Viral-Based Gene Editing System for Nutritional Improvement of Fructan Content in Lettuce
Abstract
1. Introduction
2. Results
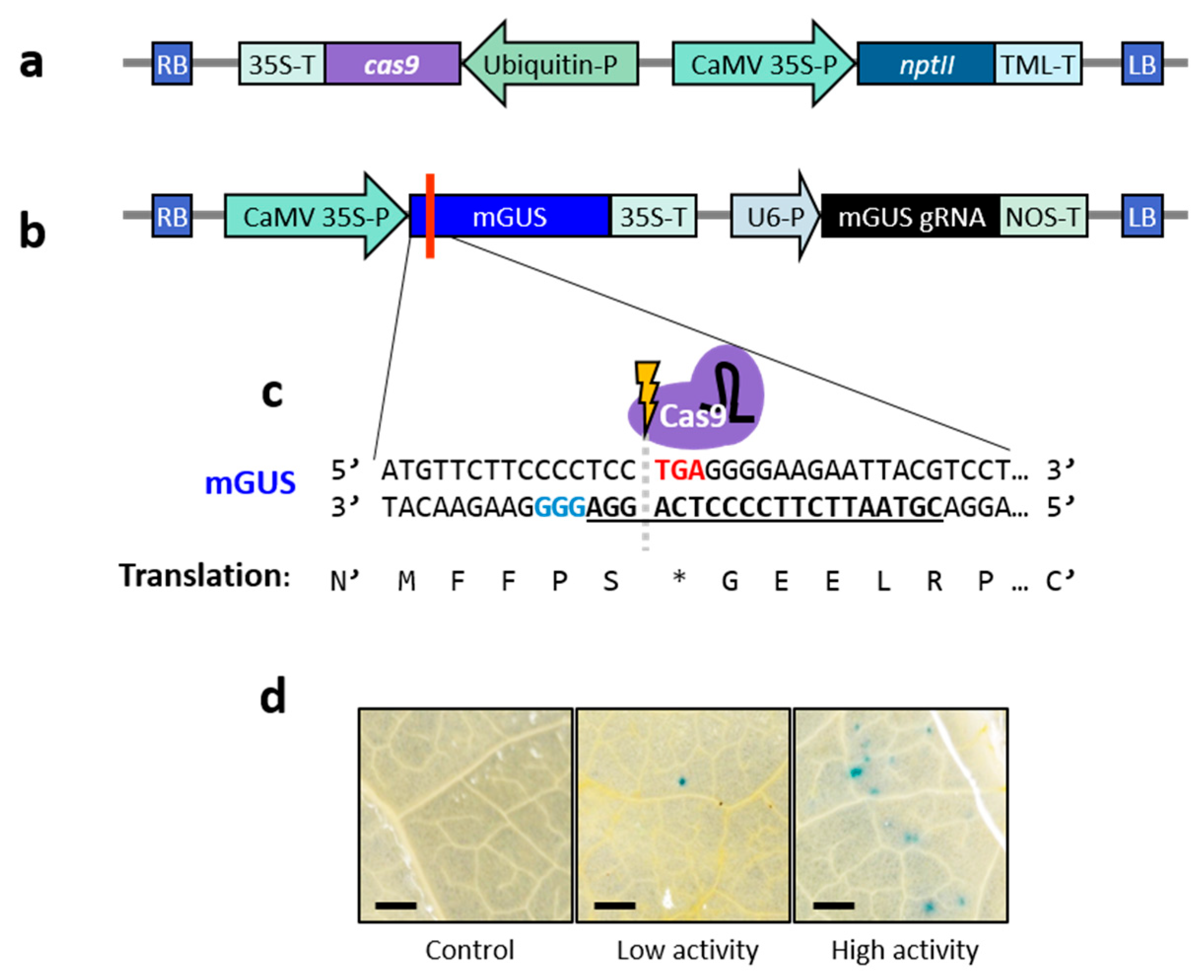
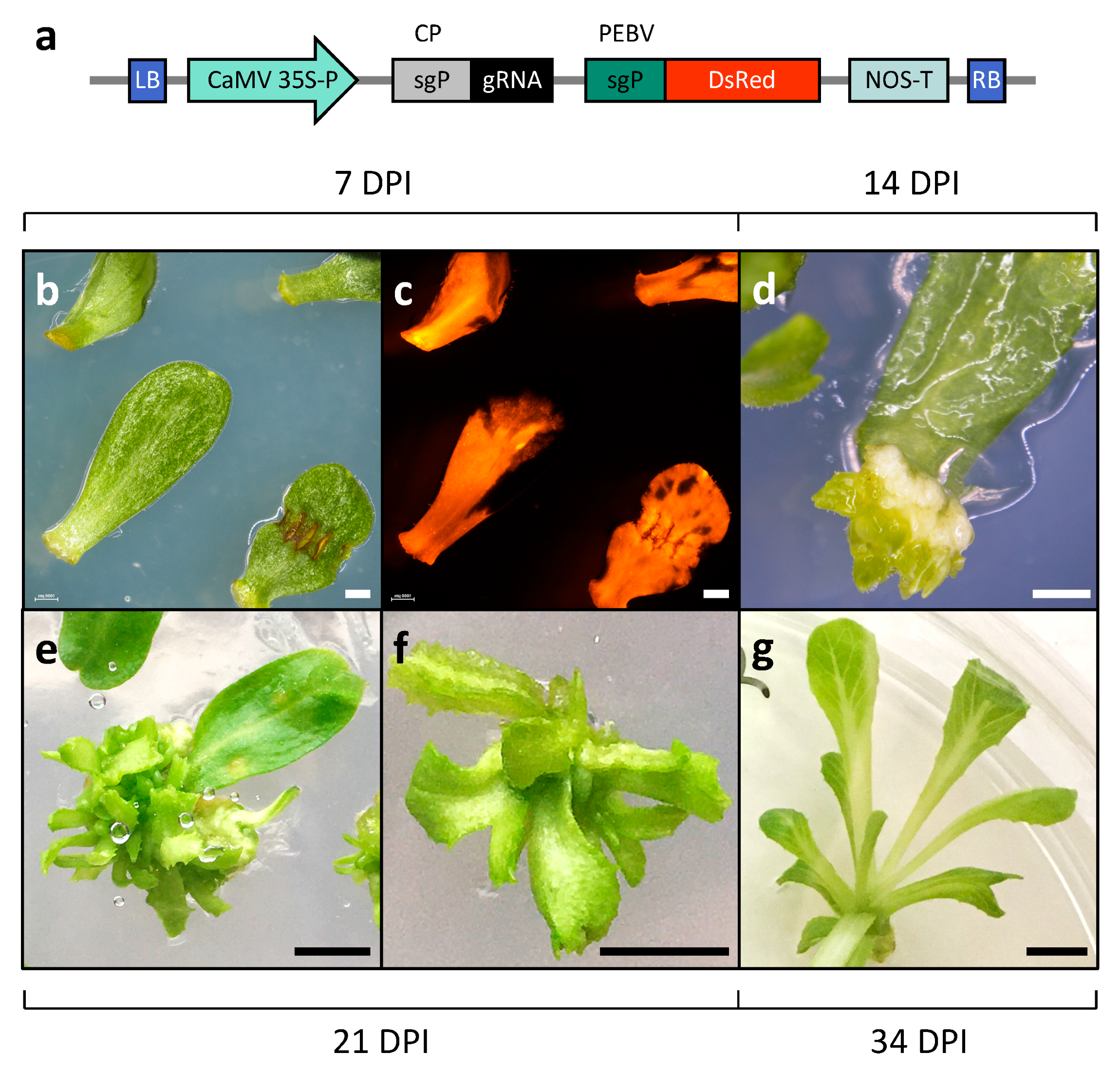
2.1. Establishment of Efficient Virus-Mediated Gene Editing System in Lettuce
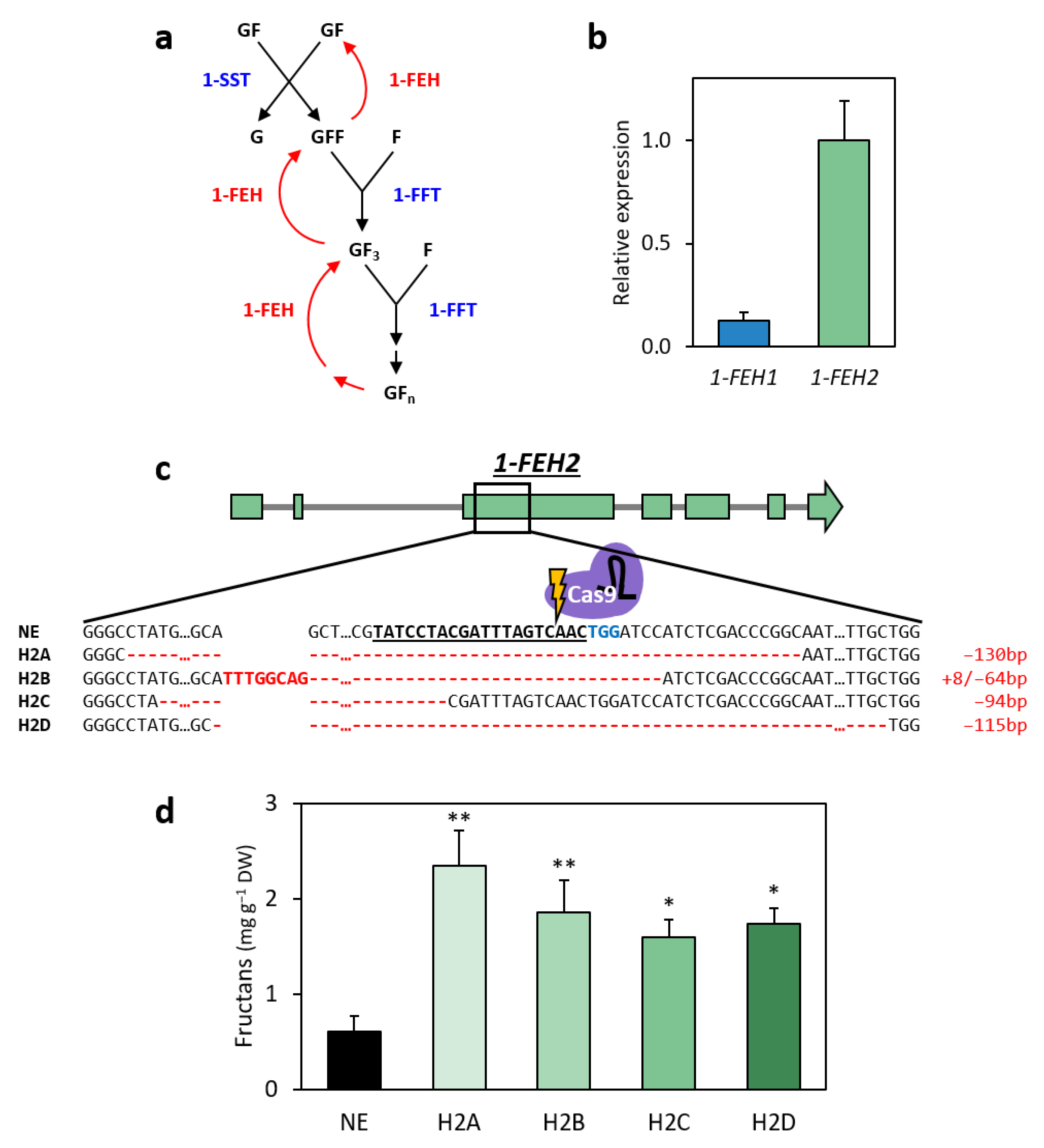
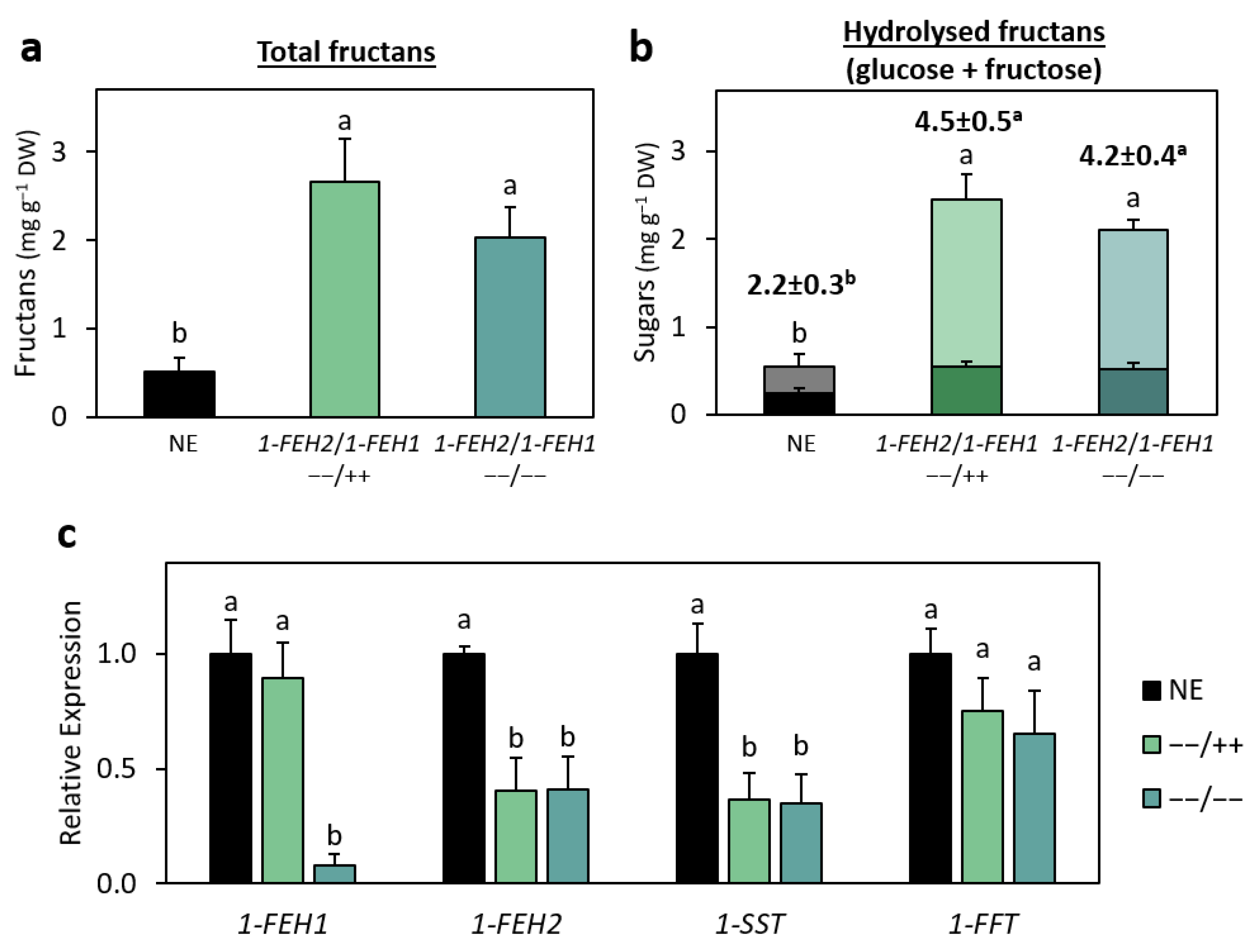
2.2. Increased Fructan Content and Mean Degree of Polymerisation in 1-FEH2 Knockout Lettuce Lines
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Plasmid Construction and gRNA Design
4.3. Tissue Culture and Transformation
4.4. Leaf Infiltration and Histochemical GUS Staining
4.5. Generation of Gene-Edited Plants
4.6. Fructan and Sugar Extraction and Quantificaiton
4.7. RNA Extraction and Quantitative Real-Time PCR (RT-qPCR)
4.8. Sequence Alignments and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rogo, U.; Simoni, S.; Fambrini, M.; Giordani, T.; Pugliesi, C.; Mascagni, F. Future-Proofing Agriculture: De Novo Domestication for Sustainable and Resilient Crops. Int. J. Mol. Sci. 2024, 25, 2374. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Chen, L.; Yan, Z.; Xu, J.; Feng, L.; He, N.; Guo, M.; Zhao, J.; Chen, Z.; Chen, H.; et al. Recent Advances of CRISPR-Based Genome Editing for Enhancing Staple Crops. Front. Plant Sci. 2024, 15, 1478398. