The Impact of a Quinone Scaffold on Thermo-TRPs Modulation by Dimethylheptyl Phytocannabinoids
Abstract
1. Introduction
2. Results
2.1. Synthesis
2.2. Biological Evaluation
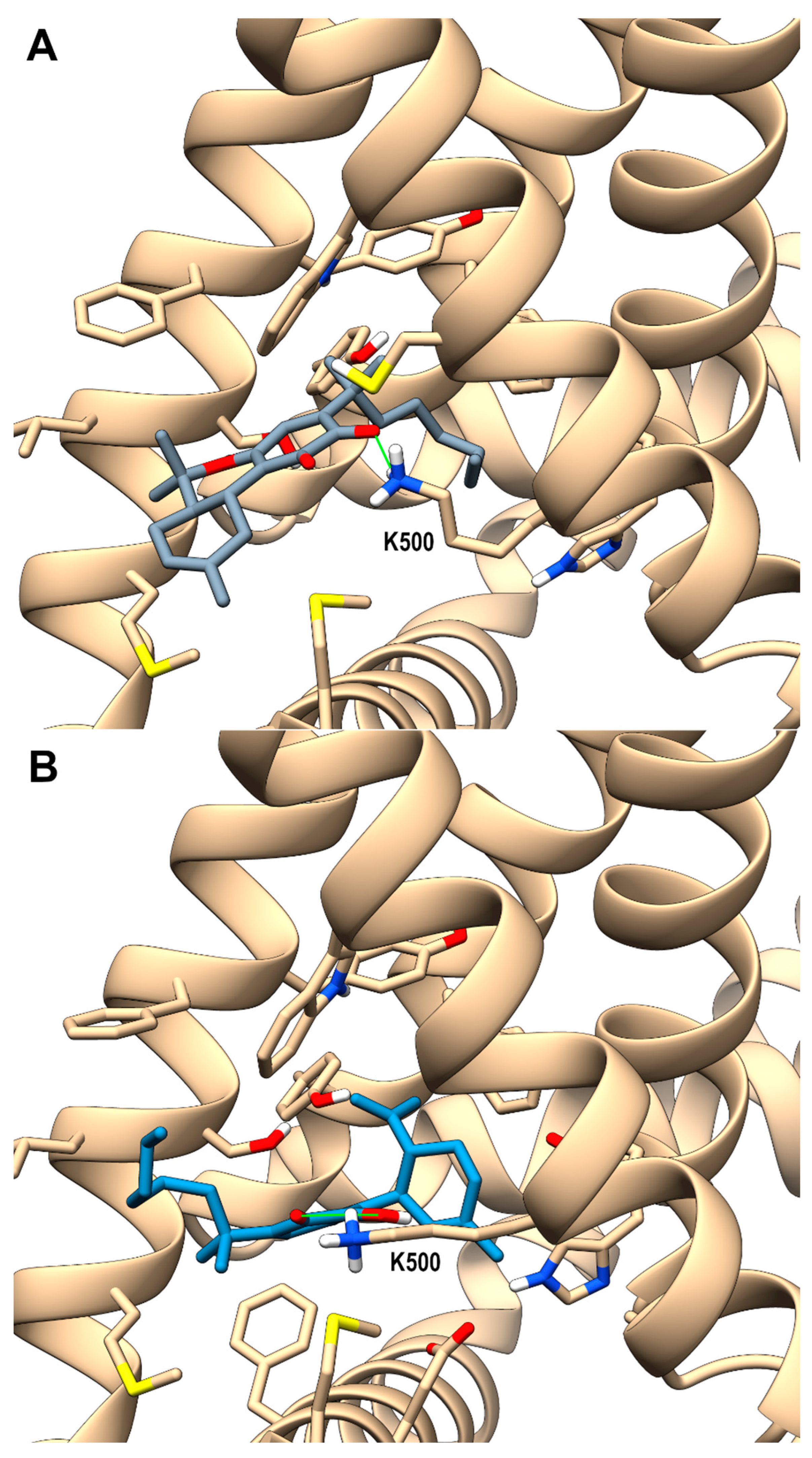
2.3. Molecular Docking Studies on Compound 5b and 6b at TRPV3 Channel
3. Discussion
4. Materials and Methods
4.1. Synthesis
4.1.1. General Experimental Procedures
4.1.2. SIBX Oxidation
4.2. Thermo-TRP Assay
4.3. Molecular Docking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanuš, L.O.; Meyer, S.M.; Muñoz, E.; Taglialatela-Scafati, O.; Appendino, G. Phytocannabinoids: A Unified Critical Inventory. Nat. Prod. Rep. 2016, 33, 1357–1392. [Google Scholar] [CrossRef]
- Reekie, T.A.; Scott, M.P.; Kassiou, M. The Evolving Science of Phytocannabinoids. Nat. Rev. Chem. 2017, 2, 0101. [Google Scholar] [CrossRef]
- Blebea, N.M.; Pricopie, A.I.; Vlad, R.-A.; Hancu, G. Phytocannabinoids: Exploring Pharmacological Profiles and Their Impact on Therapeutical Use. Int. J. Mol. Sci. 2024, 25, 4204. [Google Scholar] [CrossRef] [PubMed]
- Morales, P.; Hurst, D.P.; Reggio, P.H. Molecular Targets of the Phytocannabinoids: A Complex Picture. In Phytocannabinoids: Unraveling the Complex Chemistry and Pharmacology of Cannabis sativa; Springer: Cham, Switzerland, 2017; pp. 103–131. [Google Scholar]
- Rathod, S.S.; Agrawal, Y.O. Phytocannabinoids as Potential Multitargeting Neuroprotectants in Alzheimer’s Disease. Curr. Drug Res. Rev. 2024, 16, 94–110. [Google Scholar] [CrossRef] [PubMed]
- Vitale, R.M.; Iannotti, F.A.; Amodeo, P. The (Poly)Pharmacology of Cannabidiol in Neurological and Neuropsychiatric Disorders: Molecular Mechanisms and Targets. Int. J. Mol. Sci. 2021, 22, 4876. [Google Scholar] [CrossRef]
- Ligresti, A.; De Petrocellis, L.; Di Marzo, V. From Phytocannabinoids to Cannabinoid Receptors and Endocannabinoids: Pleiotropic Physiological and Pathological Roles Through Complex Pharmacology. Physiol. Rev. 2016, 96, 1593–1659. [Google Scholar] [CrossRef]
- Vitale, R.M.; Moriello, A.S.; De Petrocellis, L. Chapter 6. Natural Compounds and Synthetic Drugs Targeting the Ionotropic Cannabinoid Members of Transient Receptor Potential (TRP) Channels. In New Tools to Interrogate Endocannabinoid Signalling: From Natural Compounds to Synthetic Drugs; Maccarrone, M., Ed.; Royal Society of Chemistry: London, UK, 2020; pp. 201–300. ISBN 978-1-83916-075-2. [Google Scholar]
