An Assessment of the Currently Available Molecular Assay for the Diagnosis of Anisakis Sensitization
Abstract
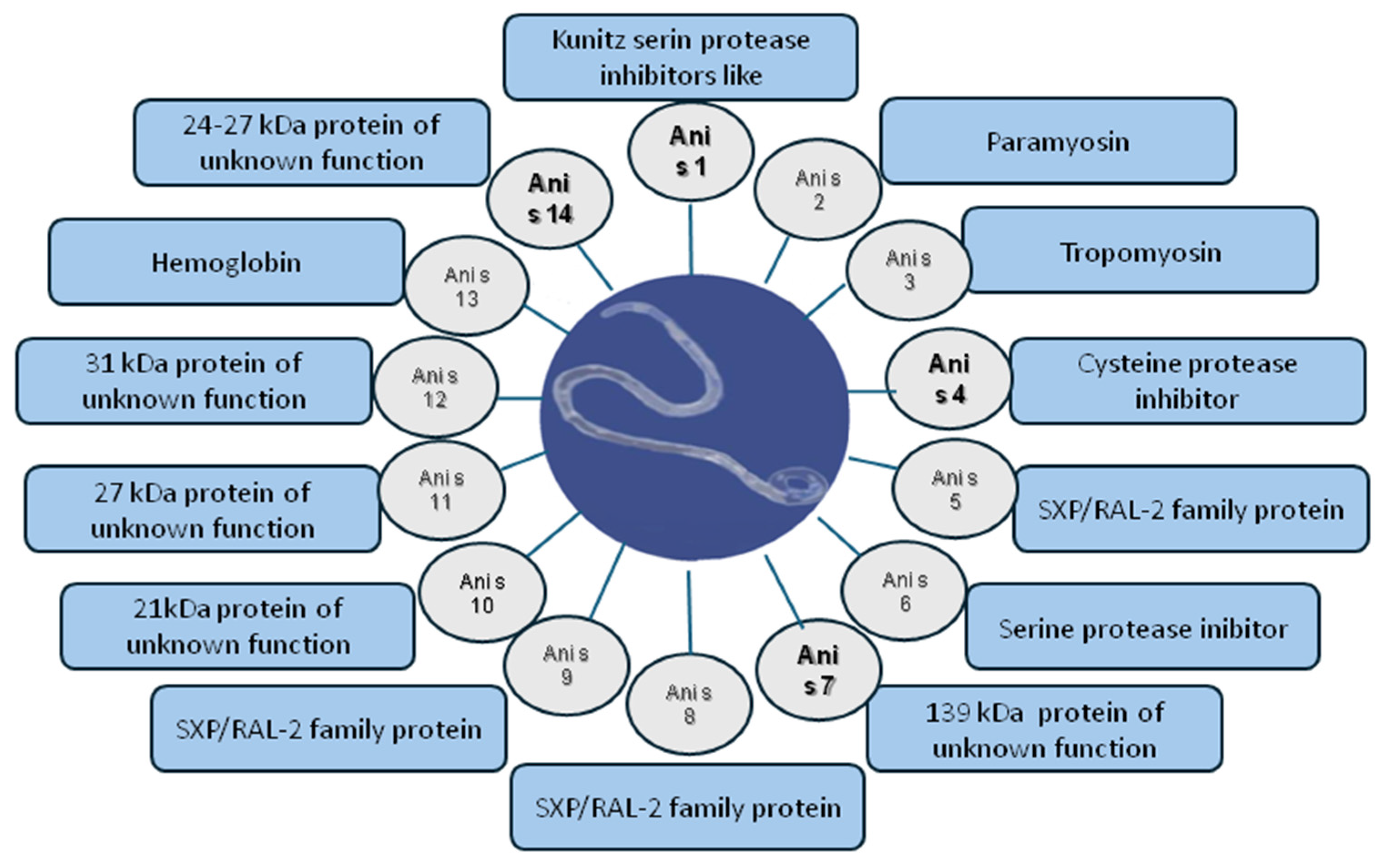
1. Introduction
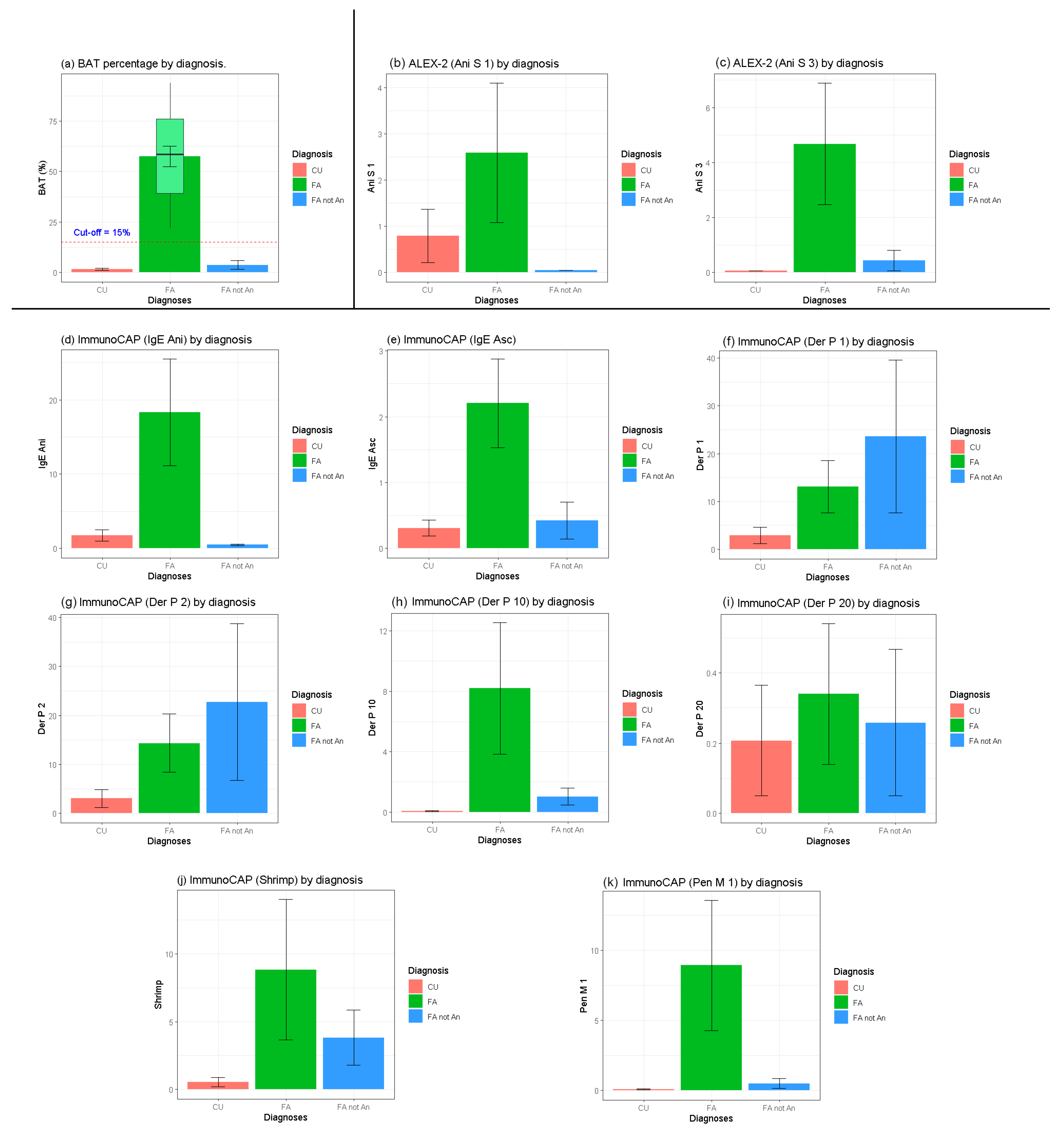
2. Results
3. Discussion
3.1. Statistical Evaluation
3.2. Diagnostic Performance of ALEX-2
3.3. Limits and Perspectives
4. Materials and Method
4.1. Study Design and Population
4.2. Laboratory Analyses
4.3. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, J.W.; Wootten, R. Anisakis and anisakiasis. Adv. Parasitol. 1987, 16, 93–163. [Google Scholar]
- World Health Organization. Soil-Transmitted Helminths. Available online: https://www.who.int/news-room/fact-sheets/detail/soil-transmitted-helminth-infections (accessed on 19 February 2025).
- Pravettoni, V.; Primavesi, L.; Piantanida, M. Anisakis simplex: Current knowledge. Eur. Ann. Allergy Clin. Immunol. 2012, 44, 150–156. [Google Scholar] [PubMed]
- Baird, F.J.; Gasser, R.B.; Jabbar, A.; Lopata, A.L. Foodborne anisakiasis and allergy. Mol. Cell Probes 2014, 28, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Tejada, M.; Olivares, F.; de las Heras, C.; Careche, M.; Solas, M.T.; García, M.L.; Fernandez, A.; Mendizábal, A.; Navas, A.; Rodríguez-Mahillo, A.I.; et al. Antigenicity of Anisakis simplex s.s. L3 in parasitized fish after heating conditions used in the canning processing. J. Sci. Food Agric. 2015, 95, 922–927. [Google Scholar] [CrossRef]
- Mazzucco, W.; Raia, D.D.; Marotta, C.; Costa, A.; Ferrantelli, V.; Vitale, F.; Casuccio, A. Anisakis sensitization in different population groups and public health impact: A systematic review. PLoS ONE 2018, 13, e0203671. [Google Scholar] [CrossRef]
- Mazzucco, W.; Lacca, G.; Cusimano, R.; Provenzani, A.; Costa, A.; Di Noto, A.M.; Massenti, M.F.; Leto-Barone, M.S.; Lorenzo, G.D.; Vitale, F. Prevalence of sensitisation to Anisakis simplex among professionally exposed populations. Arch. Environ. Occup. Health 2012, 67, 91–97. [Google Scholar]
- Carballeda-Sangiao, N.; Olivares, F.; Rodriguez-Mahillo, A.I.; Careche, M.; Tejada, M.; Moneo, I.; González-Muñuoz, M. Identification of autoclave-resistant Anisakis simplex allergens. J. Food Prot. 2014, 77, 605–609. [Google Scholar]
- Petithory, J.C. New data on anisakiasis. Bull. Acad. Natl. Med. 2007, 191, 53–65. [Google Scholar]
- Pozo, M.D.; Audicana, M.; Diez, J.M.; Munoz, D.; Ansotegui, I.J.; Fernández, E.; García, M.; Etxenagusia, M.; Moneo, I.; de Corres, L.F. Anisakis simplex, a relevant etiologic factor in acute urticaria. Allergy 1997, 52, 576–579. [Google Scholar] [CrossRef]
