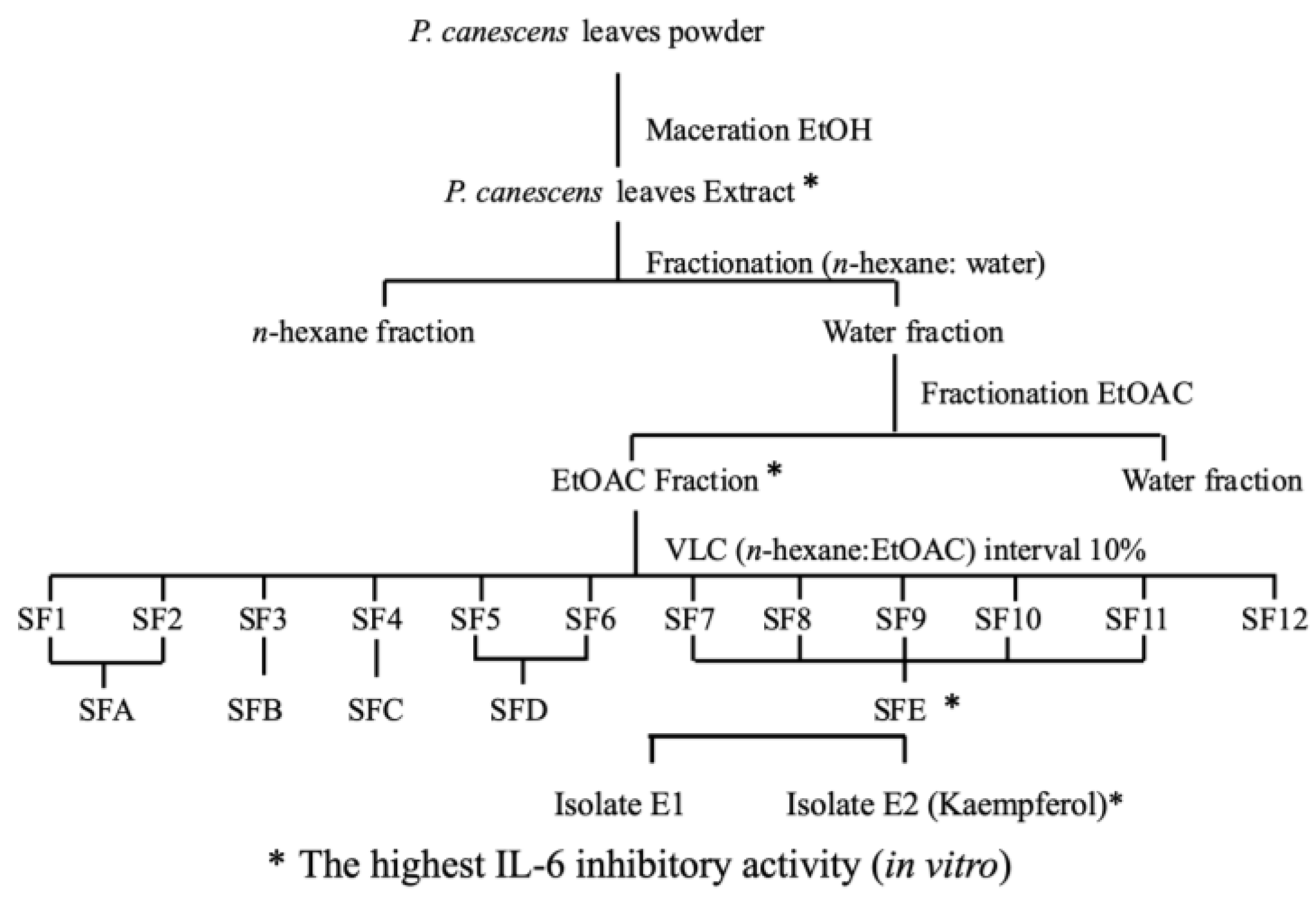
2.1. Extract Purification and Isolation
2.2. Structure Elucidation
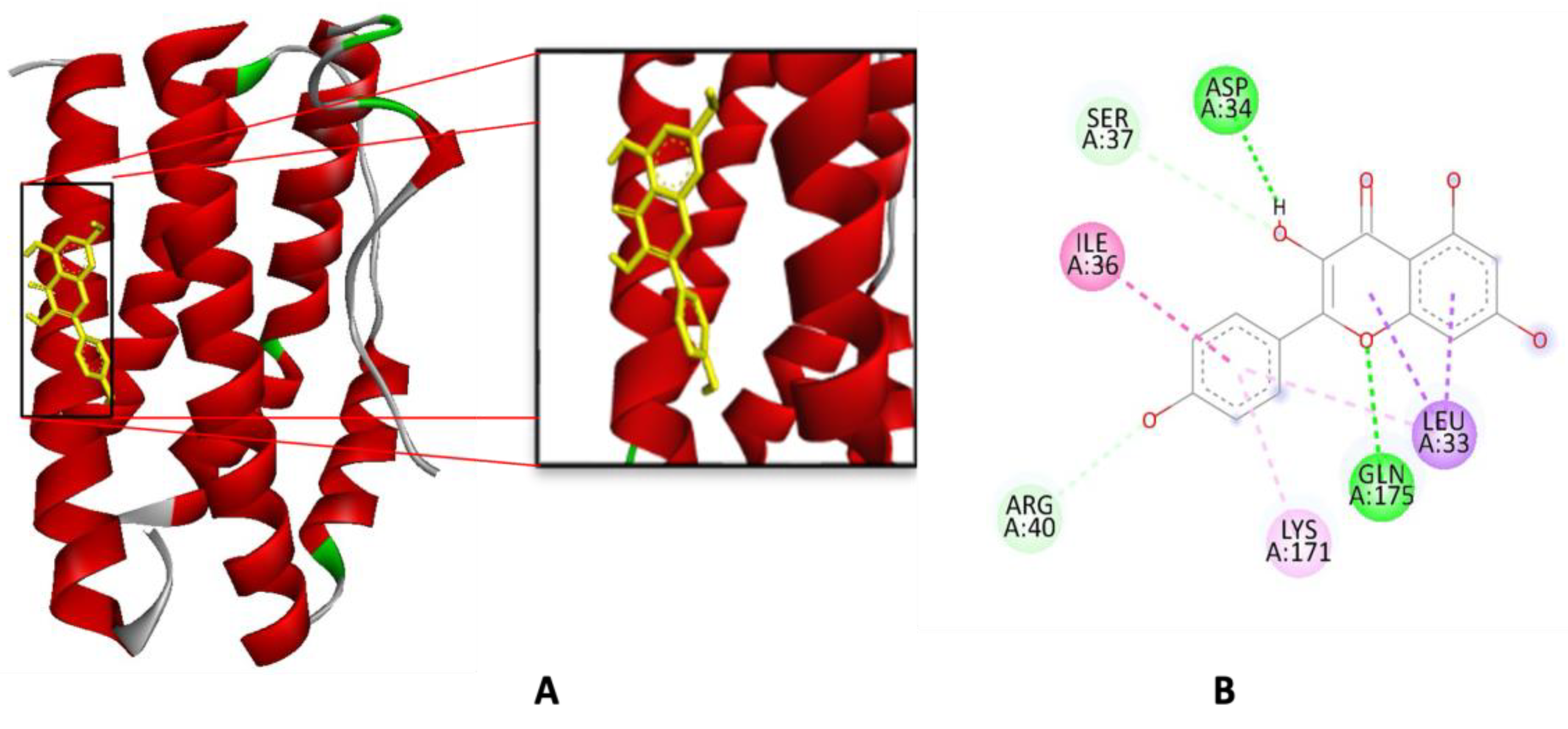
2.3. Molecular Docking Simulation
2.4. Pharmacokinetic and Toxicity Prediction
2.5. Molecular Dynamics Simulation
2.6. Immunomodulatory Activity by Inhibition of IL-6 Gene Expression
3. Discussion
3.1. Extract Purification and Isolation
3.2. Structure Elucidation
3.3. Molecular Docking Simulation
3.4. Pharmacokinetic and Toxicity Prediction
3.5. Molecular Dynamics Simulation
3.6. Immunomodulatory Activity by Inhibition of IL-6 Gene Expression
4. Materials and Methods
4.1. Sample Preparation
4.2. Extraction, Fractionation, and Isolation
4.3. Elucidation of the Compound Structure
4.4. Molecular Docking Simulation
4.5. Molecular Dynamics Simulation
4.6. Prediction of ADMET
4.7. Evaluation of Cell Viability in RAW 264.7 Macrophage Cells
4.8. Immunomodulatory Activity by Inhibition of IL-6 Gene Expression
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Behl, T.; Kumar, K.; Brisc, C.; Rus, M.; Nistor-Cseppento, D.C.; Bustea, C.; Aron, R.A.C.; Pantis, C.; Zengin, G.; Sehgal, A.; et al. Exploring the Multifocal Role of Phytochemicals as Immunomodulators. Biomed. Pharmacother. 2021, 133, 110959. [Google Scholar] [CrossRef]
- Grigore, A. Plant Phenolic Compounds as Immunomodulatory Agents. In Phenolic Compounds-Biological Activity; IntechOpen: London, UK, 2017; pp. 75–98. [Google Scholar]
- Singh, V.K.; Thakur, D.C.; Rajak, N.; Giri, R.; Garg, N. Immunomodulatory Potential of Bioactive Glycoside Syringin: A Network Pharmacology and Molecular Modeling Approach. J. Biomol. Struct. Dyn. 2024, 42, 3906–3919. [Google Scholar] [CrossRef] [PubMed]
- Baby, P.N. Immunomodulatory and Anticytokine Therapeutic Potential of Three Indian Spices Constituents and Its Hyaluronic Acid Conjugates for Prevention and Post COVID-19 Complications: A Computational Modeling Approach. J. Biomol. Struct. Dyn. 2024, 6, 1–21. [Google Scholar] [CrossRef]
- Spinelli, V.; Laurino, A.; Balducci, V.; Gencarelli, M.; Ruzzolini, J.; Nediani, C.; Mandoli, G.E.; Cameli, M.; Sacconi, L.; Sartiani, L.; et al. Interleukin-6 Modulates the Expression and Function of HCN Channels: A Link Between Inflammation and Atrial Electrogenesis. Int. J. Mol. Sci. 2024, 25, 12212. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. Immunotherapeutic Implications of IL-6 Blockade for Cytokine Storm. Immunotherapy 2016, 8, 959–970. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in Inflammation, Immunity, and Disease. Cold Spring Harb. Lab. Press 2014, 6, a016295. [Google Scholar]