- De Petrocellis, L.; Ligresti, A.; Moriello, A.S.; Allarà, M.; Bisogno, T.; Petrosino, S.; Stott, C.G.; Di Marzo, V. Effects of Cannabinoids and Cannabinoid-Enriched Cannabis Extracts on TRP Channels and Endocannabinoid Metabolic Enzymes. Br. J. Pharmacol. 2011, 163, 1479–1494. [Google Scholar] [CrossRef]
- Julius, D. TRP Channels and Pain. Annu. Rev. Cell Dev. Biol. 2013, 29, 355–384. [Google Scholar] [CrossRef]
- Zhang, M.; Ma, Y.; Ye, X.; Zhang, N.; Pan, L.; Wang, B. TRP (Transient Receptor Potential) Ion Channel Family: Structures, Biological Functions and Therapeutic Interventions for Diseases. Signal Transduct. Target. Ther. 2023, 8, 261. [Google Scholar] [CrossRef]
- Smani, T.; Shapovalov, G.; Skryma, R.; Prevarskaya, N.; Rosado, J.A. Functional and Physiopathological Implications of TRP Channels. Biochim. Biophys. Acta-Mol. Cell Res. 2015, 1853, 1772–1782. [Google Scholar] [CrossRef]
- Liao, M.; Cao, E.; Julius, D.; Cheng, Y. Structure of the TRPV1 Ion Channel Determined by Electron Cryo-Microscopy. Nature 2013, 504, 107–112. [Google Scholar] [CrossRef]
- Diver, M.M.; Lin King, J.V.; Julius, D.; Cheng, Y. Sensory TRP Channels in Three Dimensions. Annu. Rev. Biochem. 2022, 91, 629–649. [Google Scholar] [CrossRef]
- Nilius, B.; Owsianik, G. The Transient Receptor Potential Family of Ion Channels. Genome Biol. 2011, 12, 218. [Google Scholar] [CrossRef] [PubMed]
- Vay, L.; Gu, C.; McNaughton, P.A. The Thermo-TRP Ion Channel Family: Properties and Therapeutic Implications. Br. J. Pharmacol. 2012, 165, 787–801. [Google Scholar] [CrossRef] [PubMed]
- Brederson, J.-D.; Kym, P.R.; Szallasi, A. Targeting TRP Channels for Pain Relief. Eur. J. Pharmacol. 2013, 716, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Tsagareli, M. Thermo-TRP Channels in Pain Sensation. Br. J. Pharm. Res. 2015, 6, 376–384. [Google Scholar] [CrossRef]
- Kashio, M. Thermo-TRP Regulation by Endogenous Factors and Its Physiological Function at Core Body Temperature. Physiol. Rep. 2025, 13, e70164. [Google Scholar] [CrossRef]
- Caterina, M.J.; Schumacher, M.A.; Tominaga, M.; Rosen, T.A.; Levine, J.D.; Julius, D. The Capsaicin Receptor: A Heat-Activated Ion Channel in the Pain Pathway. Nature 1997, 389, 816–824. [Google Scholar] [CrossRef]
- O’Connor, B.; Robbins, N.; Koch, S.E.; Rubinstein, J. TRPV2 Channel-Based Therapies in the Cardiovascular Field. Molecular Underpinnings of Clinically Relevant Therapies. Prog. Biophys. Mol. Biol. 2021, 159, 118–125. [Google Scholar] [CrossRef]
- Gailly, P. TRP Channels in Normal and Dystrophic Skeletal Muscle. Curr. Opin. Pharmacol. 2012, 12, 326–334. [Google Scholar] [CrossRef]
