Flutamide Promotes Early Hepatocarcinogenesis Through Mitophagy in High-Fat Diet-Fed Non-Obese Steatotic Rats
Abstract
1. Introduction
2. Results
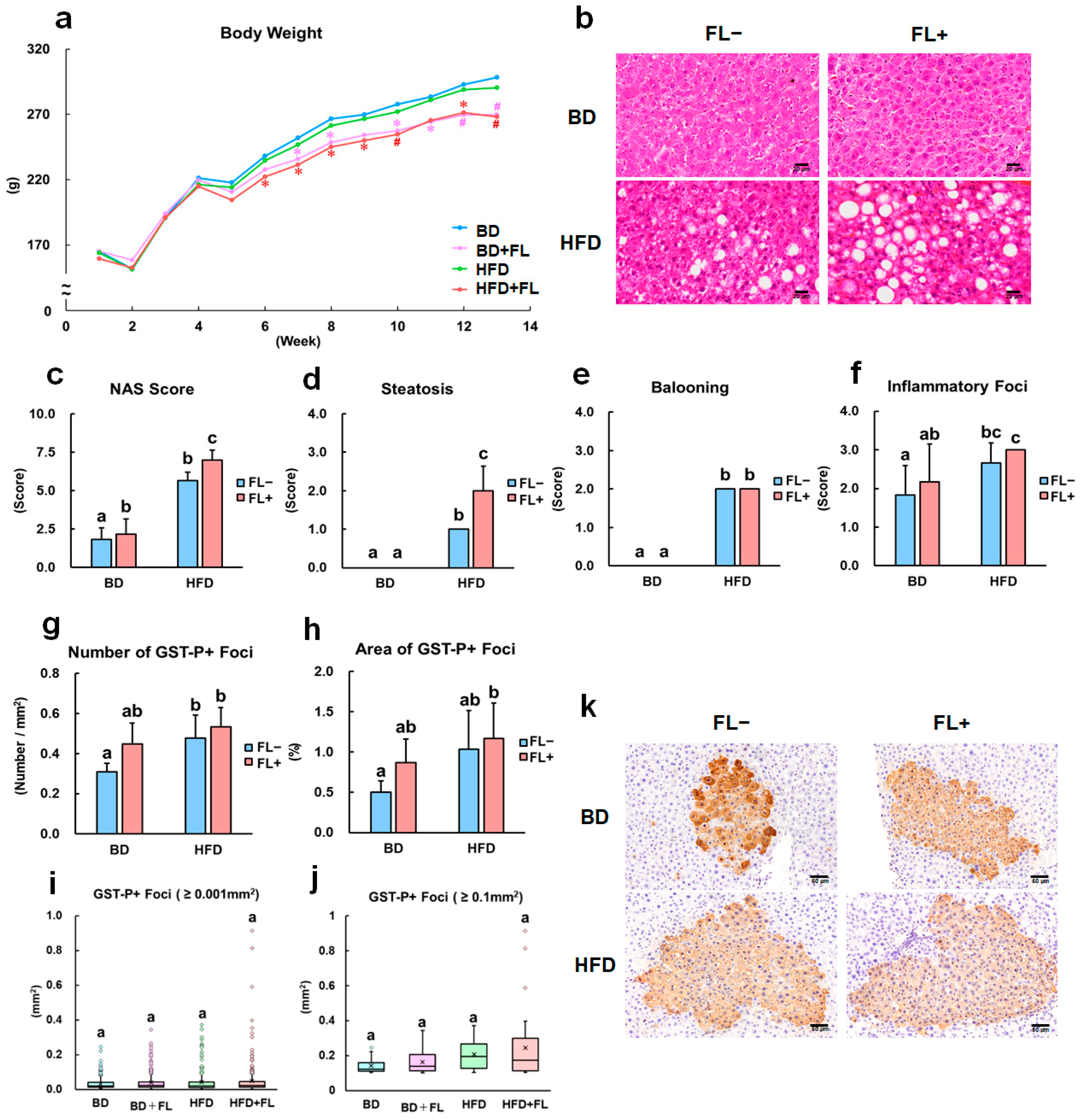
2.1. Flutamide Inhibits Body Weight Growth, Increases HFD-Induced NAFLD Score, and Enhances Preneoplastic Hepatic Lesions
2.2. Expression Analysis of Mitophagy Markers in Background Hepatocytes and Precancerous Lesions
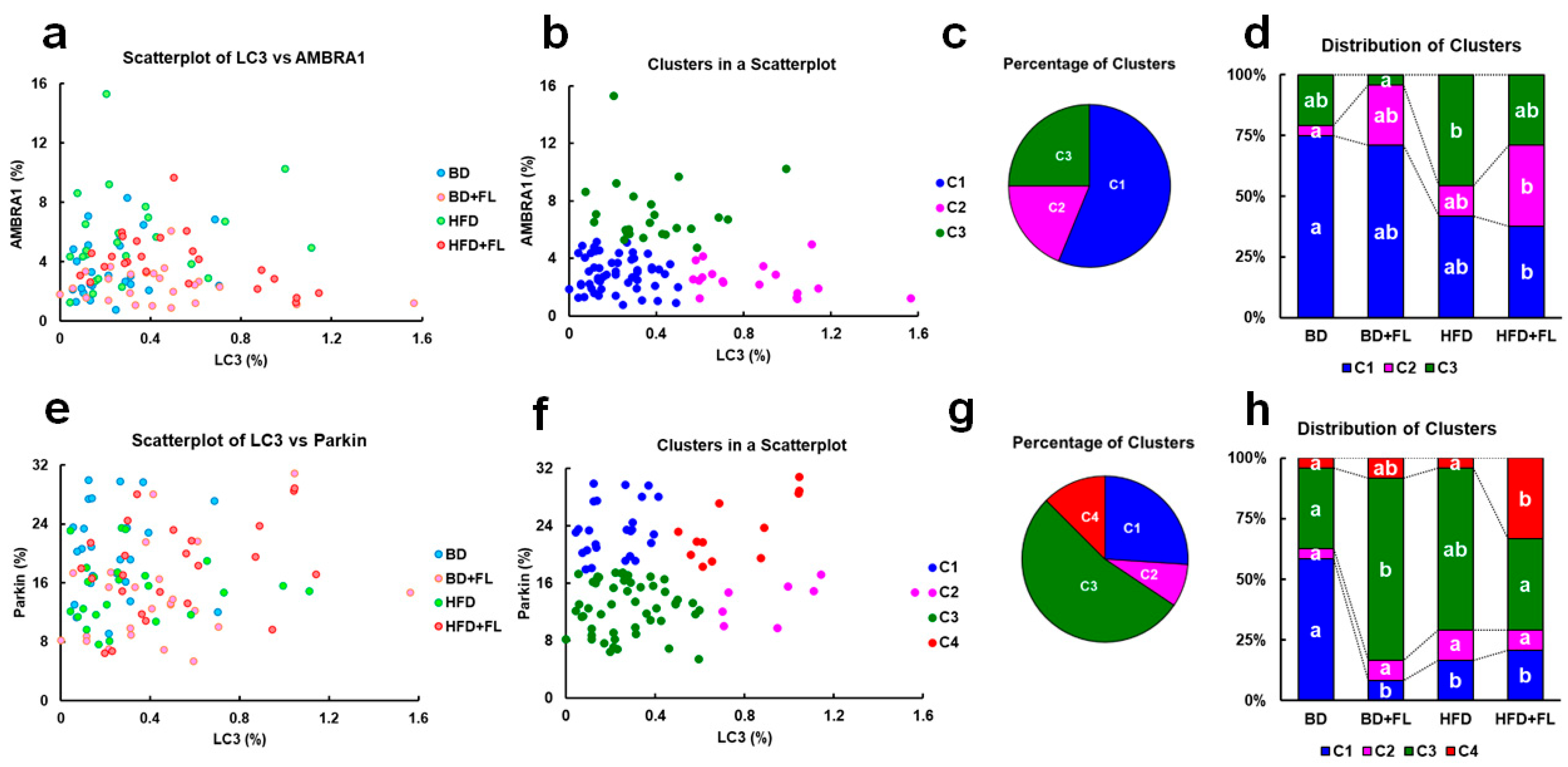
2.3. Cluster Analysis of LC3, AMBRA1, and Parkin Expression in Background Hepatocytes
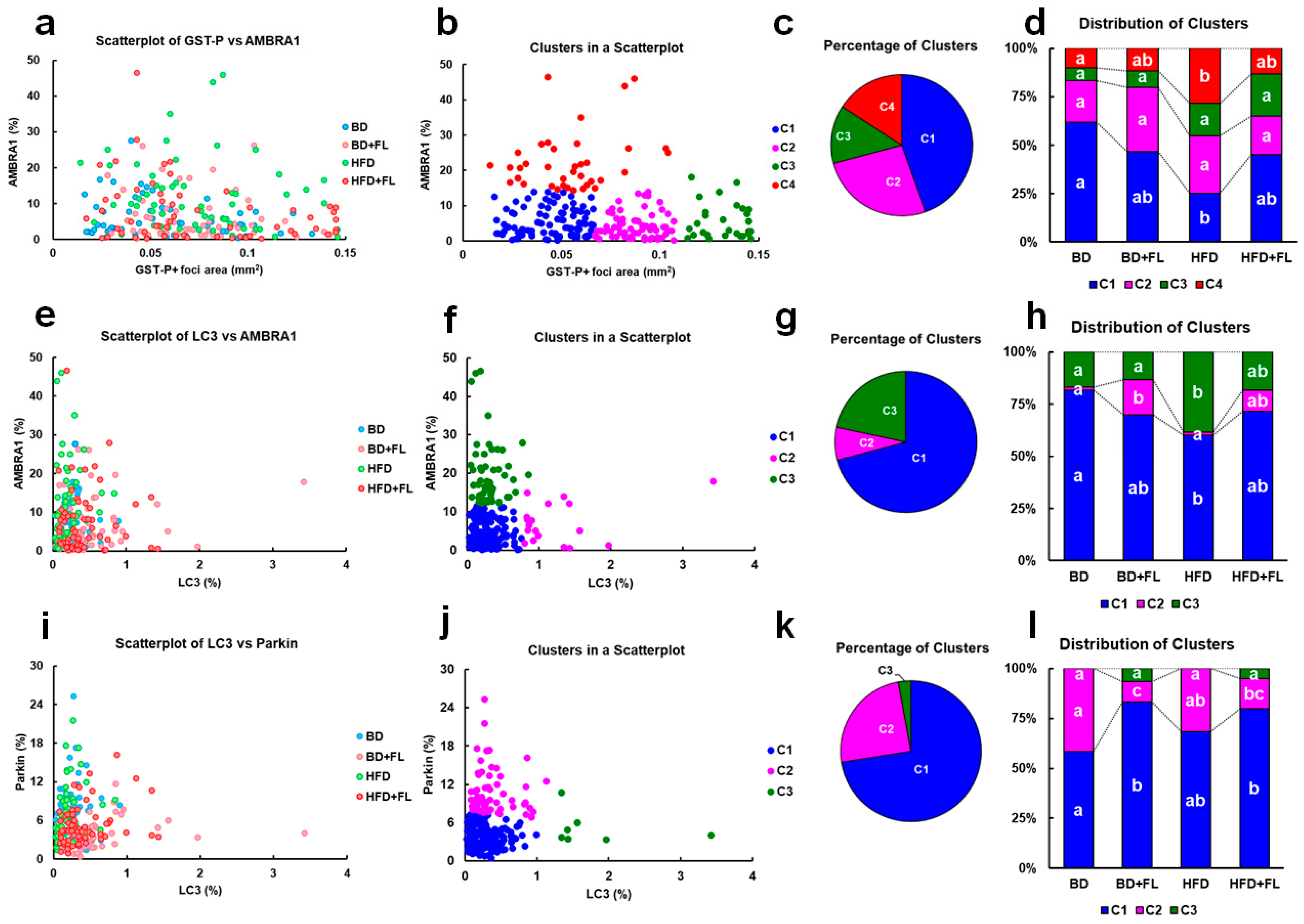
2.4. Cluster Analysis of the GST-P-Positive Foci Area and LC3, AMBRA1, and Parkin Expression in Hepatic Precancerous Lesions
2.5. Flutamide Induces Autophagy
2.6. HFD and HFD Combined with FL Alters Hepatic Gene Expression
3. Discussion
4. Methods
4.1. Chemicals
4.2. Animals and Treatments
4.3. Histopathology
4.4. Immunohistochemistry and Clustering Analysis
4.5. Ultrastructural Examination
4.6. Real-Time Reverse Transcription–Polymerase Chain Reaction Analysis
4.7. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lazarus, J.V.; Mark, H.E.; Anstee, Q.M.; Arab, J.P.; Batterham, R.L.; Castera, L.; Cortez-Pinto, H.; Crespo, J.; Cusi, K.; Dirac, M.A.; et al. NAFLD Consensus Consortium Advancing the global public health agenda for NAFLD: A consensus statement. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 60–78. [Google Scholar] [CrossRef] [PubMed]
- Begriche, K.; Igoudjil, A.; Pessayre, D.; Fromenty, B. Mitochondrial dysfunction in NASH: Causes, consequences and possible means to prevent it. Mitochondrion 2006, 6, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Rolo, A.P.; Teodoro, J.S.; Palmeira, C.M. Role of oxidative stress in the pathogenesis of nonalcoholic steatohepatitis. Free Radic. Biol. Med. 2012, 52, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, S.H.; Swerdlow, R.H.; Khan, E.M.; Iezzoni, J.C.; Hespenheide, E.E.; Parks, J.K.; Parker, W.D., Jr. Mitochondrial abnormalities in non-alcoholic steatohepatitis. J. Hepatol. 1999, 31, 430–434. [Google Scholar] [CrossRef]
- Cortez-Pinto, H.; Chatham, J.; Chacko, V.P.; Arnold, C.; Rashid, A.; Diehl, A.M. Alterations in liver ATP homeostasis in human nonalcoholic steatohepatitis: A pilot study. JAMA 1999, 282, 1659–1664. [Google Scholar] [CrossRef]
- Pérez-Carreras, M.; Del Hoyo, P.; Martín, M.A.; Rubio, J.C.; Martín, A.; Castellano, G.; Colina, F.; Arenas, J.; Solis-Herruzo, J.A. Defective hepatic mitochondrial respiratory chain in patients with nonalcoholic steatohepatitis. Hepatology 2003, 38, 999–1007. [Google Scholar] [CrossRef]
- Nassir, F.; Ibdah, J.A. Role of mitochondria in nonalcoholic fatty liver disease. Int. J. Mol. Sci. 2014, 15, 8713–8742. [Google Scholar] [CrossRef]