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Strobbe, S.; Van Der Straeten, D.; Zhang, C. Regulation of Plant Vitamin Metabolism: Backbone of Biofortification for the Alleviation of Hidden Hunger. Mol. Plant 2021, 14, 40–60. [Google Scholar] [CrossRef] [PubMed]
- Lenaerts, B.; Demont, M. The Global Burden of Chronic and Hidden Hunger Revisited: New Panel Data Evidence Spanning 1990–2017. Glob. Food Sec. 2021, 28, 100480. [Google Scholar] [CrossRef]
- Jones, J.M. CODEX-Aligned Dietary Fiber Definitions Help to Bridge the ‘Fiber Gap’. Nutr. J. 2014, 13, 34. [Google Scholar] [CrossRef]
- Puhlmann, M.L.; de Vos, W.M. Back to the Roots: Revisiting the Use of the Fiber-Rich Cichorium intybus L. Taproots. Adv. Nutr. 2020, 11, 878–889. [Google Scholar] [CrossRef]
- Mishra, A.; Pandey, V.P. CRISPR/Cas System: A Revolutionary Tool for Crop Improvement. Biotechnol. J. 2024, 19, 2300298. [Google Scholar] [CrossRef]
- Chawla, R.; Poonia, A.; Samantara, K.; Mohapatra, S.R.; Naik, S.B.; Ashwath, M.N.; Djalovic, I.G.; Prasad, P.V.V. Green Revolution to Genome Revolution: Driving Better Resilient Crops against Environmental Instability. Front. Genet. 2023, 14, 1204585. [Google Scholar] [CrossRef]
