Utilization of Crab Shell Waste for Value-Added Bioplastics by Pseudomonas-Based Microbial Cell Factories
Abstract
1. Introduction
2. Results
2.1. Effect of Individual Fermentation Factors on the Maximum Growth of the Strain
2.1.1. Time
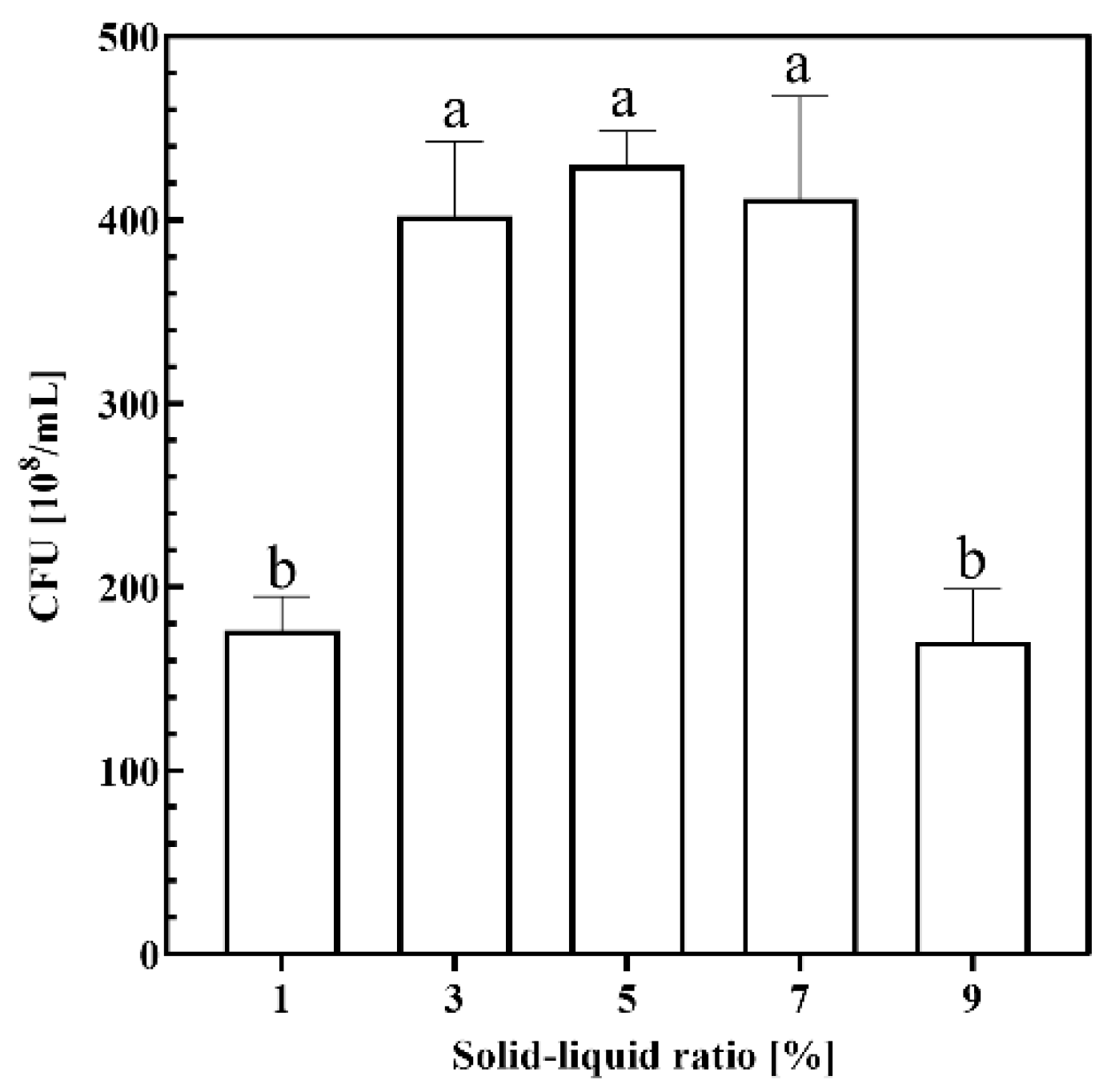
2.1.2. Solid–Liquid Ratio