- Zarghamravanbakhsh, P.; Frenkel, M.; Poretsky, L. Metabolic causes and consequences of nonalcoholic fatty liver disease (NAFLD). Metabol. Open 2021, 12, 100149, Erratum in Metabol. Open 2023, 17, 100231. [Google Scholar] [CrossRef]
- Gatica, D.; Lahiri, V.; Klionsky, D.J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 2018, 20, 233–242, Erratum in Nat. Cell Biol. 2023, 25, 1073. [Google Scholar] [CrossRef]
- Anding, A.L.; Baehrecke, E.H. Cleaning house: Selective autophagy of organelles. Dev. Cell 2017, 41, 10–22. [Google Scholar] [CrossRef]
- Ding, W.X.; Yin, X.M. Mitophagy: Mechanisms, pathophysiological roles, and analysis. Biol. Chem. 2012, 393, 547–564. [Google Scholar] [CrossRef] [PubMed]
- Shimura, H.; Hattori, N.; Kubo Si Mizuno, Y.; Asakawa, S.; Minoshima, S.; Shimizu, N.; Iwai, K.; Chiba, T.; Tanaka, K.; Suzuki, T. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 2000, 25, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Valente, E.M.; Abou-Sleiman, P.M.; Caputo, V.; Muqit, M.M.; Harvey, K.; Gispert, S.; Ali, Z.; Del Turco, D.; Bentivoglio, A.R.; Healy, D.G.; et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 2004, 304, 1158–1160. [Google Scholar] [CrossRef] [PubMed]
- Strappazzon, F.; Nazio, F.; Corrado, M.; Cianfanelli, V.; Romagnoli, A.; Fimia, G.M.; Campello, S.; Nardacci, R.; Piacentini, M.; Campanella, M.; et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015, 22, 419–432. [Google Scholar] [CrossRef]
- Narendra, D.; Tanaka, A.; Suen, D.F.; Youle, R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008, 183, 95–803. [Google Scholar] [CrossRef]
- Van Humbeeck, C.; Cornelissen, T.; Hofkens, H.; Mandemakers, W.; Gevaert, K.; De Strooper, B.; Vandenberghe, W. Parkin interacts with Ambra1 to induce mitophagy. J. Neurosci. 2011, 31, 10249–10261. [Google Scholar] [CrossRef]
- Di Rita, A.; Peschiaroli, A.; DAcunzo, P.; Strobbe, D.; Hu, Z.; Gruber, J.; Nygaard, M.; Lambrughi, M.; Melino, G.; Papaleo, E.; et al. HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKα. Nat. Commun. 2018, 9, 3755. [Google Scholar] [CrossRef]
- Wang, L.; Liu, X.; Nie, J.; Zhang, J.; Kimball, S.R.; Zhang, H.; Zhang, W.J.; Jefferson, L.S.; Cheng, Z.; Ji, Q.; et al. ALCAT1 controls mitochondrial etiology of fatty liver diseases, linking defective mitophagy to hepatosteatosis. Hepatology 2015, 61, 486–496. [Google Scholar] [CrossRef]
- Yamada, T.; Murata, D.; Adachi, Y.; Itoh, K.; Kameoka, S.; Igarashi, A.; Kato, T.; Araki, Y.; Huganir, R.L.; Dawson, T.M.; et al. Mitochondrial stasis reveals p62-mediated ubiquitination in Parkin-independent mitophagy and mitigates nonalcoholic fatty liver disease. Cell Metab. 2018, 28, 588–604. [Google Scholar] [CrossRef]