- Tran, Q.H.; Cao, H.N.; Nguyen, D.N.; Tran, T.T.N.; Le, M.T.; Nguyen, Q.T.; Tran, V.T.; Tran, V.H.; Thai, K.M. Targeting Olokizumab-Interleukin 6 Interaction Interface to Discover Novel IL-6 Inhibitors. J. Biomol. Struct. Dyn. 2023, 41, 14003–14015. [Google Scholar] [CrossRef]
- Aliyu, M.; Zohora, F.T.; Anka, A.U.; Ali, K.; Maleknia, S.; Saffarioun, M.; Azizi, G. Interleukin-6 Cytokine: An Overview of the Immune Regulation, Immune Dysregulation, and Therapeutic Approach. Int. Immunopharmacol. 2022, 111, 109130. [Google Scholar] [CrossRef]
- Tran, Q.H.; Nguyen, Q.T.; Vo, N.Q.H.; Mai, T.T.; Tran, T.T.N.; Tran, T.D.; Le, M.T.; Trinh, D.T.T.; Minh Thai, K. Structure-Based 3D-Pharmacophore Modeling to Discover Novel Interleukin 6 Inhibitors: An in silico Screening, Molecular Dynamics Simulations and Binding Free Energy Calculations. PLoS ONE 2022, 17, e0266632. [Google Scholar] [CrossRef]
- Hayati, H.; Razi, R. Comparing the Cost-Effectiveness of Herbal Medicines with Chemical Medicines (A Review Article). Herb. Med. J. 2024, 9, 1–6. [Google Scholar]
- Posadzki, P.; Watson, L.K.; Ernst, E. Adverse Effects of Herbal Medicines: An Overview of Systematic Reviews. Clin. Med. J. R. Coll. Physicians Lond. 2013, 13, 7–12. [Google Scholar] [CrossRef]
- Ekor, M. The Growing Use of Herbal Medicines: Issues Relating to Adverse Reactions and Challenges in Monitoring Safety. Front. Neurol 2014, 4, 177. [Google Scholar] [CrossRef]
- Karimi, A.; Majlesi, M.; Rafieian-Kopaei, M. Herbal versus Synthetic Drugs; Beliefs and Facts. J. Nephropharmacol. 2015, 4, 27–30. [Google Scholar]
- Rahardhian, M.R.R.; Susilawati, Y.; Sumiwi, S.A.; Muktiwardoyo, M.; Muchtaridi, M. A Review of Sungkai (Peronema canescens): Traditional Usage, Phytoconstituent, And Pharmacological Activities. Int. J. Appl. Pharm. 2022, 14, 15–23. [Google Scholar]
- Dillasamola, D.; Aldi, Y.; Wahyuni, F.S.; Rita, R.S.; Dachriyanus; Umar, S.; Rivai, H. Study of Sungkai (Peronema canescens, Jack) Leaf Extract Activity as an Immunostimulators with in vivo and in vitro Methods. Pharmacogn. J. 2021, 13, 1397–1407. [Google Scholar] [CrossRef]
- Mazura, M.P.; Ling, S.K. Preliminary Studies on Lipoxygenase Inhibitory Activity of Selected Malaysian Medicinal Plants. Pharmacogn. Res. 2009, 1, 1. [Google Scholar]
- Rahardhian, M.R.R.; Susilawati, Y.; Musfiroh, I.; Febriyanti, R.M.; Muchtaridi; Sumiwi, S.A. In silico Study of Bioactive Compounds From Sungkai (Peronema canescens) As Immunomodulator. Int. J. Appl. Pharm. 2022, 14, 135–141. [Google Scholar] [CrossRef]