- Daschner, A.; De Frutos, C.; Valls, A.; Vega, F. Anisakis simplex sensitization-associated urticaria: Short-lived immediate type or prolonged acute urticaria. Arch. Dermatol. Res. 2010, 302, 625–629. [Google Scholar] [CrossRef]
- Sastre, J.; Lluch-Bernal, M.; Quirce, S.; Arrieta, I.; Lahoz, C.; Del Amo, A.; Fernández-Caldas, E.; Marañón, F. A double blind, placebo-controlled oral challenge study with lyophilized larvae and antigen of the fish parasite, Anisakis simplex. Allergy 2000, 55, 560–564. [Google Scholar] [PubMed]
- Ventura, M.T.; Napolitano, S.; Menga, R.; Cecere, R.; Asero, R. Anisakis simplex hypersensitivity is associated with chronic urticaria in endemic areas. Int. Arch. Allergy Immunol. 2013, 160, 297–300. [Google Scholar] [PubMed]
- Kobayashi, Y.; Ikeda, K.; Shiomi, K. Elucidation of IgE-binding epitopes of Ani s 1: The major Anisakis simplex allergen. J. Mol. Biochem. Parasitol. 2010, 174, 128–131. [Google Scholar] [CrossRef]
- Caballero, M.L.; Asero, R.; Antonicelli, L.; Kamberi, E.; Colangelo, C.; Fazii, P.; De Burgos, C.; Rodriguez-Perez, R. Anisakis allergy component-resolved diagnosis: Clinical and immunologic differences between patients from Italy and Spain. Int. Arch. Allergy Immunol. 2013, 162, 39–44. [Google Scholar] [PubMed]
- Lin, A.H.; Nepstad, I.; Florvaag, E.; Egaas, E.; van Do, T. An extended study of seroprevalence of anti-Anisakis simplex IgE antibodies in Norwegian blood donors. J. Scand. J. Immunol. 2014, 79, 61–67. [Google Scholar]
- Cuéllar, C.; Daschner, A.; Valls, A.; De Frutos, C.; Fernández-Fígares, V.; Anadón, A.M.; Rodríguez, E.; Gárate, T.; Rodero, M.; Ubeira, F.M. Ani s 1 and Ani s 7 recombinant allergens are able to differentiate distinct Anisakis simplex-associated allergic clinical disorders. Arch. Dermatol. Res. 2012, 304, 283–288. [Google Scholar] [CrossRef]
- Moneo, I.; Caballero, M.L.; Gómez, F.; Ortega, E.; Alonso, M.J. Isolation and characterization of a major allergen from the fish parasite Anisakis simplex. J. Allergy Clin. Immunol. 2000, 106, 177–182. [Google Scholar]
- Toro, C.; Caballero, M.L.; Baquero, M.; Garcia-Samaniego, J.; Casado, I.; Rubio, M.; Moneo, I. High Prevalence of Seropositivity to a Major Allergen of Anisakis simplex, Ani s 1, in Dyspeptic Patients. Clin. Diagn. Lab. Immunol. 2004, 11, 115–118. [Google Scholar]