- Zhou, T.; Chang, L.; Luo, Y.; Zhou, Y.; Zhang, J. Mst1 inhibition attenuates non-alcoholic fatty liver disease via reversing Parkin-related mitophagy. Redox Biol. 2019, 21, 101120, Erratum in Redox Biol. 2020, 28, 101299 . [Google Scholar] [CrossRef]
- Li, X.; Shi, Z.; Zhu, Y.; Shen, T.; Wang, H.; Shui, G.; Loor, J.J.; Fang, Z.; Chen, M.; Wang, X.; et al. Cyanidin-3-O-glucoside improves non-alcoholic fatty liver disease by promoting PINK1-mediated mitophagy in mice. Br. J. Pharmacol. 2020, 177, 3591–3607. [Google Scholar] [CrossRef] [PubMed]
- Okahara, A.; Nomura, A.; Tanioka, H.; Sakamoto, H.; Yamashita, K.; Ikuse, T. Collaborative work to evaluate toxicity on male reproductive organs by repeated dose studies in rats 5). Effects of repeated doses of flutamide for 2 and 4 weeks. J. Toxicol. Sci. 2000, 25, 63–70. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, L.S.; Franco-Belussi, L.; Gomes, F.R.; de Oliveira, C. Flutamide effects on morphology of reproductive organs and liver of Neotropical Anura, hinelRla schneideri. Aquat. Toxicol. 2016, 176, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Della-Flora, A.; Wilde, M.L.; Pinto, I.D.F.; Lima, É.C.; Sirtori, C. Degradation of the anticancer drug flutamide by solar photo-Fenton treatment at near-neutral pH: Identification of transformation products and in silico (Q)SAR risk assessment. Environ. Res. 2020, 183, 109223. [Google Scholar] [CrossRef]
- Coe, K.J.; Jia, Y.; Ho, H.K.; Rademacher, P.; Bammler, T.K.; Beyer, R.P.; Farin, F.M.; Woodke, L.; Plymate, S.R.; Fausto, N.; et al. Comparison of the cytotoxicity of the nitroaromatic drug flutamide to its cyano analogue in the hepatocyte cell line TAMH: Evidence for complex I inhibition and mitochondrial dysfunction using toxicogenomic screening. Chem. Res. Toxicol. 2007, 20, 1277–1290. [Google Scholar] [CrossRef]
- Ball, A.L.; Kamalian, L.; Alfirevic, A.; Lyon, J.J.; Chadwick, A.E. Identification of the additional mitochondrial liabilities of 2-hydroxyflutamide when compared with its parent compound, flutamide in HepG2 Cells. Toxicol. Sci. 2016, 153, 41–351. [Google Scholar] [CrossRef]
- Tawfeeq, M.M.; Hayashi, H.; Shimamoto, K.; Suzuki, K.; Shibutani, M.; Inokuma, H.; Mitsumori, K. Mechanistic study on liver tumor promoting effects of flutamide in rats. Arch. Toxicol. 2012, 86, 497–507. [Google Scholar] [CrossRef]
- Murayama, H.; Eguchi, A.; Nakamura, M.; Kawashima, M.; Nagahara, R.; Mizukami, S.; Kimura, M.; Makino, E.; Takahashi, N.; Ohtsuka, R.; et al. Spironolactone in combination with α-glycosyl isoquercitrin prevents steatosis-related early hepatocarcinogenesis in rats through the Observed NADPH Oxidase Modulation. Toxicol. Pathol. 2018, 46, 530–539. [Google Scholar] [CrossRef]