- Afnuhazi, R.; Sari, F.S.; Hendrawati; Sarli, D.; Poddar, R. The Effect of Stewed Sungkai Leaves (Peronema canescens Jack) on the Decrease in Inflammatory Symptoms of COVID-19 Patients. Res. J. Pharm. Technol. 2022, 15, 5464–5466. [Google Scholar] [CrossRef]
- Yahfoufi, N.; Alsadi, N.; Jambi, M.; Matar, C. The Immunomodulatory and Anti-Inflammatory Role of Polyphenols. Nutrients 2018, 10, 1618. [Google Scholar] [CrossRef]
- Kitagawa, I.; Simanjuntak, P.; Hori, K.; Nagami, N.; Mahmud, T.; Kobayashi, M.; Shibuya, H. Indonesian Medicinal Plants. VII. Seven New Clerodane-Type Diterpenoids, Peronemins A2, A3, B1, B2, B3, C1, and D1, from the Leaves of Peronema canescens (Verbenaceae). Chem. Pharm. Bull. 1994, 42, 1050–1055. [Google Scholar] [CrossRef]
- Meng, X.Y.; Zhang, H.X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Current Computer-Aided Drug Design. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [PubMed]
- Calderón-Montaño, J.M.; Burgos-Morón, E.; Pérez-Guerrero, C.; López-Lázaro, M. A Review on the Dietary Flavonoid Kaempferol|BenthamScience. Mini Rev. Med. Chem. 2011, 11, 298–344. [Google Scholar]
- Alonso, H.; Bliznyuk, A.A.; Gready, J.E. Combining Docking and Molecular Dynamic Simulations in Drug Design. Med. Res. Rev. 2006, 26, 531–568. [Google Scholar] [CrossRef] [PubMed]
- El-Zahabi, M.A.; Elkady, H.; Sakr, H.; Abdelraheem, A.S.; Eissa, S.I.; El-Adl, K. Design, Synthesis, Anticancer Evaluation, in Silico Docking and ADMET Analysis of Novel Indole-Based Thalidomide Analogs as Promising Immunomodulatory Agents. J. Biomol. Struct. Dyn. 2023, 41, 15106–15123. [Google Scholar] [CrossRef] [PubMed]
- Duistermaat, J.J.; Kolk, J.A.C. Extraction, Isolation and Characterization of Bioactive Compounds from Plants’ Extracts. Afr. J. Tradit. Complement. Altern. Med. 2000, 8, 93–130. [Google Scholar] [CrossRef]
- Reynolds, W.F. Natural Product Structure Elucidation by NMR Spectroscopy; Elsevier Inc.: Amsterdam, The Netherlands, 2017; ISBN 9780128020999. [Google Scholar]
- Xiao, Z.P.; Wu, H.K.; Wu, T.; Shi, H.; Hang, B.; Aisa, H.A. Kaempferol and Quercetin Flavonoids from Rosa rugosa. Chem. Nat. Compd. 2006, 42, 736–737. [Google Scholar] [CrossRef]
- Behera, B.; Meher, R.K.; Mir, S.A.; Nayak, B.; Satapathy, K.B. Phytochemical Profiling, in vitro Analysis for Anti-Inflammatory, Immunomodulatory Activities, Structural Elucidation and in silico Evaluation of Potential Selective COX-2 and TNF-α Inhibitor from Hydrilla verticillata (L.f.) Royle. J. Biomol. Struct. Dyn. 2025, 43, 859–873. [Google Scholar] [CrossRef]