- Altpeter, F.; Springer, N.M.; Bartley, L.E.; Blechl, A.E.; Brutnell, T.P.; Citovsky, V.; Conrad, L.J.; Gelvin, S.B.; Jackson, D.P.; Kausch, A.P.; et al. Advancing Crop Transformation in the Era of Genome Editing. Plant Cell 2016, 28, 1510–1520. [Google Scholar] [CrossRef]
- Kausch, A.P.; Nelson-vasilchik, K.; Hague, J.; Mookkan, M.; Quemada, H.; Dellaporta, S.; Fragoso, C.; Zhang, Z.J. Plant Science Edit at Will: Genotype Independent Plant Transformation in the Era of Advanced Genomics and Genome Editing. Plant Sci. 2019, 281, 186–205. [Google Scholar] [CrossRef]
- Honig, A.; Marton, I.; Rosenthal, M.; Smith, J.J.; Nicholson, M.G.; Jantz, D.; Zuker, A.; Vainstein, A. Transient Expression of Virally Delivered Meganuclease In Planta Generates Inherited Genomic Deletions. Mol. Plant 2015, 8, 1292–1294. [Google Scholar] [CrossRef] [PubMed]
- Ellison, E.E.; Chamness, J.C.; Voytas, D.F. Viruses as Vectors for the Delivery of Gene-Editing Reagents. In Genome Editing for Precision Crop Breeding; Burleigh Dodds Science Publishing: Cambridge, UK, 2021; pp. 97–122. ISBN 9781003048237. [Google Scholar]