- Masuda, S.; Mizukami, S.; Eguchi, A.; Ichikawa, R.; Nakamura, M.; Nakamura, K.; Okada, R.; Tanaka, T.; Shibutani, M.; Yoshida, T. Immunohistochemical expression of autophagosome markers LC3 and p62 in preneoplastic liver foci in high fat diet-fed rats. J. Toxicol. Sci. 2019, 44, 565–574. [Google Scholar] [CrossRef]
- Nakamura, M.; Eguchi, A.; Inohana, M.; Nagahara, R.; Murayama, H.; Kawashima, M.; Mizukami, S.; Koyanagi, M.; Hayashi, S.M.; Maronpot, R.R.; et al. Differential impacts of mineralocorticoid receptor antagonist potassium canrenoate on liver and renal changes in high fat diet-mediated early hepatocarcinogenesis model rats. J. Toxicol. Sci. 2018, 43, 611–621. [Google Scholar] [CrossRef]
- Eguchi, A.; Mizukami, S.; Nakamura, M.; Masuda, S.; Murayama, H.; Kawashima, M.; Inohana, M.; Nagahara, R.; Kobayashi, M.; Yamashita, R.; et al. Metronidazole enhances steatosis-related early-stage hepatocarcinogenesis in high fat diet-fed rats through DNA double-strand breaks and modulation of autophagy. Environ. Sci. Pollut. Res. Int. 2022, 29, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Ito, N.; Tamano, S.; Shirai, T. A medium-term rat liver bioassay for rapid in vivo detection of carcinogenic potential of chemicals. Cancer Sci. 2003, 94, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, R.; Masuda, S.; Nakahara, J.; Kobayashi, M.; Yamashita, R.; Uomoto, S.; Kanami, O.; Hara, E.; Ito, Y.; Shibutani, M.; et al. Inhibition of autophagy with expression of NADPH oxidase subunit p22phox in preneoplastic lesions in a high-fat diet and streptozotocin-related hepatocarcinogenesis rat model. J. Toxicol. Sci. 2022, 47, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Uomoto, S.; Takesue, K.; Shimizu, S.; Maeda, N.; Oshima, K.; Hara, E.; Kobayashi, M.; Takahashi, Y.; Shibutani, M.; Yoshida, T. Phenobarbital, a hepatic metabolic enzyme inducer, inhibits preneoplastic hepatic lesions with expression of selective autophagy receptor p62 and ER-phagy receptor FAM134B in high-fat diet-fed rats through the inhibition of ER stress. Food Chem. Toxicol. 2023, 173, 113607. [Google Scholar] [CrossRef]
- Kashimshetty, R.; Desai, V.G.; Kale, V.M.; Lee, T.; Moland, C.L.; Branham, W.S.; New, L.S.; Chan, E.C.; Younis, H.; Boelsterli, U.A. Underlying mitochondrial dysfunction triggers flutamide-induced oxidative liver injury in a mouse model of idiosyncratic drug toxicity. Toxicol. Appl. Pharmacol. 2009, 238, 150–159. [Google Scholar] [CrossRef]
- Ke, P.Y. Mitophagy in the pathogenesis of liver diseases. Cells 2020, 9, 831. [Google Scholar] [CrossRef]
- Coe, K.J.; Nelson, S.D.; Ulrich, R.G.; He, Y.; Dai, X.; Cheng, O.; Caguyong, M.; Roberts, C.J.; Slatter, J.G. Profiling the hepatic effects of flutamide in rats: A microarray comparison with classical aryl hydrocarbon receptor ligands and atypical CYP1A inducers. Drug Metab. Dispos. 2006, 34, 1266–1275. [Google Scholar] [CrossRef]