- Shimakura, K.; Miura, H.; Ikeda, K.; Ishizaki, S.; Nagashima, Y.; Shirai, T.; Kasuya, S.; Shiomi, K. Purification and molecular cloning of a major allergen from Anisakis simplex. Mol. Biochem. Parasitol. 2004, 135, 69–75. [Google Scholar]
- Moneo, I.; Caballero, M.L.; Gonzalez-Munoz, M.; Rodriguez-Mahillo, A.I.; Rodriguez-Perez, R.; Silva, A. Isolation of a heat-resistant allergen from the fish parasite Anisakis simplex. Parasitol. Res. 2005, 96, 285–289. [Google Scholar]
- Rodríguez, E.; Anadón, A.M.; García-Bodas, E.; Romarís, F.; Iglesias, R.; Gárate, T.; Ubeira, F.M. Novel sequences and epitopes of diagnostic value derived from the Anisakis simplex Ani s 7 major allergen. Allergy 2008, 63, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Anadon, A.M.; Romaris, F.; Escalante, M.; Rodriguez, E.; Garate, T.; Cuellar, C.; Ubeira, F.M. The Anisakis simplex Ani s 7 major allergen as an indicator of true Anisakis infections. Clin. Exp. Immunol. 2009, 156, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kakemoto, S.; Shimakura, K.; Shiomi, K. Molecular cloning and expression of a new major allergen, Ani s 14, from Anisakis simplex. Shokuhin Eiseigaku Zasshi 2015, 56, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Perez-Perez, J.; Fernandez-Caldas, E.; Maranon, F.; Sastre, J.; Bernal, M.L.; Rodriguez, J. Molecular cloning of paramyosin, a new allergen of Anisakis simplex. Int. Arch. Allergy Immunol. 2000, 123, 120–129. [Google Scholar] [CrossRef]
- Kochanowski, M.; Różycki, M.; Dąbrowska, J.; Bełcik, A.; Karamon, J.; Sroka, J.; Cencek, T. Proteomic and Bioinformatic Investigations of Heat-Treated Anisakis simplex Third-Stage Larvae. Biomolecules 2020, 10, 1066. [Google Scholar] [CrossRef]
- Fæste, C.K.; Jonscher, K.R.; Dooper, M.M.; Egge-Jacobsen, W.; Moen, A.; Daschner, A.; Egaas, E.; Christians, U. Characterisation of potential novel allergens in the fish parasite Anisakis simplex. EuPA Open Proteom. 2014, 4, 140–155. [Google Scholar] [CrossRef]
- González-Fernández, J.; Daschner, A.; Nieuwenhuizen, N.E.; Lopata, A.L.; De Frutos, C.; Valls, A.; Cuéllar, C. Haemoglobin, a new major allergen of Anisakis simplex. Int. J. Parasitol. 2015, 45, 399–407. [Google Scholar] [CrossRef]
- Fitzsimmons, C.M.; Falcone, F.H.; Dunne, D.W. Helminth allergens, parasite-specific IgE, and its protective role in human immunity. Front. Immunol. 2014, 5, 61. [Google Scholar] [CrossRef]
- Gamboa, P.M.; Asturias, J.; Martínez, R.; Antépara, I.; Jáuregui, I.; Urrutia, I.; Fernández, J.; Sanz, M.L. Diagnostic utility of components in allergy to Anisakis simplex. J. Investig. Allergol. Clin. Immunol. 2012, 22, 13–19. [Google Scholar]