- Spitzer, B.; Zvi, M.M.B.; Ovadis, M.; Marhevka, E.; Barkai, O.; Edelbaum, O.; Marton, I.; Masci, T.; Alon, M.; Morin, S.; et al. Reverse Genetics of Floral Scent: Application of Tobacco Rattle Virus-Based Gene Silencing in Petunia. Plant Physiol. 2007, 145, 1241–1250. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Abul-Faraj, A.; Li, L.; Ghosh, N.; Piatek, M.; Mahjoub, A.; Aouida, M.; Piatek, A.; Baltes, N.J.; Voytas, D.F.; et al. Efficient Virus-Mediated Genome Editing in Plants Using the CRISPR/Cas9 System. Mol. Plant 2015, 8, 1288–1291. [Google Scholar] [CrossRef] [PubMed]
- Skaliter, O.; Bednarczyk, D.; Shor, E.; Shklarman, E.; Manasherova, E.; Aravena-Calvo, J.; Kerzner, S.; Cna’ani, A.; Jasinska, W.; Masci, T.; et al. The R2R3-MYB Transcription Factor EVER Controls the Emission of Petunia Floral Volatiles by Regulating Epicuticular Wax Biosynthesis in the Petal Epidermis. Plant Cell 2024, 36, 174–193. [Google Scholar] [CrossRef]
- Visser, P.B.; Mathis, A.; Linthorst, H.J. Tobraviruses. In Encyclopedia of Virology, 2nd ed.; Academic Press: New York, NY, USA, 1999; pp. 1784–1789. [Google Scholar]
- FAOSTAT. Statistics of the Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/faostat/en/#data/QCL (accessed on 21 October 2024).