- Hall, A.P.; Elcombe, C.R.; Foster, J.R.; Harada, T.; Kaufmann, W.; Knippel, A.; Küttler, K.; Malarkey, D.E.; Maronpot, R.R.; Nishikawa, A.; et al. Liver hypertrophy: A review of adaptive (adverse and non-adverse) changes-conclusions from the 3rd International ESTP Expert Workshop. Toxicol. Pathol. 2012, 40, 971–994. [Google Scholar] [CrossRef]
- Zhang, X.J.; Chen, S.; Huang, K.X.; Le, W.D. Why should autophagic flux be assessed? Acta Pharmacol. Sin. 2013, 34, 595–599. [Google Scholar] [CrossRef]
- Yoshida, G.J. Therapeutic strategies of drug repositioning targeting autophagy to induce cancer cell death: From pathophysiology to treatment. J. Hematol. Oncol. 2017, 10, 67. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Levine, B. Methods in mammalian autophagy research. Cell 2010, 140, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Dawson, T.M.; Dawson, V.L. The role of parkin in familial and sporadic Parkinson’s disease. Mov. Disord. 2010, 25, S32–S39. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.A.; Ni, H.M.; Ding, Y.; Ding, W.X. Parkin regulates mitophagy and mitochondrial function to protect against alcohol-induced liver injury and steatosis in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, G324–G340, Erratum in Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, G142. [Google Scholar] [CrossRef] [PubMed]
- Fukuo, Y.; Yamashina, S.; Sonoue, H.; Arakawa, A.; Nakadera, E.; Aoyama, T.; Uchiyama, A.; Kon, K.; Ikejima, K.; Watanabe, S. Abnormality of autophagic function and cathepsin expression in the liver from patients with non-alcoholic fatty liver disease. Hepatol. Res. 2014, 44, 1026–1036. [Google Scholar] [CrossRef]
- González-Rodríguez, A.; Mayoral, R.; Agra, N.; Valdecantos, M.P.; Pardo, V.; Miquilena-Colina, M.E.; Vargas-Castrillón, J.; Lo Iacono, O.; Corazzari, M.; Fimia, G.M.; et al. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis. 2014, 5, e1179. [Google Scholar] [CrossRef]
- Begriche, K.; Massart, J.; Robin, M.A.; Bonnet, F.; Fromenty, B. Mitochondrial adaptations and dysfunctions in nonalcoholic fatty liver disease. Hepatology 2013, 58, 1497–1507. [Google Scholar] [CrossRef]
- Kowalik, M.A.; Perra, A.; Ledda-Columbano, G.M.; Ippolito, G.; Piacentini, M.; Columbano, A.; Falasca, L. Induction of autophagy promotes the growth of early preneoplastic rat liver nodules. Oncotarget 2016, 7, 5788–5799. [Google Scholar] [CrossRef]
- Panigrahi, D.P.; Praharaj, P.P.; Bhol, C.S.; Mahapatra, K.K.; Patra, S.; Behera, B.P.; Mishra, S.R.; Bhutia, S.K. The emerging, multifaceted role of mitophagy in cancer and cancer therapeutics. Semin. Cancer Biol. 2020, 66, 45–58. [Google Scholar] [CrossRef]