- Gonzalez-Muñoz, M.; Luque, R.; Nauwelaers, F.; Moneo, I. Detection of Anisakis simplex-induced basophil activation by flow cytometry. Cytometry B Clin. Cytom. 2005, 68, 31–36. [Google Scholar] [CrossRef]
- Frezzolini, A.; Cadoni, S.; De Pita, O. Usefulness of the CD63 basophil activation test in detecting Anisakis hypersensitivity in patients with chronic urticaria: Diagnosis and follow-up. Clin. Exp. Dermatol. 2010, 35, 765–770. [Google Scholar] [PubMed]
- Brusca, I.; Graci, S.; Barrale, M.; Cammilleri, G.; Zarcone, M.; Onida, R.; Costa, A.; Ferrantelli, V.; Buscemi, M.D.; Uasuf, C.G.; et al. Use of a comprehensive diagnostic algorithm for Anisakis allergy in a high seroprevalence Mediterranean setting. Eur. Ann. Allergy Clin. Immunol. 2020, 52, 131–141. [Google Scholar] [PubMed]
- Brusca, I.; Barrale, M.; Zarcone, M.; Fruscione, S.; Onida, R.; De Bella, D.D.; Alba, D.; Belluzzo, M.; Uasuf, C.G.; Cammilleri, G.; et al. Basophil Activation Test in the Diagnosis of Anisakis Allergy: An Observational Study from an Area of High Seafood Consumption in Italy. Pathogens 2023, 12, 777. [Google Scholar] [CrossRef] [PubMed]
- Lanfranchi, A.L.; Sardella, N.H. Anisakis survival after microwaving, freezing and salting fish from Argentina. Food Sci. Technol. Res. 2010, 16, 499–504. [Google Scholar]
- Audicana, M.T.; Ansotegui, I.J.; de Corres, L.F.; Kennedy, M.W. Anisakis simplex: Dangerous—Dead and alive? Trends Parasitol. 2002, 18, 20–25. [Google Scholar] [CrossRef]
- FAO/WHO. Evaluation of Allergenicity of Genetically Modified Foods. Report of a Joint FAO/WHO Expert Consultation on Allergenicity of Foods from Biotechnology; FAO/WHO: Rome, Italy, 2002. [Google Scholar]
- Ruethers, T.; Taki, A.C.; Johnston, E.B.; Nugraha, R.; Le, T.T.; Kalic, T.; McLean, T.R.; Kamath, S.D.; Lopata, A.L. Seafood allergy: A comprehensive review of fish and shellfish allergens. Mol. Immunol. 2018, 100, 28–57. [Google Scholar]
- Peixoto, S.; Monteiro, T.; Carvalho, M.; Santos, M.; Matos, C.; Bartolomé, B.; Labrador-Horrillo, M.; Quaresma, M. Vertebrate Tropomyosin as an Allergen. J. Investig. Allergol. Clin. Immunol. 2018, 28, 51–53. [Google Scholar]
- Liu, R.; Holck, A.L.; Yang, E.; Liu, C.; Xue, W. Tropomyosin from tilapia (Oreochromis mossambicus) as an allergen. Clin. Exp. Allergy 2013, 43, 365–377. [Google Scholar]
- Ruethers, T.; Taki, A.C.; Karnaneedi, S.; Nie, S.; Kalic, T.; Dai, D.; Daduang, S.; Leeming, M.; Williamson, N.A.; Breiteneder, H.; et al. Expanding the allergen repertoire of salmon and catfish. Allergy 2021, 76, 1443–1453. [Google Scholar]