- De Moura Cipriano, T.; Pedroso, M.T.M.; de Paula Nunes, I.A.; Queiroz, L.N.; Aragão, F.J.L. Public Perception of Folate-Biofortified Genetically Modified Lettuce Varieties in Brazil. Transgenic Res. 2024, 33, 359–368. [Google Scholar] [CrossRef]
- Marlett, J.A.; Vollendorf, N.W. Dietary Fiber Content and Composition of Vegetables Determined by Two Methods of Analysis. J. Agric. Food Chem. 1993, 41, 1608–1612. [Google Scholar] [CrossRef]
- Khanam, U.K.S.; Oba, S.; Yanase, E.; Murakami, Y. Phenolic Acids, Flavonoids and Total Antioxidant Capacity of Selected Leafy Vegetables. J. Funct. Foods 2012, 4, 979–987. [Google Scholar] [CrossRef]
- Rowe, S.; Carr, A.C. Factors Affecting Vitamin C Status and Prevalence of Deficiency: A Global Health Perspective. Nutrients 2020, 12, 1963. [Google Scholar] [CrossRef]
- Carazo, A.; Macáková, K.; Matoušová, K.; Krčmová, L.K.; Protti, M.; Mladěnka, P. Vitamin a Update: Forms, Sources, Kinetics, Detection, Function, Deficiency, Therapeutic Use and Toxicity. Nutrients 2021, 13, 1703. [Google Scholar] [CrossRef]
- Van Laere, A.; Van den Ende, W. Inulin Metabolism in Dicots: Chicory as a Model System. Plant Cell Environ. 2002, 25, 803–813. [Google Scholar] [CrossRef]
- Hernández, L.; Plou, F.J. Chapter 1—Fructans: The Terminology. In The Book of Fructans; den Ende, W.V., Öner, E.T., Eds.; Academic Press: New York, NY, USA, 2023; pp. 3–10. ISBN 978-0-323-85410-8. [Google Scholar]
- Van Arkel, J.; Vergauwen, R.; Sévenier, R.; Hakkert, J.C.; van Laere, A.; Bouwmeester, H.J.; Koops, A.J.; van der Meer, I.M. Sink Filling, Inulin Metabolizing Enzymes and Carbohydrate Status in Field Grown Chicory (Cichorium intybus L.). J. Plant Physiol. 2012, 169, 1520–1529. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.M.; Bauer, L.L.; Fahey, G.C.; Hogarth, A.J.C.L.; Wolf, B.W.; Hunter, D.E. Selected Fructooligosaccharide (1-Kestose, Nystose, and 1F-Beta-Fructofuranosylnystose) Composition of Foods and Feeds. J. Agric. Food Chem. 1997, 45, 3076–3082. [Google Scholar] [CrossRef]
- Sobolev, A.P.; Brosio, E.; Gianferri, R.; Segre, A.L. Metabolic Profile of Lettuce Leaves by High-Field NMR Spectra. Magn. Reson. Chem. 2005, 43, 625–638. [Google Scholar] [CrossRef] [PubMed]
- Blanch, M.; Alvarez, M.D.; Sanchez-Ballesta, M.T.; Escribano, M.I.; Merodio, C. Water Relations, Short-Chain Oligosaccharides and Rheological Properties in Lettuces Subjected to Limited Water Supply and Low Temperature Stress. Sci. Hortic. 2017, 225, 726–735. [Google Scholar] [CrossRef]
- Pool-Zobel, B.L. Inulin-Type Fructans and Reduction in Colon Cancer Risk: Review of Experimental and Human Data. Br. J. Nutr. 2005, 93, S73–S90. [Google Scholar] [CrossRef]
- Chen, G.; Li, C.; Chen, K. Fructooligosaccharides: A Review on Their Mechanisms of Action and Effects. Stud. Nat. Prod. Chem. 2016, 48, 209–229. [Google Scholar] [CrossRef]
- Le Bastard, Q.; Chapelet, G.; Javaudin, F.; Lepelletier, D.; Batard, E.; Montassier, E. The Effects of Inulin on Gut Microbial Composition: A Systematic Review of Evidence from Human Studies. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 403–413. [Google Scholar] [CrossRef]
- Dou, Y.; Yu, X.; Luo, Y.; Chen, B.; Ma, D.; Zhu, J. Effect of Fructooligosaccharides Supplementation on the Gut Microbiota in Human: A Systematic Review and Meta-Analysis. Nutrients 2022, 14, 3289. [Google Scholar] [CrossRef]
- Fernández-Lainez, C.; López-Velázquez, G.; de Vos, P. Chapter 13—Health Benefits of Inulin and Agavin-Type Fructans in Food: Impact on Microbiota, Immune and Gut Barrier Function. In The Book of Fructans; den Ende, W.V., Öner, E.T., Eds.; Academic Press: New York, NY, USA, 2023; pp. 211–234. ISBN 978-0-323-85410-8. [Google Scholar]
- Huazano-García, A.; Silva-Adame, M.B.; López, M.G. Chapter 14—Preclinical and Clinical Fructan Studies. In The Book of Fructans; den Ende, W.V., Öner, E.T., Eds.; Academic Press: New York, NY, USA, 2023; pp. 235–256. ISBN 978-0-323-85410-8. [Google Scholar]