- Kumari, G.; Nigam, V.K.; Pandey, D.M. The Molecular Docking and Molecular Dynamics Study of Flavonol Synthase and Flavonoid 3’-Monooxygenase Enzymes Involved for the Enrichment of Kaempferol. J. Biomol. Struct. Dyn. 2023, 41, 2478–2491. [Google Scholar] [CrossRef]
- Somers, W.; Stahl, M.; Seehra, J.S. 1.9 Å Crystal Structure of Interleukin 6: Implications for a Novel Mode of Receptor Dimerization and Signaling. EMBO J. 1997, 16, 989–997. [Google Scholar]
- Arifian, H.; Maharani, R.; Megantara, S.; Ikram, N.K.K.; Muchtaridi, M. Glycine-Conjugated α-Mangostins as Potential Estrogen Receptor Alpha (ERα) Antagonists through Pharmacophore Modeling, Docking Analysis, and Molecular Dynamics Simulations. Appl. Sci. 2024, 14, 5549. [Google Scholar] [CrossRef]
- Yasir, M.; Park, J.; Han, E.T.; Han, J.H.; Park, W.S.; Chun, W. Identification of Malaria-Selective Proteasome Β5 Inhibitors Through Pharmacophore Modeling, Molecular Docking, and Molecular Dynamics Simulation. Int. J. Mol. Sci. 2024, 25, 11881. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, S.M. A Multi-Target Mechanism of Withania somnifera Bioactive Compounds in Autism Spectrum Disorder (ASD) Treatment: Network Pharmacology, Molecular Docking, and Molecular Dynamics Simulations Studies. Arab. J. Chem. 2024, 17, 105772. [Google Scholar] [CrossRef]
- MacArron, R.; Banks, M.N.; Bojanic, D.; Burns, D.J.; Cirovic, D.A.; Garyantes, T.; Green, D.V.S.; Hertzberg, R.P.; Janzen, W.P.; Paslay, J.W.; et al. Impact of High-Throughput Screening in Biomedical Research. Nat. Rev. Drug Discov. 2011, 10, 188–195. [Google Scholar] [CrossRef]
- Myung, Y.; de Sá, A.G.C.; Ascher, D.B. Deep-PK: Deep Learning for Small Molecule Pharmacokinetic and Toxicity Prediction. Nucleic Acids Res. 2024, 52, W469–W475. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Musfiroh, I.; Ifaya, M.; Sahidin, I.; Herawati, D.M.D.; Tjitraresmi, A.; Abdurrahman, S.; Muchtaridi, M.; Khairul Ikram, N.K. Isolation of Phenolic Compound from Lawsonia inermis and Its Prediction as Anti-Diabetic Agent Using Molecular Docking and Molecular Dynamic Simulation. J. Biomol. Struct. Dyn. 2024, 42, 11415–11424. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Cerutti, D.S.; Cisneros, G.A.; Cruzeiro, V.W.D.; Forouzesh, N.; Giese, T.J.; Götz, A.W.; Gohlke, H.; et al. AmberTools. J. Chem. Inf. Model. 2023, 63, 6183–6191. [Google Scholar] [CrossRef]
- Stank, A.; Kokh, D.B.; Fuller, J.C.; Wade, R.C. Protein Binding Pocket Dynamics. Acc. Chem. Res. 2016, 49, 809–815. [Google Scholar] [CrossRef]
- Malik, A.; Naz, A.; Ahmad, S.; Hafeez, M.; Awan, F.M.; Jafar, T.H.; Zahid, A.; Ikram, A.; Rauff, B.; Hassan, M. Inhibitory Potential of Phytochemicals on Interleukin-6-Mediated T-Cell Reduction in COVID-19 Patients: A Computational Approach. Bioinform. Biol. Insights 2021, 15, 544–546. [Google Scholar] [CrossRef]