- Celi, G.; Brusca, I.; Scala, E.; Villalta, D.; Pastorello, E.; Farioli, L.; Cortellini, G.; Deleonardi, G.; Galati, P.; Losappio, L.; et al. House dust mite allergy and shrimp allergy: A complex interaction. Eur. Ann. Allergy Clin. Immunol. 2020, 52, 205–209. [Google Scholar]
- de Las Vecillas, L.; Muñoz-Cacho, P.; López-Hoyos, M.; Monttecchiani, V.; Martínez-Sernández, V.; Ubeira, F.M.; Rodríguez-Fernández, F. Analysis of Ani s 7 and Ani s 1 allergens as biomarkers of sensitization and allergy severity in human anisakiasis. Sci. Rep. 2020, 10, 11275. [Google Scholar] [CrossRef] [PubMed]
- Cardona, V.; Ansotegui, I.J. Component-resolved diagnosis in anaphylaxis. Curr. Opin. Allergy Clin. Immunol. 2016, 16, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Aibinu, I.E.; Smooker, P.M.; Lopata, A.L. Anisakis nematodes in fish and shellfish—From infection to allergies. Int. J. Parasitol. Parasites Wildl. 2019, 9, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Armentia, A.; Santos, J.; Serrano, Z.; Martín, B.; Martín, S.; Barrio, J.; Fernández, S.; González-Sagrado, M.; Pineda, F.; Palacios, R. Molecular diagnosis of allergy to Anisakis simplex and Gymnorhynchus gigas fish parasites. Allergol. Immunopathol. 2017, 45, 463–472. [Google Scholar] [CrossRef]
- González-Fernández, J.; Rivas, L.; Luque-Ortega, J.R.; Núñez-Ramírez, R.; Campioli, P.; Gárate, T.; Perteguer, M.J.; Daschner, A.; Cuéllar, C. Recombinant vs. native Anisakis haemoglobin (Ani s 13): Its appraisal as a new gold standard for the diagnosis of allergy. Exp. Parasitol. 2017, 181, 119–129. [Google Scholar] [CrossRef]
- Yang, A.C.; Arruda, L.K.; Santos, A.B.R.; Barbosa, M.C.; Chapman, M.D.; Galvão, C.E.; Kalil, J.; Morato-Castro, F.F. Measurement of IgE antibodies to shrimp tropomyosin is superior to skin prick testing with commercial extract and measurement of IgE to shrimp for predicting clinically relevant allergic reactions after shrimp ingestion. J. Allergy Clin. Immunol. 2010, 125, 872–878. [Google Scholar] [CrossRef]
- Sampson, H.A. Utility of food-specific IgE concentrations in predicting symptomatic food allergy. J. Allergy Clin. Immunol. 2001, 107, 891–896. [Google Scholar] [CrossRef]
- Sampson, H.A.; Aceves, S.; Bock, S.A.; James, J.; Jones, S.; Lang, D.; Nadeau, K.; Nowak-Wegrzyn, A.; Oppenheimer, J.; Perry, T.T.; et al. Food allergy: A practice parameter update—2014. J. Allergy Clin. Immunol. 2014, 134, 1016–1025. [Google Scholar] [CrossRef]