- Dauchot, N.; Raulier, P.; Maudoux, O.; Notté, C.; Draye, X.; Van Cutsem, P. Loss of Function of 1-FEH IIb Has More Impact on Post-Harvest Inulin Degradation in Cichorium Intybus than Copy Number Variation of Its Close Paralog 1-FEH IIa. Front. Plant Sci. 2015, 6, 455. [Google Scholar] [CrossRef][Green Version]
- Sobolev, A.P.; Segre, A.L.; Giannino, D.; Mariotti, D.; Nicolodi, C.; Brosio, E.; Amato, M.E. Strong Increase of Foliar Inulin Occurs in Transgenic Lettuce Plants (Lactuca sativa L.) Overexpressing the Asparagine Synthetase A Gene from Escherichia Coli. J. Agric. Food Chem. 2007, 55, 10827–10831. [Google Scholar] [CrossRef]
- Sobolev, A.P.; Testone, G.; Santoro, F.; Nicolodi, C.; Iannelli, M.A.; Amato, M.E.; Ianniello, A.; Brosio, E.; Giannino, D.; Mannina, L. Quality Traits of Conventional and Transgenic Lettuce (Lactuca sativa L.) at Harvesting by NMR Metabolic Profiling. J. Agric. Food Chem. 2010, 58, 6928–6936. [Google Scholar] [CrossRef] [PubMed]
- Marton, I.; Zuker, A.; Shklarman, E.; Zeevi, V.; Tovkach, A.; Roffe, S.; Ovadis, M.; Tzfira, T.; Vainstein, A. Nontransgenic Genome Modification in Plant Cells. Plant Physiol. 2010, 154, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Van den Ende, W.; Michiels, A.; Van Wonterghem, D.; Clerens, S.P.; De Roover, J.; Van Laere, A.J. Defoliation Induces Fructan 1-Exohydrolase II in Witloof Chicory Roots. Cloning and Purification of Two Isoforms, Fructan 1-Exohydrolase IIa and Fructan 1-Exohydrolase IIb. Mass Fingerprint of the Fructan 1-Exohydrolase II Enzymes. Plant Physiol. 2001, 126, 1186–1195. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Oda-Yamamizo, C.; Sage-Ono, K.; Ohmiya, A.; Ono, M. Alteration of Flower Colour in Ipomoea Nil through CRISPR/Cas9-Mediated Mutagenesis of Carotenoid Cleavage Dioxygenase 4. Transgenic Res. 2018, 27, 25–38. [Google Scholar] [CrossRef]
- Yu, S.; Li, M.; Dubcovsky, J.; Tian, L. Mutant Combinations of Lycopene Ɛ-Cyclase and β-Carotene Hydroxylase 2 Homoeologs Increased β-Carotene Accumulation in Endosperm of Tetraploid Wheat (Triticum turgidum L.) Grains. Plant Biotechnol. J. 2022, 20, 564–576. [Google Scholar] [CrossRef]
- Wang, J.; Yu, Y.; Zhang, Z.; Quan, R.; Zhang, H.; Ma, L.; Deng, X.W.; Huang, R. Arabidopsis CSN5B Interacts with VTC1 and Modulates Ascorbic Acid Synthesis. Plant Cell 2013, 25, 625–636. [Google Scholar] [CrossRef]
- Zhang, H.; Si, X.; Ji, X.; Fan, R.; Liu, J.; Chen, K.; Wang, D.; Gao, C. Genome Editing of Upstream Open Reading Frames Enables Translational Control in Plants. Nat. Biotechnol. 2018, 36, 894–900. [Google Scholar] [CrossRef]
- Concordet, J.P.; Haeussler, M. CRISPOR: Intuitive Guide Selection for CRISPR/Cas9 Genome Editing Experiments and Screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef]
- Van Den Ende, W.; Michiels, A.; De Roover, J.; Verhaert, P.; Van Laere, A. Cloning and Functional Analysis of Chicory Root Fructan 1-Exohydrolase I (1-FEH I): A Vacuolar Enzyme Derived from a Cell-Wall Invertase Ancestor? Mass Fingerprint of the 1-FEH I Enzyme. Plant J. 2000, 24, 447–456. [Google Scholar] [CrossRef]
- Dauchot, N.; Raulier, P.; Maudoux, O.; Notté, C.; Bertin, P.; Draye, X.; Van Cutsem, P. Mutations in Chicory FEH Genes Are Statistically Associated with Enhanced Resistance to Post-Harvest Inulin Depolymerization. Theor. Appl. Genet. 2014, 127, 125–135. [Google Scholar] [CrossRef]
- Ricachenevsky, F.K.; Vasconcelos, M.W.; Shou, H.; Johnson, A.A.T.; Sperotto, R.A. Editorial: Improving the Nutritional Content and Quality of Crops: Promises, Achievements, and Future Challenges. Front. Plant Sci. 2023, 10, 738. [Google Scholar] [CrossRef] [PubMed]
- Combie, J. Chapter 12—The Fructan Industry. In The Book of Fructans; den Ende, W.V., Öner, E.T., Eds.; Academic Press: New York, NY, USA, 2023; pp. 201–209. ISBN 978-0-323-85410-8. [Google Scholar]