- Shoji, K.F.; Bayet, E.; Leverrier-Penna, S.; Le Devedec, D.; Mallavialle, A.; Marionneau-Lambot, S.; Rambow, F.; Perret, R.; Joussaume, A.; Viel, R.; et al. The Mechanosensitive TRPV2 Calcium Channel Promotes Human Melanoma Invasiveness and Metastatic Potential. EMBO Rep. 2023, 24, e55069. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Shi, Y.; Wu, H.; Niu, C.; Sun, X.; Wang, K. Inhibition of Temperature-Sensitive TRPV3 Channel by Two Natural Isochlorogenic Acid Isomers for Alleviation of Dermatitis and Chronic Pruritus. Acta Pharm. Sin. B 2022, 12, 723–734. [Google Scholar] [CrossRef]
- Han, Y.; Luo, A.; Kamau, P.M.; Takomthong, P.; Hu, J.; Boonyarat, C.; Luo, L.; Lai, R. A Plant-derived TRPV3 Inhibitor Suppresses Pain and Itch. Br. J. Pharmacol. 2021, 178, 1669–1683. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, K. The Ca2+-Permeable Cation Transient Receptor Potential TRPV3 Channel: An Emerging Pivotal Target for Itch and Skin Diseases. Mol. Pharmacol. 2017, 92, 193–200. [Google Scholar] [CrossRef]
- Ni, C.; Yan, M.; Zhang, J.; Cheng, R.; Liang, J.; Deng, D.; Wang, Z.; Li, M.; Yao, Z. A Novel Mutation in TRPV3 Gene Causes Atypical Familial Olmsted Syndrome. Sci. Rep. 2016, 6, 21815. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.D.; Itoh, S.G.; Okumura, H.; Tominaga, M. Structural Basis for Promiscuous Action of Monoterpenes on TRP Channels. Commun. Biol. 2021, 4, 293. [Google Scholar] [CrossRef]
- Sonkusare, S.K.; Laubach, V.E. Endothelial TRPV4 Channels in Lung Edema and Injury. In Current Topics in Membranes; Elsevier: Amsterdam, The Netherlands, 2022; pp. 43–62. [Google Scholar]
- Bandell, M.; Story, G.M.; Hwang, S.W.; Viswanath, V.; Eid, S.R.; Petrus, M.J.; Earley, T.J.; Patapoutian, A. Noxious Cold Ion Channel TRPA1 Is Activated by Pungent Compounds and Bradykinin. Neuron 2004, 41, 849–857. [Google Scholar] [CrossRef]
- Vitale, R.M.; de Petrocellis, L.; Amodeo, P. An Updated Patent Review of TRPA1 Antagonists (2020–Present). Expert Opin. Ther. Pat. 2024, 34, 315–332. [Google Scholar] [CrossRef]
- Talavera, K.; Startek, J.B.; Alvarez-Collazo, J.; Boonen, B.; Alpizar, Y.A.; Sanchez, A.; Naert, R.; Nilius, B. Mammalian Transient Receptor Potential TRPA1 Channels: From Structure to Disease. Physiol. Rev. 2020, 100, 725–803. [Google Scholar] [CrossRef]
- Liu, B.; Fan, L.; Balakrishna, S.; Sui, A.; Morris, J.B.; Jordt, S.-E. TRPM8 Is the Principal Mediator of Menthol-Induced Analgesia of Acute and Inflammatory Pain. Pain 2013, 154, 2169–2177. [Google Scholar] [CrossRef]