- Labuschagne, C.F.; Cheung, E.C.; Blagih, J.; Domart, M.C.; Vousden, K.H. Cell clustering promotes a metabolic switch that supports metastatic ccolonization. Cell Metab. 2019, 30, 720–734. [Google Scholar] [CrossRef]
- Maiani, E.; Milletti, G.; Nazio, F.; Holdgaard, S.G.; Bartkova, J.; Rizza, S.; Cianfanelli, V.; Lorente, M.; Simoneschi, D.; Di Marco, M.; et al. AMBRA1 regulates cyclin D to guard S-phase entry and genomic integrity. Nature 2021, 592, 799–803. [Google Scholar] [CrossRef]
- Cianfanelli, V.; Fuoco, C.; Lorente, M.; Salazar, M.; Quondamatteo, F.; Gherardini, P.F.; De Zio, D.; Nazio, F.; Antonioli, M.; D’Orazio, M.; et al. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat. Cell Biol. 2015, 17, 20–30, Erratum in Nat. Cell Biol. 2015, 17, 706. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.M.; Lee, W.K.; Lee, Y.H.; Kang, E.S.; Cha, B.S.; Lee, B.W. Metformin restores Parkin-mediated mitophagy, suppressed by cytosolic p53. Int. J. Mol. Sci. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Hao, M.; Liu, Y.; Ma, X.; Lin, W.; Xu, Q.; Zhou, H.; Shao, N.; Kuang, H. Liraglutide ameliorates non-alcoholic steatohepatitis by inhibiting NLRP3 inflammasome and pyroptosis activation via mitophagy. Eur. J. Pharmacol. 2019, 864, 172715. [Google Scholar] [CrossRef]
- Boland, M.L.; Laker, R.C.; Mather, K.; Nawrocki, A.; Oldham, S.; Boland, B.B.; Lewis, H.; Conway, J.; Naylor, J.; Guionaud, S.; et al. Resolution of NASH and hepatic fibrosis by the GLP-1R/GcgR dual-agonist Cotadutide via modulating mitochondrial function and lipogenesis. Nat. Metab. 2020, 2, 413–431. [Google Scholar] [CrossRef]
- Guan, Y.; Wang, Y.; Li, B.; Shen, K.; Li, Q.; Ni, Y.; Huang, L. Mitophagy in carcinogenesis, drug resistance and anticancer therapeutics. Cancer Cell Int. 2021, 21, 350. [Google Scholar] [CrossRef]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, K.; Hara, E.; Takimoto, M.; Bai, Y.; Hirata, M.; Zeng, W.; Uomoto, S.; Todoroki, M.; Kobayashi, M.; Kozono, T.; et al. Peroxisome proliferator activator α agonist clofibrate induces pexophagy in coconut oil-based high-fat diet-fed rats. Biology 2024, 13, 1027. [Google Scholar] [CrossRef]
| Group | BD | BD + FL | HFD | HFD + FL |
|---|---|---|---|---|
| No. of animals | 6 | 6 | 6 | 6 |
| Final body weight (g) | 301.5 ± 15.15 a | 273.7 ± 6.96 bc | 293.4 ± 15.13 ab | 271.8 ± 12.05 c |
| Food intake (g/kg) | 47.1 ± 16.8 a | 46.0 ± 15.5 a | 34.1 ± 15.0 a | 33.3 ± 13.9 a |
| Water intake (g/kg) | 66.6 ± 19.3 a | 64.4 ± 17.3 a | 53.8 ± 17.6 a | 51.3 ± 16.5 a |
| Absolute liver weight (g) | 7.68 ± 0.72 ab | 7.91 ± 0.32 a | 6.89 ± 0.86 b | 6.90 ± 0.38 b |
| Relative liver weight (%BW) | 2.54 ± 0.13 a | 2.89 ± 0.07 b | 2.34 ± 0.19 a | 2.54 ± 0.16 a |
| Absolute intraperitoneal fat weight (g) | 6.27 ± 1.47 a | 4.40 ± 0.40 b | 6.78 ± 0.86 a | 5.04 ± 1.33 ab |