- Baňas, Š.; Tvrdá, E.; Benko, F.; Ďuračka, M.; Čmiková, N.; Lukáč, N.; Kačániová, M. Kaempferol as an Alternative Cryosupplement for Bovine Spermatozoa: Cytoprotective and Membrane-Stabilizing Effects. Int. J. Mol. Sci. 2024, 25, 4129. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An Overview. J. Nutr. Sci. 2016, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Shiyan, S.; Ramadona, N.; Utami, W.F.; Depriyanti, N.; Mukafi, A.; Noviandhani, W. Preparation and FTIR-ATR Combined with Chemometrics Analysis of Self-Emulsifying Loaded Sungkai Extract from Peronema canescens. Res. J. Pharm. Technol. 2023, 16, 79–85. [Google Scholar] [CrossRef]
- Norouzi, H.; Dastan, D.; Abdullah, F.O.; Al-Qaaneh, A.M. Recent Advances in Methods of Extraction, Pre-Concentration, Purification, Identification, and Quantification of Kaempferol. J. Chromatogr. A 2024, 1735, 465297. [Google Scholar] [CrossRef]
- Aisyah, L.S.; Yun, Y.F.; Herlina, T.; Julaeha, E.; Zainuddin, A.; Nurfarida, I.; Hidayat, A.T.; Supratman, U.; Shiono, Y. Flavonoid Compounds from the Leaves of Kalanchoe prolifera and Their Cytotoxic Activity against P-388 Murine Leukimia Cells. Nat. Prod. Sci. 2017, 23, 139–145. [Google Scholar] [CrossRef]
- Hong, C.; Chang, C.; Zhang, H.; Jin, Q.; Wu, G.; Wang, X. Identification and Characterization of Polyphenols in Different Varieties of Camellia Oleifera Seed Cakes by UPLC-QTOF-MS. Food Res. Int. 2019, 126, 108614. [Google Scholar] [CrossRef]
- Parvez, A.; Malik, K.; Grover, S.; Farooqi, H. Fermentation Alters the Bioactive Properties of Kaempferol Derived from Horse gram Extract. Phytomed. Plus 2024, 4, 100647. [Google Scholar] [CrossRef]
- Ram, J.; Calzada, F.; Garc, N.; Barbosa, E.; Vel, C.; Valdes, M. Caryophyllene Oxide, a Bicyclic Terpenoid Isolated from Annona Macroprophyllata with Antitumor Activity: In vivo, In vitro, and In silico Studies. Int. J. Mol. Sci. 2024, 25, 13355. [Google Scholar] [CrossRef]
- Pagadala, N.S.; Syed, K.; Tuszynski, J. Software for Molecular Docking: A Review. Biophys. Rev. 2017, 9, 91–102. [Google Scholar] [CrossRef]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef]
- Lan, T.; Wang, J.; Zeng, R.; Gao, C.; Liu, X.; Luo, L.; Liang, Y.; Guo, Z.; Wang, W.; Hong, M. Therapeutic Targets and Molecular Mechanisms of Huangqin Decoction in Liver Cancer: A Network Pharmacology and Molecular Docking Approach. J. Herb. Med. 2024, 43, 100822. [Google Scholar] [CrossRef]