- Zhang, W.; Qiao, X.; Li, Q.; Cui, C.; Qiao, C.; Shen, Y.; Zhao, W. Comprehensive Pan-Cancer Analysis of TRPM8 in Tumor Metabolism and Immune Escape. Front. Oncol. 2022, 12, 914060. [Google Scholar] [CrossRef]
- Bouma, J.; Broekhuis, J.D.; van der Horst, C.; Kumar, P.; Ligresti, A.; van der Stelt, M.; Heitman, L.H. Dual Allosteric and Orthosteric Pharmacology of Synthetic Analog Cannabidiol-Dimethylheptyl, but Not Cannabidiol, on the Cannabinoid CB2 Receptor. Biochem. Pharmacol. 2023, 218, 115924. [Google Scholar] [CrossRef] [PubMed]
- Mattoteia, D.; Schiano Moriello, A.; Taglialatela-Scafati, O.; Amodeo, P.; De Petrocellis, L.; Appendino, G.; Vitale, R.M.; Caprioglio, D. The Combined Effect of Branching and Elongation on the Bioactivity Profile of Phytocannabinoids. Part I: Thermo-TRPs. Biomedicines 2021, 9, 1070. [Google Scholar] [CrossRef]
- Kogan, N.M.; Peters, M.; Mechoulam, R. Cannabinoid Quinones—A Review and Novel Observations. Molecules 2021, 26, 1761. [Google Scholar] [CrossRef] [PubMed]
- Cores, Á.; Carmona-Zafra, N.; Clerigué, J.; Villacampa, M.; Menéndez, J.C. Quinones as Neuroprotective Agents. Antioxidants 2023, 12, 1464. [Google Scholar] [CrossRef]
- Zucchi, R.; Danesi, R. Cardiac Toxicity of Antineoplastic Anthracyclines. Curr. Med. Chem.-Anti-Cancer Agents 2003, 3, 151–171. [Google Scholar] [CrossRef]
- Peters, M.; Kogan, N.M. HU-331: A Cannabinoid Quinone, with Uncommon Cytotoxic Properties and Low Toxicity. Expert Opin. Investig. Drugs 2007, 16, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- Casares, L.; Unciti-Broceta, J.D.; Prados, M.E.; Caprioglio, D.; Mattoteia, D.; Higgins, M.; Apendino, G.; Dinkova-Kostova, A.T.; Muñoz, E.; de la Vega, L. Isomeric O-Methyl Cannabidiolquinones with Dual BACH1/NRF2 Activity. Redox Biol. 2020, 37, 101689. [Google Scholar] [CrossRef]
- Harrington, P.E.; Stergiades, I.A.; Erickson, J.; Makriyannis, A.; Tius, M.A. Synthesis of Functionalized Cannabinoids. J. Org. Chem. 2000, 65, 6576–6582. [Google Scholar] [CrossRef]
- Bloemendal, V.R.L.J.; van Hest, J.C.M.; Rutjes, F.P.J.T. Synthetic Pathways to Tetrahydrocannabinol (THC): An Overview. Org. Biomol. Chem. 2020, 18, 3203–3215. [Google Scholar] [CrossRef]
- Baek, S.-H.; Han, D.S.; Yook, C.N.; Kim, Y.C.; Kwak, J.S. Synthesis and Antitumor Activity of Cannabigerol. Arch. Pharm. Res. 1996, 19, 228–230. [Google Scholar] [CrossRef]
- Pollastro, F.; Caprioglio, D.; Del Prete, D.; Rogati, F.; Minassi, A.; Taglialatela-Scafati, O.; Munoz, E.; Appendino, G. Cannabichromene. Nat. Prod. Commun. 2018, 13, 1189–1194. [Google Scholar] [CrossRef]