- Ukleja-Sokołowska, N.; Lis, K.; Żbikowska-Gotz, M.; Adamczak, R.; Kuźmiński, A.; Bartuzi, Z. Clinical utility of immunological methods based on the singleplex and multiplex ImmunoCap systems for diagnosis of shrimp allergy. J. Int. Med. Res. 2021, 49, 3000605211006597. [Google Scholar] [CrossRef]
- Foong, R.X.; Roberts, G.; Fox, A.T.; du Toit, G. Pilot study: Assessing the clinical diagnosis of allergy in atopic children using a macroarray assay in addition to skin prick testing and serum specific IgE. Clin. Mol. Allergy 2016, 14, 8. [Google Scholar] [CrossRef]
- Buzzulini, F.; Da Re, M.; Scala, E.; Martelli, P.; Conte, M.; Brusca, I.; Villalta, D. Evaluation of a new multiplex assay for allergy diagnosis. Clin. Chim. Acta 2019, 493, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Cañas, J.A.; Pérez-Sánchez, N.; Lopera-Doblas, L.; Palomares, F.; Molina, A.; Bartra, J.; Torres, M.J.; Gómez, F.; Mayorga, C. Basophil Activation Test Utility as a Diagnostic Tool in LTP Allergy. Int. J. Mol. Sci. 2022, 23, 4979. [Google Scholar] [CrossRef] [PubMed]
- Fruscione, S.; Barrale, M.; Zarcone, M.; Alba, D.; Ravazzolo, B.; Belluzzo, M.; Onida, R.; Cammilleri, G.; Costa, A.; Ferrantelli, V.; et al. Screening of Anisakis-Related Allergies and Associated Factors in a Mediterranean Community Characterized by High Seafood Consumption. Foods 2024, 13, 2821. [Google Scholar] [CrossRef] [PubMed]
| Specific IgE kIU/L | ALEX 2 kIU/L | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | SPT | Anisakis | Ascaris | Dermat. P | Dermat. F | Tropom.Mites | Shrimp | Tropom.Shrimp | BAT Anisakis | Diagnosis | Grade | Ani s 1 | Ani s 3 | Der p 20 |
| 1 | + | 1.91 | 0.15 | 3.56 | 3.73 | 0.08 | 0 | 0.83 | 24.44 | FA | 1 | <0.10 | <0.10 | <0.10 |
| 2 | − | 0.42 | 0.84 | 0.07 | 0.05 | 0.01 | 0 | 0 | 3.15 | CU | 0 | <0.10 | <0.10 | <0.10 |
| 3 | + | 2.29 | 0.18 | 1.05 | 1.39 | 0.03 | 0.84 | 0 | 2.36 | CU | 0 | <0.10 | <0.10 | <0.10 |
| 4 | − | 0.19 | 0.04 | 97.8 | 100 | 0.07 | 0.04 | 0.03 | 1.46 | FA not An | 1 | <0.10 | <0.10 | <0.10 |
| 5 | + | 100 | 7.27 | 0.03 | 0.05 | 0.05 | 0.09 | 0.03 | 93.72 | FA | 1 | 5.35 | <0.10 | <0.10 |
| 6 | + | 2.31 | 3.01 | 6.21 | 5.47 | 9.87 | 12.5 | 10.3 | 36.58 | FA | 1 | <0.10 | 9.76 | 3.26 |
| 7 | + | 5.4 | 0.47 | 0.86 | 0.47 | 0.1 | 0.26 | 0.1 | 0.69 | CU | 0 | 1.19 | <0.10 | <0.10 |
| 8 | + | 98.3 | 6 | 0.03 | 0.02 | 0.1 | 0.09 | 0.1 | 84.43 | FA | 2 | <0.10 | <0.10 | <0.10 |