- Javaid, N.; Choi, S. CRISPR/Cas System and Factors Affecting Its Precision and Efficiency. Front. Cell Dev. Biol. 2021, 9, 761709. [Google Scholar] [CrossRef] [PubMed]
- Darqui, F.S.; Radonic, L.M.; Beracochea, V.C.; Hopp, H.E.; López Bilbao, M. Peculiarities of the Transformation of Asteraceae Family Species: The Cases of Sunflower and Lettuce. Front. Plant Sci. 2021, 12, 767459. [Google Scholar] [CrossRef] [PubMed]
- Bertier, L.D.; Ron, M.; Huo, H.; Bradford, K.J.; Britt, A.B.; Michelmore, R.W. High-Resolution Analysis of the Efficiency, Heritability, and Editing Outcomes of CRISPR/Cas9-Induced Modifications of NCED4 in Lettuce (Lactuca sativa). G3 Genes Genomes Genet. 2018, 8, 1513–1521. [Google Scholar] [CrossRef]
- Luo, C.; Wang, S.; Ning, K.; Chen, Z.; Wang, Y.; Yang, J.; Qi, M.; Wang, Q. The APETALA2 Transcription Factor LsAP2 Regulates Seed Shape in Lettuce. J. Exp. Bot. 2021, 72, 2463–2476. [Google Scholar] [CrossRef]
- Pan, W.; Liu, X.; Li, D.; Zhang, H. Establishment of an Efficient Genome Editing System in Lettuce Without Sacrificing Specificity. Front. Plant Sci. 2022, 13, 930592. [Google Scholar] [CrossRef]
- Gerszberg, A.; Grzegorczyk-Karolak, I. Influence of Selected Antibiotics on the Tomato Regeneration in in Vitro Cultures. Not. Bot. Horti Agrobot. Cluj-Napoca 2019, 47, 558–564. [Google Scholar] [CrossRef]
- Verma, S.; Kumar, A.; Modgil, M. Impact of Cefotaxime and Kanamycin on in Vitro Regeneration via Agrobacterium Mediated Transformation in Apple Cv. Red Chief. Plant Physiol. Rep. 2023, 28, 34–42. [Google Scholar] [CrossRef]
- Riu, Y.S.; Kim, G.H.; Chung, K.W.; Kong, S.G. Enhancement of the CRISPR/Cas9-Based Genome Editing System in Lettuce (Lactuca sativa L.) Using the Endogenous U6 Promoter. Plants 2023, 12, 878. [Google Scholar] [CrossRef]
- Liu, H.; Chen, W.; Li, Y.; Sun, L.; Chai, Y.; Chen, H.; Nie, H.; Huang, C. CRISPR/Cas9 Technology and Its Utility for Crop Improvement. Int. J. Mol. Sci. 2022, 23, 442. [Google Scholar] [CrossRef]
- Medina-Lozano, I.; Díaz, A. Applications of Genomic Tools in Plant Breeding: Crop Biofortification. Int. J. Mol. Sci. 2022, 23, 3086. [Google Scholar] [CrossRef] [PubMed]
- Tachikawa, M.; Matsuo, M. Global Regulatory Trends of Genome Editing Technology in Agriculture and Food. Breed. Sci. 2024, 74, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Sevenier, R.; Arkel, J.; Hakkert, J.C.; Koops, A. Fructan: Nutritional Significance, Application, Biosynthesis, Molecular Biology and Genetic Engineering. In Plant Genetic Engineering, Volume 7, Metabolic Engineering and Molecular Farming-I; Wageningen University&Research: Wageningen, The Netherlands, 2005. [Google Scholar]
- Valluru, R.; Lammens, W.; Claupein, W.; Van den Ende, W. Freezing Tolerance by Vesicle-Mediated Fructan Transport. Trends Plant Sci. 2008, 13, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Livingston, D.P.; Hincha, D.K.; Heyer, A.G. Fructan and Its Relationship to Abiotic Stress Tolerance in Plants. Cell. Mol. Life Sci. 2009, 66, 2007–2023. [Google Scholar] [CrossRef]
- Livingston, D.P., III; Heyer, A.G.; Kırtel, O. Chapter 7—The Role of Fructans in Stress Responses. In The Book of Fructans; den Ende, W.V., Öner, E.T., Eds.; Academic Press: New York, NY, USA, 2023; pp. 109–126. ISBN 978-0-323-85410-8. [Google Scholar]
- Kusch, U.; Greiner, S.; Steininger, H.; Meyer, A.D.; Corbière-Divialle, H.; Harms, K.; Rausch, T. Dissecting the Regulation of Fructan Metabolism in Chicory (Cichorium intybus) Hairy Roots. New Phytol. 2009, 184, 127–140. [Google Scholar] [CrossRef]
- Li, H.; Yang, A.; Zhang, X.; Gao, F.; Zhang, J.-R. Improving Freezing Tolerance of Transgenic Tobacco Expressing Sucrose: Sucrose 1-Fructosyltransferase Gene from Lactuca Sativa. Plant Cell Tissue Organ Cult. 2007, 89, 37–48. [Google Scholar] [CrossRef]
- Bosscher, D.; Van Loo, J.; Franck, A. Inulin and Oligofructose as Prebiotics in the Prevention of Intestinal Infections and Diseases. Nutr. Res. Rev. 2006, 19, 216–226. [Google Scholar] [CrossRef]