- Hossain, M.A.; Sohel, M.; Sultana, T.; Hasan, M.I.; Khan, M.S.; Kibria, K.M.K.; Mahmud, S.M.H.; Rahman, M.H. Study of Kaempferol in the Treatment of COVID-19 Combined with Chikungunya Co-Infection by Network Pharmacology and Molecular Docking Technology. Inf. Med. Unlocked 2023, 40, 101289. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.L.; Ai, S.M.; Liang, J.; Sang, P.; Ji, X.L.; Liu, S.Q. Insights into Protein–Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Ren, R.; Zhang, Z.; Li, X.; Liu, L.; Liu, H.; Xia, Y.; Masumuzzaman, M. Antihypertensive Mechanism of the Medicine Food Homology Compound Solution with High ACE Inhibition Rate Based on Network Pharmacology and Molecular Docking. Food Biosci. 2024, 62, 105077. [Google Scholar] [CrossRef]
- van de Waterbeemd, H.; Gifford, E. ADMET in silico Modelling: Towards Prediction Paradise? Nat. Rev. Drug Discov. 2003, 2, 192–204. [Google Scholar] [CrossRef]
- Zheng, L.; Zhu, L.; Zhao, M.; Shi, J.; Li, Y.; Yu, J.; Jiang, H.; Wu, J.; Tong, Y.; Liu, Y.; et al. In Vivo Exposure of Kaempferol Is Driven by Phase II Metabolic Enzymes and Efflux Transporters. AAPS J. 2016, 18, 1289–1299. [Google Scholar] [CrossRef]
- Merisko-Liversidge, E.M.; Liversidge, G.G. Drug Nanoparticles: Formulating Poorly Water-Soluble Compounds. Toxicol. Pathol. 2008, 36, 43–48. [Google Scholar] [CrossRef]
- He, X.; Cui, J.; Li, H.; Zhou, Y.; Wu, X.; Jiang, C.; Xu, Z.; Wang, R.; Xiong, L. Antipyretic Effects of Xiangqin Jiere Granules on Febrile Young Rats Revealed by Combining Pharmacodynamics, Metabolomics, Network Pharmacology, Molecular Biology Experiments and Molecular Docking Strategies. J. Biomol. Struct. Dyn. 2024, 43, 4183–4200. [Google Scholar]
- Santos, L.H.S.; Ferreira, R.S.; Caffarena, E.R. Integrating Molecular Docking and Molecular Dynamics Simulations. Methods Mol. Biol. 2019, 2053, 13–34. [Google Scholar] [CrossRef]
- Sargsyan, K.; Grauffel, C.; Lim, C. How Molecular Size Impacts RMSD Applications in Molecular Dynamics Simulations. J. Chem. Theory Comput. 2017, 13, 1518–1524. [Google Scholar] [CrossRef]
- Watanabe, K.; Saito, K.; Kinjo, M.; Matsuda, T.; Tamura, M.; Kon, S.; Miyazaki, T.; Uede, T. Molecular Dynamics of STAT3 on IL-6 Signaling Pathway in Living Cells. Biochem. Biophys. Res. Commun. 2004, 324, 1264–1273. [Google Scholar] [CrossRef]
- Elkazzaz, M.; Ullah, S.; Gao, T.; Shamkh, I.M.; Ahmed, A.; Wu, T.X.; Elsharayidi, M.S.; Khan, M.S.; Rehman, A.U.; Lotfy, M.M.; et al. Inhibition of Colorectal Cancer Targets IL-6, CTLA-4, & B7-2 by Tislelizumab: Molecular Docking, Dynamics, & STRING Protein-Protein Network Analysis. Inf. Med. Unlocked 2023, 41, 101323. [Google Scholar] [CrossRef]