| 9 | − | 1.04 | 1.8 | 1.66 | 2.4 | 0.1 | 7.52 | 0.1 | 0.21 | FA not An | 1 | <0.10 | <0.10 | <0.10 |
| 10 | + | 3.99 | 6.16 | 41.5 | 43.4 | 31.4 | 27.9 | 30.27 | 74.07 | FA | 2 | <0.10 | 21.07 | <0.10 |
| 11 | + | 1.92 | 0.15 | 0.53 | 0.57 | 0.01 | 0.79 | 0.01 | 21.57 | FA | 1 | <0.10 | <0.10 | 2.63 |
| 12 | − | 0.21 | 0.55 | 6.78 | 6.69 | 0.11 | 2.53 | 0.14 | 0.2 | CU | 0 | <0.10 | <0.10 | <0.10 |
| 13 | − | 0.06 | 0.06 | 0.36 | 0.07 | 0.01 | 0.6 | 0.01 | 39.52 | FA | 1 | <0.10 | <0.10 | <0.10 |
| 14 | − | 0.67 | 3.44 | 12.4 | 10.25 | 0.03 | 0.83 | 0.05 | 30.63 | FA | 1 | <0.10 | <0.10 | <0.10 |
| 15 | − | 0.53 | 0.16 | 37.3 | 27.5 | 0.75 | 0.32 | 0.69 | 0.97 | FA not An | 1 | <0.10 | <0.10 | <0.10 |
| 16 | + | 7.3 | 0.58 | 16.9 | 21 | 39.2 | 29.2 | 51 | 41.24 | FA | 1 | 0.26 | 27.92 | <0.10 |
| 17 | − | 0.43 | 2.24 | 0.07 | 0.09 | 0.01 | 0.01 | 0.03 | 48 | FA | 1 | <0.10 | <0.10 | <0.10 |
| 18 | − | 0.32 | 0.05 | 0.9 | 0.77 | 0.01 | 0.91 | 0.03 | 13.4 | FA not An | 1 | <0.10 | <0.10 | 1.3 |
| 19 | − | 0.49 | 0.37 | 0.88 | 0.91 | 1.89 | 12.3 | 2.09 | 5.19 | FA not An | 1 | <0.10 | <0.10 | <0.10 |
| 20 | + | 3.2 | 0.07 | 0.04 | 0.09 | 0.01 | 0.03 | 0.05 | 3.15 | CU | 0 | 4.09 | <0.10 | <0.10 |
| 21 | − | 0.24 | 0.09 | 3.11 | 4.71 | 3.42 | 1.81 | <0.10 | 1.01 | FA not An | 1 | <0.10 | 2.34 | <0.10 |
| 22 | − | 0.36 | 0.02 | 5.53 | 4.02 | 0.05 | 0.05 | 4.18 | 42.4 | FA | 1 | 4.09 | <0.10 | <0.10 |
| 23 | + | 19.1 | 0.72 | 0.02 | 0.03 | 0 | 0 | 0.04 | 74.8 | FA | 1 | <0.10 | <0.10 | <0.10 |
| 24 | − | 0.38 | 0.05 | 0.04 | 0.03 | 0.05 | 0.05 | 0.02 | 0.35 | CU | 0 | <0.10 | <0.10 | <0.10 |
| 25 | + | 4.09 | 10.5 | 100 | 100 | 76.6 | 100 | 78.3 | 81.3 | FA | 2 | 4.56 | 31.14 | <0.10 |
| 26 | − | 0.35 | 0 | 11.6 | 12.5 | 0.28 | 0.14 | 0.23 | 1.12 | CU | 0 | <0.10 | <0.10 | 1.15 |
| 27 | + | 15 | 1.89 | 0.01 | 0.01 | 0.01 | 0.06 | 0.01 | 74.6 | FA | 2 | <0.10 | <0.10 | <0.10 |
| 28 | + | 74.2 | 0.29 | 0.06 | 0.03 | 0.06 | 0.05 | 0.01 | 58.9 | FA | 2 | 29.67 | <0.10 | <0.10 |
| 29 | + | 2.35 | 0.08 | 35.58 | 32.14 | 0.01 | 2.58 | 0.01 | 61.27 | FA | 1 | <0.10 | <0.10 | <0.10 |
| 30 | + | 12.4 | 0.11 | 0.01 | 0.01 | 0 | 0 | 0.01 | 58 | FA | 1 | <0.10 | <0.10 | <0.10 |
| 31 | − | 0.36 | 1.01 | 3.24 | 2.97 | 6.25 | 1.92 | 3.17 | 79.67 | FA | 1 | <0.10 | 2.93 | <0.10 |
| 32 | + | 14 | 0.2 | 0.14 | 0.16 | 0 | 0.01 | 0 | 85.3 | FA | 1 | 7.19 | <0.10 | <0.10 |
| 33 | + | 7.35 | 0.19 | 35.9 | 62.2 | 0.01 | 0.05 | 0.05 | 37.67 | FA | 1 | <0.10 | <0.10 | <0.10 |
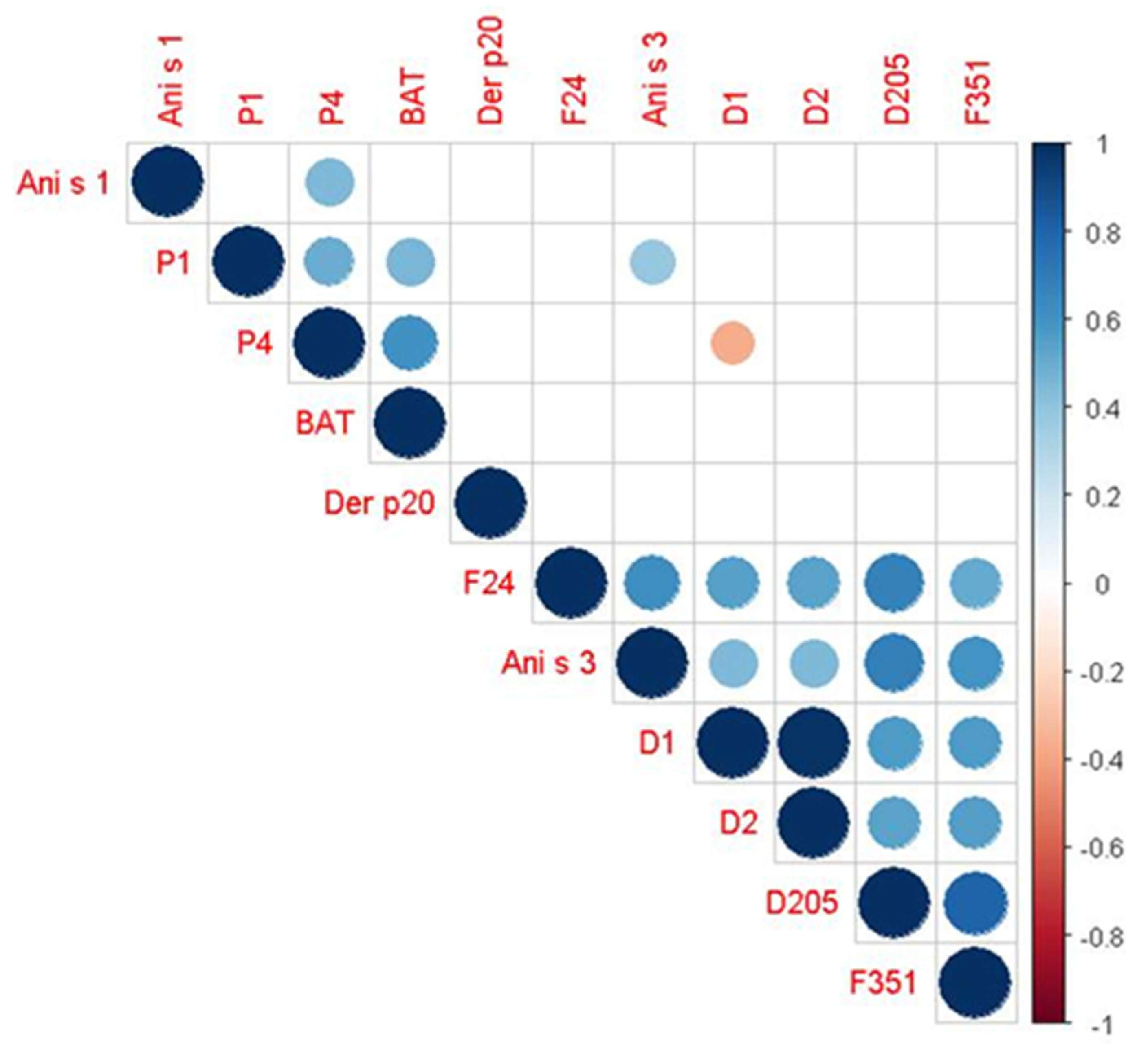