- Mattoteia, D.; Taglialatela-Scafati, O.; Muñoz, E.; de la Vega, L.; Caprioglio, D.; Appendino, G. Regiodivergent Synthesis of Ortho- and Para-Cannabinoquinones. Eur. J. Org. Chem. 2020, 2020, 7429–7434. [Google Scholar] [CrossRef]
- Nadezhdin, K.D.; Neuberger, A.; Khosrof, L.S.; Talyzina, I.A.; Khau, J.; Yelshanskaya, M.V.; Sobolevsky, A.I. TRPV3 Activation by Different Agonists Accompanied by Lipid Dissociation from the Vanilloid Site. Sci. Adv. 2024, 10, eadn2453. [Google Scholar] [CrossRef]
- Neuberger, A.; Nadezhdin, K.D.; Zakharian, E.; Sobolevsky, A.I. Structural Mechanism of TRPV3 Channel Inhibition by the Plant-derived Coumarin Osthole. EMBO Rep. 2021, 22, e53233. [Google Scholar] [CrossRef]
- Caprioglio, D.; Mattoteia, D.; Minassi, A.; Pollastro, F.; Lopatriello, A.; Muňoz, E.; Taglialatela-Scafati, O.; Appendino, G. One-Pot Total Synthesis of Cannabinol via Iodine-Mediated Deconstructive Annulation. Org. Lett. 2019, 21, 6122–6125. [Google Scholar] [CrossRef]
- Moriello, A.S.; De Petrocellis, L.; Vitale, R.M. Fluorescence-Based Assay for TRPV1 Channels. In Endocannabinoid Signaling. Methods in Molecular Biology; Maccarrone, M., Ed.; Springer: New York, NY, USA, 2023; pp. 119–131. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Schmidt, M.W.; Baldridge, K.K.; Boatz, J.A.; Elbert, S.T.; Gordon, M.S.; Jensen, J.H.; Koseki, S.; Matsunaga, N.; Nguyen, K.A.; Su, S.; et al. General Atomic and Molecular Electronic Structure System. J. Comput. Chem. 1993, 14, 1347–1363. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Goetz, A.W.; Homeyer, N.; Izadi, S.; et al. Amber16; University of California, San Francisco: San Francisco, CA, USA, 2016. [Google Scholar]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]



| TRPA1 | TRPV1 | TRPV4 | TRPM8 | |||
|---|---|---|---|---|---|---|
| Efficacy (% AITC 100 μM) | Potency EC50 μM | IC50 a TRPA1 μM | IC50 b TRPV1 μM | IC50 c TRPV4 μM | IC50 d TRPM8 μM | |
| 5b | 120.4 ± 3.0 | 5.2 ± 0.8 | 3.5 ± 0.20 | >100 | >100 | 1.2 ± 0.1 |
| 5a [36] | 133.4 ± 5.3 | 0.40 ± 0.15 | 0.51 ± 0.03 | >100 | >100 | 13.4 ± 2.1 |
| 6b | 147.3 ± 7.3 | 4.4 ± 1.25 | 1.8 ± 0.3 | >100 | >100 | >100 |
| 6a [36] | 124.2 ± 4.0 | 4.8 ± 1.1 | 6.0 ± 0.9 | >100 | >100 | 39.9 ± 6.9 |
| 7b | 114.2 ± 4.1 | 1.1 ± 0.2 | 1.1 ± 0.1 | >100 | >50 | 14.9 ± 3.9 |
| 7a [36] | 99.2 ± 4.5 | 9.1 ± 2.0 | 1.7 ± 0.15 | >100 | 35.4 ± 3.2 | 9.2 ± 0.8 |
| 8b | 111.2 ± 2.1 | 0.19 ± 0.03 | 0.35 ± 0.02 | >100 | >100 | >100 |
| 8a [36] | 120.4 ± 2.8 | 0.76 ± 0.12 | 0.32 ± 0.01 | >100 | >100 | >100 |