- Rahardhian, M.R.R.; Yuniarti, N.; Ariani, L.W.; Suharsanti, R. In vitro Determination of Antioxidant Activity, Total Phenolics, Total Flavonoid, Anti-Cholesterol of Extracts Saffron (Crocus sativus). J. Glob. Pharma Technol. 2020, 12, 223–230. [Google Scholar]
- Suharsanti, R.; Sugihartini, N.; Lukitaningsih, E.; Rahardhian, M.R.R. Effect of Different Solvent on Total Phenolic, Total Flavonoid, and Sun Protection Factor of Belimbing Wuluh (Averrhoa bilimbi Linn.) Fruits Fraction. J. Glob. Pharma Technol. 2019, 11, 154–162. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2786–2791. [Google Scholar] [CrossRef]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Duellman, S.; Benink, H.; Worzella, T.; Minor, L. The Assay Guidance Manual: Cell Viability Assays; National Center for Biotechnology of Medicine: Bethesda, MD, USA, 2016. [Google Scholar]
- Beurel, E.; Jope, R.S. Lipopolysaccharide-Induced Interleukin-6 Production Is Controlled by Glycogen Synthase Kinase-3 and STAT3 in the Brain. J. Neuroinflamm. 2009, 6, 9. [Google Scholar] [CrossRef]




| No | Isolated Compound | Kaempferol [28] | ||
|---|---|---|---|---|
| δC, Type | δH, Mult. (J in Hz) | δC, Type | δH, Mult. (J in Hz) | |
| 2 | 146.7, C | 146.9, C | ||
| 3 | 135.6, C | 136.5, C | ||
| 4 | 175.8, C | 176.5, C | ||
| 5 | 160.6, C | 157.7, C | ||
| 6 | 98.1, CH | 6.19, d (2.0) | 98.9, CH | 6.21, d (2.0) |
| 7 | 163.8, C | 165.0, C | ||
| 8 | 93.4, CH | 6.44, d (2.0) | 94.3, CH | 6.43, d (2.0) |
| 9 | 156.0, C | 160.1, C | ||
| 10 | 102.9, C | 103.9, C | ||
| 1′ | 121.6, C | 123.1, C | ||
| 2′/6′ | 129.4, CH | 8.05, d (9.0) | 130.3, CH | 8.11, d (8.5) |
| 3′/5′ | 115.3, CH | 6.93, d (9.0) | 116.1, CH | 6.93, d (8.5) |
| 4′ | 159.1, C | 161.9, C | ||
| 3-OH | 9.43, s | 9.30, s | ||
| 5-OH | 12.50, s | 12.60, s | ||
| 7-OH | 10.80, s | 10.90, s | ||
| 4′-OH | 10.13, s | 10.20, s | ||
| No | Compound | ∆G kcal/mol | Ki (μM) | Types of Ligan Bonds and Interactions with Amino Acids | |||
|---|---|---|---|---|---|---|---|
| Hydrogen | Pi-Sigma | Pi-Alkyl | Pi-Amides | ||||
| 1 | TLA | −5.90 | 47.35 | Gln175, Arg182, Arg179 | - | - | - |
| 2 | Kaempferol | −5.98 | 41.28 | Gln175, Asp34 | Leu33 | Lys171 | Ile36 |
| Compound | Pharmacokinetic Prediction | Toxicity Prediction | |||
|---|---|---|---|---|---|
| Absorption Caco-2 log Papp in 10−6 cm/s) | Distribution BBB (BB log) | Metabolism (CYP2D6) (Yes/No) | Excretion (Total Clearance) (Log mL/min/kg) | Ames (Yes/No) | |
| Kaempferol | −5.26 | −2.75 | Not | 5.81 | Not |