| Spearman’s ρ (p-Value) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Variables | P1 | P4 | Ani S3 | D1 | D2 | D205 | F24 | F351 |
| P1 | – | +49.7% (0.003) | – | – | – | – | – | – |
| BAT | +45.0% (0.009) | +60.7% (<0.001) | – | – | – | – | – | – |
| Ani S1 | – | +44.3% (0.010) | – | – | – | – | – | – |
| Ani S3 | +38.9% (0.025) | – | – | +44.3% (0.010) | +44.3% (0.010) | +68.0% (<0.001) | +61.6% (<0.001) | +59.5% (<0.001) |
| D1 | – | −36.1% (0.038) | +44.3% (0.010) | – | +98.6% (<0.001) | +56.8% (<0.001) | 54.8% (<0.001) | +56.1% (<0.001) |
| D2 | – | – | +44.3% (0.010) | +98.6% (<0.001) | – | +53.8% (0.001) | +53.6% (0.001) | +55.7% (<0.001) |
| D205 | – | – | +68.0% (<0.001) | +56.8% (<0.001) | +53.8% (0.001) | – | +67.7% (<0.001) | +80.5% (<0.001) |
| F24 | – | – | +61.6% (<0.001) | 54.8% (<0.001) | +53.6% (0.001) | +67.7% (<0.001) | – | +50.6% (0.003) |
| F351 | – | – | +59.5% (<0.001) | +56.1% (<0.001) | +55.7% (<0.001) | +80.5% (<0.001) | +50.6% (0.003) | – |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrale, M.; Mazzucco, W.; Fruscione, S.; Zarcone, M.; Cantisano, V.; Cammilleri, G.; Costa, A.; Ferrantelli, V.; Onida, R.; Scala, E.; et al. An Assessment of the Currently Available Molecular Assay for the Diagnosis of Anisakis Sensitization. Int. J. Mol. Sci. 2025, 26, 3033. https://doi.org/10.3390/ijms26073033
Barrale M, Mazzucco W, Fruscione S, Zarcone M, Cantisano V, Cammilleri G, Costa A, Ferrantelli V, Onida R, Scala E, et al. An Assessment of the Currently Available Molecular Assay for the Diagnosis of Anisakis Sensitization. International Journal of Molecular Sciences. 2025; 26(7):3033. https://doi.org/10.3390/ijms26073033
Chicago/Turabian StyleBarrale, Maria, Walter Mazzucco, Santo Fruscione, Maurizio Zarcone, Vincenzo Cantisano, Gaetano Cammilleri, Antonella Costa, Vincenzo Ferrantelli, Rosa Onida, Enrico Scala, and et al. 2025. "An Assessment of the Currently Available Molecular Assay for the Diagnosis of Anisakis Sensitization" International Journal of Molecular Sciences 26, no. 7: 3033. https://doi.org/10.3390/ijms26073033
APA StyleBarrale, M., Mazzucco, W., Fruscione, S., Zarcone, M., Cantisano, V., Cammilleri, G., Costa, A., Ferrantelli, V., Onida, R., Scala, E., Villalta, D., Uasuf, C. G., & Brusca, I. (2025). An Assessment of the Currently Available Molecular Assay for the Diagnosis of Anisakis Sensitization. International Journal of Molecular Sciences, 26(7), 3033. https://doi.org/10.3390/ijms26073033