| Relative intraperitoneal fat weight (%BW) | 2.07 ± 0.46 ab | 1.61 ± 0.13 b | 2.30 ± 0.18 a | 1.85 ± 0.46 ab |
| Absolute testis weight (g) | 3.00 ± 0.07 a | 2.77 ± 0.14 a | 2.94 ± 0.15 a | 2.94 ± 0.18 a |
| Relative testis weight (%BW) | 1.00 ± 0.04 a | 1.01 ± 0.04 a | 1.00 ± 0.03 a | 1.08 ± 0.07 a |
| Absolute epididymis weight (g) | 1.04 ± 0.06 a | 0.48 ± 0.09 c | 0.90 ± 0.09 b | 0.56 ± 0.08 c |
| Relative epididymis weight (%BW) | 0.34 ± 0.02 a | 0.17 ± 0.03 b | 0.31 ± 0.03 a | 0.21 ± 0.04 b |
| Absolute SC/CG weight (g) | 1.16 ± 0.18 a | 0.18 ± 0.05 b | 1.05 ± 0.13 a | 0.24 ± 0.08 b |
| Relative SC/CG weight (%BW) | 0.39 ± 0.07 a | 0.07 ± 0.02 b | 0.36 ± 0.03 a | 0.09 ± 0.03 ab |
| Absolute prostate weight (g) | 0.37 ± 0.03 a | 0.07 ± 0.03 b | 0.33 ± 0.07 a | 0.07 ± 0.04 b |
| Relative prostate weight (%BW) | 0.12 ± 0.01 a | 0.03 ± 0.01 b | 0.11 ± 0.02 a | 0.03 ± 0.01 b |
| Group | BD | BD + FL | HFD | HFD + FL | |
|---|---|---|---|---|---|
| No. of animals | 6 | 6 | 6 | 6 | |
| Autophagy-related genes | |||||
| Atg3 | 1.05 ± 0.34 a | 1.69 ± 0.40 a | 2.58 ± 1.23 a | 1.60 ± 0.32 a | |
| Atg5 | 1.10 ± 0.51 a | 1.77 ± 0.72 ab | 2.61 ± 0.91 b | 2.27 ± 0.83 ab | |
| Atg7 | 1.03 ± 0.26 a | 1.10 ± 0.23 a | 2.08 ± 0.64 b | 1.98 ± 1.02 ab | |
| Lamp1 | 1.06 ± 0.40 a | 1.50 ± 0.41 ab | 2.08 ± 0.72 b | 1.54 ± 0.59 ab | |
| Lamp2 | 1.05 ± 0.35 a | 1.57 ± 0.36 ab | 2.13 ± 0.93 b | 1.19 ± 0.39 a | |
| Lc3 | 1.00 ± 0.08 a | 1.24 ± 0.38 ab | 2.06 ± 0.83 b | 1.58 ± 1.04 ab | |
| p62 | 1.03 ± 0.26 a | 1.30 ± 0.57 a | 1.82 ± 0.72 a | 1.67 ± 1.02 a | |
| Mitochondria/Mitophagy-related genes | |||||
| Ambta1 | 1.14 ± 0.66 a | 1.98 ± 0.75 a | 3.95 ± 3.18 a | 4.64 ± 5.32 a | |
| Parkin | 1.17 ± 0.64 a | 2.54 ± 0.99 ab | 3.27 ± 2.12 ab | 3.57 ± 1.59 b | |
| Pink1 | 1.03 ± 0.24 a | 1.09 ± 0.22 a | 1.81 ± 0.65 a | 1.12 ± 0.29 a | |
| Bnip3 | 1.01 ± 0.18 a | 1.48 ± 0.41 a | 1.43 ± 0.47 a | 1.05 ± 0.34 a | |
| Nadh | 1.03 ± 0.26 a | 1.32 ± 0.26 a | 2.21 ± 0.97 a | 1.17 ± 0.32 a | |
| Sdhd | 1.04 ± 0.30 a | 1.67 ± 0.40 ab | 2.17 ± 0.86 b | 1.51 ± 0.51 ab | |
| ATP synthase | 1.03 ± 0.29 a | 1.47 ± 0.45 a | 1.79 ± 0.75 a | 1.24 ± 0.30 a | |
| Lipid metabolism-related genes | |||||
| Abca1 | 1.09 ± 0.40 a | 1.89 ± 0.79 ab | 3.26 ± 1.07 c | 2.74 ± 0.89 bc | |
| Acox1 | 1.14 ± 0.59 a | 1.80 ± 0.54 a | 2.74 ± 1.36 a | 1.89 ± 0.34 a | |
| Apob | 1.05 ± 0.30 a | 0.82 ± 0.21 a | 1.49 ± 0.76 a | 1.31 ± 0.32 a | |
| Dgat2 | 1.06 ± 0.39 a | 1.36 ± 0.41 ab | 1.77 ± 0.51 b | 1.70 ± 0.35 ab | |
| Fasn | 1.27 ± 0.87 a | 3.04 ± 1.50 a | 1.72 ± 0.96 a | 2.43 ± 2.25 a | |
| Hmgcs1 | 1.09 ± 0.47 a | 0.72 ± 0.36 a | 1.51 ± 0.77 a | 1.18 ± 0.36 a | |
| Hsd3 b1 | 1.10 ± 0.48 a | 0.38 ± 0.40 a | 2.17 ± 0.90 b | 0.83 ± 0.60 a | |
| Lpl | 1.06 ± 0.37 a | 0.97 ± 0.27 a | 1.50 ± 0.80 a | 1.17 ± 0.46 a | |