| System | H-Bond Acceptors (res@atom) | H-Bond Donor (res@atom) | Fraction | Avg. Distance (Å) | Avg. Angle (◦) |
|---|---|---|---|---|---|
| TLA | TLA_158@O | Arg_155@H | 0.10 | 28.07 | 1.586.07 |
| TLA_158@O | Arg_155@H | 0.09 | 28.02 | 1.565.14 | |
| TLA_158@O | Arg_12@H | 0.09 | 28.03 | 1.606.04 | |
| TLA_158@O | Arg_155@H | 0.08 | 28.39 | 1.576.96 | |
| TLA_158@O | Arg_152@H | 0.07 | 28.48 | 1.574.45 | |
| TLA_158@O | Arg_152@H | 0.07 | 28.43 | 1.570.53 | |
| Kaempferol | Phe_98@O | Kaempferol@H | 0.20 | 28.60 | 1.501.65 |
| Kaempferol@O | Arg_6@H | 0.01 | 29.16 | 1.492.99 | |
| Arg_6@O | Kaempferol@H | 0.00 | 28.70 | 1.569.58 | |
| Kaempferol@O | Ser_3@H | 0.00 | 28.93 | 1.492.76 | |
| Kaempferol@O | Leu_1@H | 0.00 | 28.72 | 1.469.966 | |
| Gln_97@O | Kaempferol@H | 0.0023 | 28.66 | 1.544.905 | |
| Kaempferol@O | Gln_10@H | 0.0023 | 28.68 | 1.493.293 | |
| Leu_1@O | Kaempferol@H | 0.0022 | 28.62 | 1.434.219 | |
| Kaempferol@O | Leu_1@H | 0.0022 | 28.69 | 1.464.555 | |
| Kaempferol@O | Leu_1@H | 0.0020 | 28.68 | 1.468.561 | |
| Kaempferol@O | Lys_101@H | 0.0018 | 28.63 | 1.552.401 |
| Energy Component | Bond Energy (kcal/mol) | ||||||
|---|---|---|---|---|---|---|---|
| ∆GVDW | ∆GEL | ∆EGB | ∆ESURF | ∆GGAS | ∆GSOLV | ∆GTOTAL | |
| TLA | −0.44 | −37.06 | 36.68 | −0.19 | −37.50 | 36.50 | −1.00 |
| Kaempferol | −17.02 | −293.16 | 295.06 | −2.73 | −310.18 | 292.33 | −17.85 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahardhian, M.R.R.; Sumiwi, S.A.; Susilawati, Y.; Muchtaridi, M. Immunomodulatory Potential of Kaempferol Isolated from Peronema canescens Jack. Leaves Through Inhibition of IL-6 Expression. Int. J. Mol. Sci. 2025, 26, 3068. https://doi.org/10.3390/ijms26073068
Rahardhian MRR, Sumiwi SA, Susilawati Y, Muchtaridi M. Immunomodulatory Potential of Kaempferol Isolated from Peronema canescens Jack. Leaves Through Inhibition of IL-6 Expression. International Journal of Molecular Sciences. 2025; 26(7):3068. https://doi.org/10.3390/ijms26073068
Chicago/Turabian StyleRahardhian, Muhammad Ryan Radix, Sri Adi Sumiwi, Yasmiwar Susilawati, and Muchtaridi Muchtaridi. 2025. "Immunomodulatory Potential of Kaempferol Isolated from Peronema canescens Jack. Leaves Through Inhibition of IL-6 Expression" International Journal of Molecular Sciences 26, no. 7: 3068. https://doi.org/10.3390/ijms26073068
APA StyleRahardhian, M. R. R., Sumiwi, S. A., Susilawati, Y., & Muchtaridi, M. (2025). Immunomodulatory Potential of Kaempferol Isolated from Peronema canescens Jack. Leaves Through Inhibition of IL-6 Expression. International Journal of Molecular Sciences, 26(7), 3068. https://doi.org/10.3390/ijms26073068