- Van De Wiele, T.; Boon, N.; Possemiers, S.; Jacobs, H.; Verstraete, W. Inulin-Type Fructans of Longer Degree of Polymerization Exert More Pronounced in vitro Prebiotic Effects. J. Appl. Microbiol. 2007, 102, 452–460. [Google Scholar] [CrossRef]
- Van Arkel, J.; Sévenier, R.; Hakkert, J.C.; Bouwmeester, H.J.; Koops, A.J.; Van Der Meer, I.M. Tailor-Made Fructan Synthesis in Plants: A Review. In Carbohydrate Polymers; Elsevier: Amsterdam, The Netherlands, 2013; Volume 93, pp. 48–56. [Google Scholar]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Wang, X.; Xu, L.; Liu, X.; Xin, L.; Wu, S.; Chen, X. Development of Potent Promoters That Drive the Efficient Expression of Genes in Apple Protoplasts. Hortic. Res. 2021, 8, 211. [Google Scholar] [CrossRef] [PubMed]
- Zvi, M.M.B.; Negre-Zakharov, F.; Masci, T.; Ovadis, M.; Shklarman, E.; Ben-meir, H.; Tzfira, T.; Dudareva, N.; Vainstein, A. Interlinking Showy Traits: Co-Engineering of Scent and Colour Biosynthesis in Flowers. Plant Biotechnol. J. 2008, 6, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Verwaaijen, B.; Wibberg, D.; Nelkner, J.; Gordin, M.; Rupp, O.; Winkler, A.; Bremges, A.; Blom, J.; Grosch, R.; Pühler, A.; et al. Assembly of the Lactuca sativa L. Cv. Tizian Draft Genome Sequence Reveals Differences within Major Resistance Complex 1 as Compared to the Cv. Salinas Reference Genome. J. Biotechnol. 2018, 267, 12–18. [Google Scholar] [CrossRef]
- Dinesh-Kumar, S.P.; Anandalakshmi, R.; Marathe, R.; Schiff, M.; Liu, Y. Virus-Induced Gene Silencing. Methods Mol. Biol. 2003, 236, 287–294. [Google Scholar] [CrossRef]
- Lazo, G.R.; Stein, P.A.; Ludwig, R.A. A DNA Transformation–Competent Arabidopsis Genomic Library in Agrobacterium. Biotechnology 1991, 9, 963–967. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS Fusions: Beta-Glucuronidase as a Sensitive and Versatile Gene Fusion Marker in Higher Plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Sgamma, T.; Pape, J.; Massiah, A.; Jackson, S. Selection of Reference Genes for Diurnal and Developmental Time-Course Real-Time PCR Expression Analyses in Lettuce. Plant Methods 2016, 12, 21. [Google Scholar] [CrossRef]
- Nozue, K.; Covington, M.F.; Duek, P.D.; Lorrain, S.; Fankhauser, C.; Harmer, S.L.; Maloof, J.N. Rhythmic Growth Explained by Coincidence between Internal and External Cues. Nature 2007, 448, 358–361. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher Sequence Analysis Tools Framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
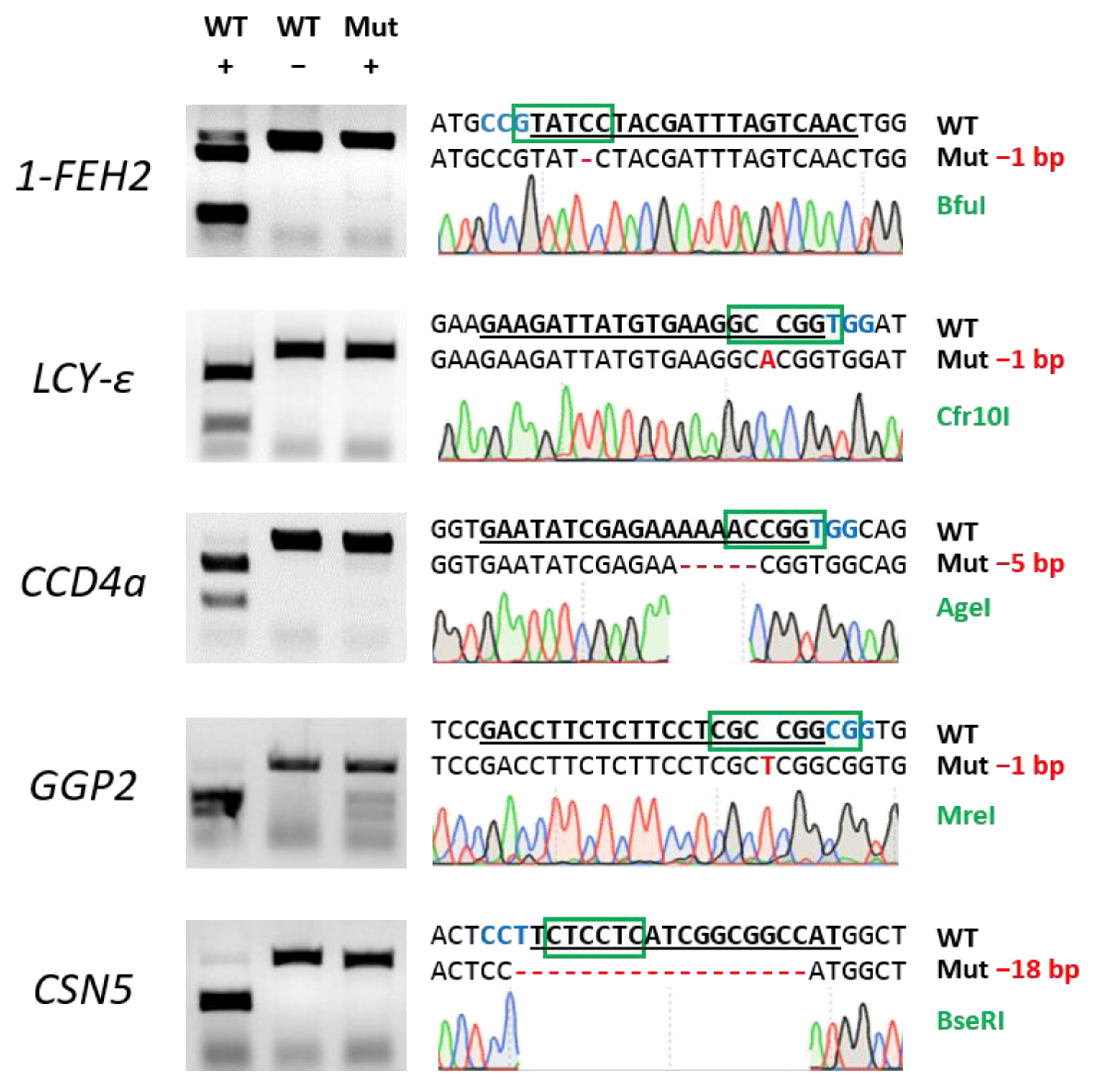
| Target Gene | Regeneration Efficiency 1 | Editing Efficiency (%/Regenerated) 2 | Editing Efficiency (%/Initial Explants) 3 |
|---|---|---|---|
| 1-FEH2 | 81.0% | 32.4% | 26.2% |
| LCY-ε | 48.8% | 38.5% | 18.8% |
| CCD4a | 58.0% | 35.0% | 20.3% |
| GGP2 | 64.2% | 58.8% | 37.7% |
| CSN5 | 95.0% | 78.9% | 75.0% |
| Average | 69.4% | 48.7% | 35.6% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Livneh, Y.; Agmon, D.; Leor-Librach, E.; Vainstein, A. Viral-Based Gene Editing System for Nutritional Improvement of Fructan Content in Lettuce. Int. J. Mol. Sci. 2025, 26, 2594. https://doi.org/10.3390/ijms26062594
Livneh Y, Agmon D, Leor-Librach E, Vainstein A. Viral-Based Gene Editing System for Nutritional Improvement of Fructan Content in Lettuce. International Journal of Molecular Sciences. 2025; 26(6):2594. https://doi.org/10.3390/ijms26062594
Chicago/Turabian StyleLivneh, Yarin, Dor Agmon, Ehud Leor-Librach, and Alexander Vainstein. 2025. "Viral-Based Gene Editing System for Nutritional Improvement of Fructan Content in Lettuce" International Journal of Molecular Sciences 26, no. 6: 2594. https://doi.org/10.3390/ijms26062594
APA StyleLivneh, Y., Agmon, D., Leor-Librach, E., & Vainstein, A. (2025). Viral-Based Gene Editing System for Nutritional Improvement of Fructan Content in Lettuce. International Journal of Molecular Sciences, 26(6), 2594. https://doi.org/10.3390/ijms26062594