| Lss | 1.11 ± 0.53 a | 1.46 ± 0.72 a | 1.69 ± 0.66 a | 2.30 ± 1.19 a | |
| Ppara | 1.19 ± 0.69 a | 1.81 ± 0.97 a | 4.55 ± 1.44 b | 2.08 ± 0.71 a | |
| Pparg | 1.05 ± 0.36 a | 1.16 ± 0.98 a | 0.61 ± 0.26 a | 1.91 ± 1.47 a | |
| Scd1 | 1.03 ± 0.26 a | 1.27 ± 0.38 a | 1.69 ± 0.46 a | 1.25 ± 0.62 a | |
| Srebf1 | 1.17 ± 0.73 a | 0.84 ± 0.43 a | 1.59 ± 1.26 a | 1.58 ± 1.02 a | |
| Srebf2 | 1.08 ± 0.41 a | 1.53 ± 0.44 ab | 2.49 ± 0.95 b | 2.33 ± 0.88 b | |
| Drug metabolism-related genes | |||||
| Cyp1a1 | 1.27 ± 1.09 a | 73.20 ± 45.16 b | 1.90 ± 0.94 a | 96.14 ± 93.51 b | |
| Cyp2b1 | 1.64 ± 1.91 a | 1.97 ± 0.95 a | 1.84 ± 0.91 a | 2.51 ± 1.10 a | |
| Cyp3a1 | 1.05 ± 0.34 a | 0.65 ± 0.25 a | 1.40 ± 0.54 a | 0.89 ± 0.79 a | |
| Inflammation related genes | |||||
| Tnf-a | 1.11 ± 0.51 a | 1.29 ± 0.64 a | 1.81 ± 1.11 a | 3.33 ± 3.87 a | |
| Oxidative stress-related genes | |||||
| Catalase | 1.04 ± 0.33 a | 1.75 ± 0.58 ab | 2.70 ± 1.22 b | 2.18 ± 0.98 ab | |
| Gpx1 | 1.16 ± 0.70 a | 1.68 ± 0.67 a | 1.92 ± 0.75 a | 2.05 ± 0.95 a | |
| Gpx2 | 1.08 ± 0.47 a | 5.36 ± 1.91 b | 3.44 ± 2.86 ab | 4.98 ± 2.16 b | |
| Mn-SOD | 1.04 ± 0.32 a | 1.23 ± 0.38 a | 1.65 ± 0.73 a | 1.54 ± 0.56 a | |
| Sod1 | 1.11 ± 0.53 a | 1.32 ± 0.54 a | 1.18 ± 0.55 a | 1.00 ± 0.40 a | |
| Sod2 | 1.13 ± 0.63 a | 1.41 ± 0.23 a | 2.07 ± 0.87 a | 1.33 ± 0.35 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hara, E.; Ohshima, K.; Takimoto, M.; Bai, Y.; Hirata, M.; Zeng, W.; Uomoto, S.; Todoroki, M.; Kobayashi, M.; Kozono, T.; et al. Flutamide Promotes Early Hepatocarcinogenesis Through Mitophagy in High-Fat Diet-Fed Non-Obese Steatotic Rats. Int. J. Mol. Sci. 2025, 26, 2709. https://doi.org/10.3390/ijms26062709
Hara E, Ohshima K, Takimoto M, Bai Y, Hirata M, Zeng W, Uomoto S, Todoroki M, Kobayashi M, Kozono T, et al. Flutamide Promotes Early Hepatocarcinogenesis Through Mitophagy in High-Fat Diet-Fed Non-Obese Steatotic Rats. International Journal of Molecular Sciences. 2025; 26(6):2709. https://doi.org/10.3390/ijms26062709
Chicago/Turabian StyleHara, Emika, Kanami Ohshima, Mio Takimoto, Yidan Bai, Mai Hirata, Wen Zeng, Suzuka Uomoto, Mai Todoroki, Mio Kobayashi, Takuma Kozono, and et al. 2025. "Flutamide Promotes Early Hepatocarcinogenesis Through Mitophagy in High-Fat Diet-Fed Non-Obese Steatotic Rats" International Journal of Molecular Sciences 26, no. 6: 2709. https://doi.org/10.3390/ijms26062709
APA StyleHara, E., Ohshima, K., Takimoto, M., Bai, Y., Hirata, M., Zeng, W., Uomoto, S., Todoroki, M., Kobayashi, M., Kozono, T., Kigata, T., Shibutani, M., & Yoshida, T. (2025). Flutamide Promotes Early Hepatocarcinogenesis Through Mitophagy in High-Fat Diet-Fed Non-Obese Steatotic Rats. International Journal of Molecular Sciences, 26(6), 2709. https://doi.org/10.3390/ijms26062709