| 9b | 20.5 | NA | 15.1 ± 2.4 | >100 | >100 | >100 |
| 9a [36] | 132.3 ± 7.9 | 2.1 ± 0.9 | 3.2 ± 0.6 | >100 | >100 | 0.98 ± 0.12 |
| TRPV2 | TRPV3 | |||||
|---|---|---|---|---|---|---|
| Efficacy (% Ionomycin 4 μM) | Potency EC50 μM | IC50 a TRPV2 μM | Efficacy (% Ionomycin 4 μM) | Potency EC50 μM | IC50 b TRPV3 μM | |
| 5b | 30.6 ± 6.5 | 12.2 ± 9.7 | 5.45 ± 0.5 | 64.3 ± 0.9 | 0.46 ± 0.02 | 0.76 ± 0.01 |
| 5a [36] | <10 | NA | 16.8± 0.2 | 15.8 ± 0.4 | 11.0 ± 1.1 | 32.6 ± 5.1 |
| 6b | 78.1 ± 4.0 | >50 | 8.5 ± 0.5 | 66.8 ± 2.2 | 0.50 ± 0.1 | 0.72 ± 0.08 |
| 6a [36] | <10 | NA | 45.8 ± 3.9 | 53.8 ± 1.6 | 0.14 ± 0.03 | 2.1 ± 0.6 |
| 7b | 47.8 ± 1.4 | 30.5 ± 3.9 | 23.7 ± 2.9 | 51.1 ± 1.0 | 1.5 ± 0.2 | 8.6 ± 0.2 |
| 7a [36] | 65.3 ± 0.7 | 9.8 ±0.6 | 39.4 ± 4.5 | 64.6 ± 4.8 | 39.7 ± 0.03 | 35.4 ± 3 |
| 8b | 31.5 ± 2.9 | >50 | 6.9 ± 0.1 | 37.55 ± 0.85 | 13.6 ± 1.8 | 38.3 ± 2.0 |
| 8a [36] | 76.1 ± 0.6 | >50 | 22.6 ± 0.2 | 41.6 ± 1.8 | >50 | >50 |
| 9b | 23.4 ± 3.7 | 2.0 ± 1.6 | 5.2 ± 0.6 | 32.5 ± 0.65 | 1.15 ± 0.15 | 19.4 ± 3.5 |
| 9a [36] | 10.5 ± 0.02 | 0.25 ± 0.03 | >50 | 20.9 ± 3.0 | 41.4 ± 21.6 | >100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schiano Moriello, A.; Bossoni, A.; Mattoteia, D.; Caprioglio, D.; Minassi, A.; Appendino, G.; De Petrocellis, L.; Amodeo, P.; Vitale, R.M. The Impact of a Quinone Scaffold on Thermo-TRPs Modulation by Dimethylheptyl Phytocannabinoids. Int. J. Mol. Sci. 2025, 26, 2682. https://doi.org/10.3390/ijms26062682
Schiano Moriello A, Bossoni A, Mattoteia D, Caprioglio D, Minassi A, Appendino G, De Petrocellis L, Amodeo P, Vitale RM. The Impact of a Quinone Scaffold on Thermo-TRPs Modulation by Dimethylheptyl Phytocannabinoids. International Journal of Molecular Sciences. 2025; 26(6):2682. https://doi.org/10.3390/ijms26062682
Chicago/Turabian StyleSchiano Moriello, Aniello, Aurora Bossoni, Daiana Mattoteia, Diego Caprioglio, Alberto Minassi, Giovanni Appendino, Luciano De Petrocellis, Pietro Amodeo, and Rosa Maria Vitale. 2025. "The Impact of a Quinone Scaffold on Thermo-TRPs Modulation by Dimethylheptyl Phytocannabinoids" International Journal of Molecular Sciences 26, no. 6: 2682. https://doi.org/10.3390/ijms26062682
APA StyleSchiano Moriello, A., Bossoni, A., Mattoteia, D., Caprioglio, D., Minassi, A., Appendino, G., De Petrocellis, L., Amodeo, P., & Vitale, R. M. (2025). The Impact of a Quinone Scaffold on Thermo-TRPs Modulation by Dimethylheptyl Phytocannabinoids. International Journal of Molecular Sciences, 26(6), 2682. https://doi.org/10.3390/ijms26062682