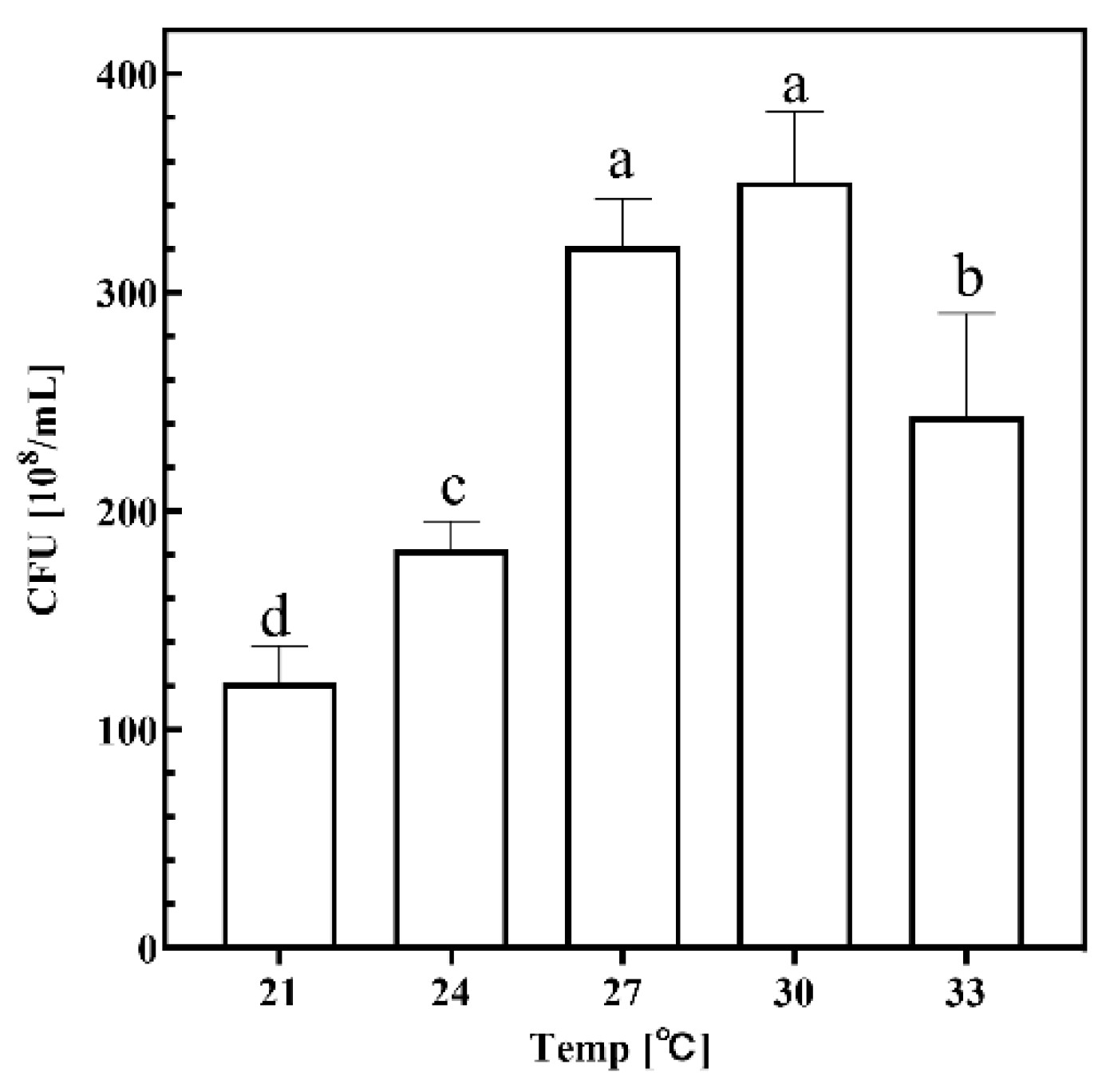
2.1.3. Temperature
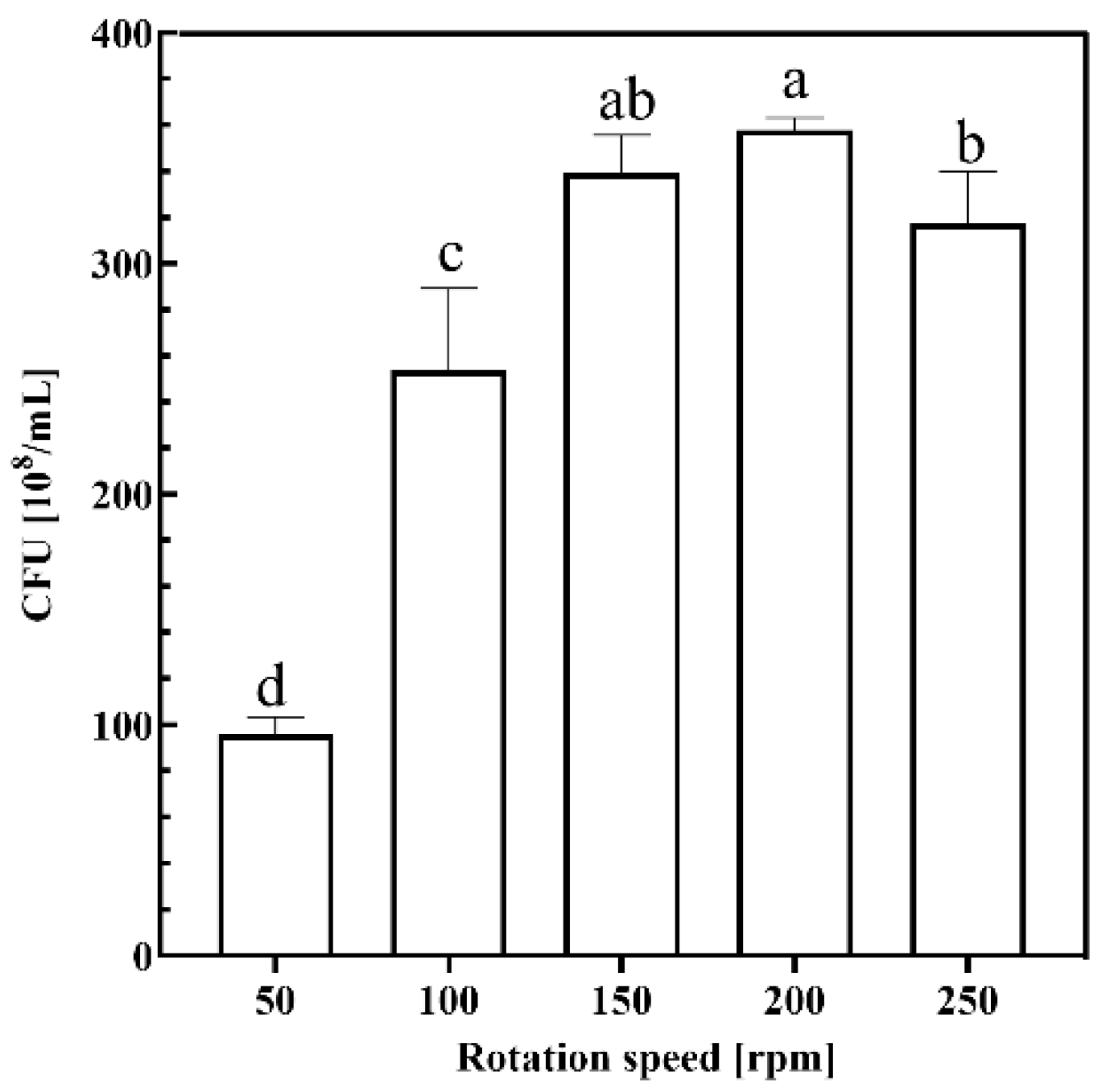
2.1.4. Rotation Speed
2.2. Optimal Fermentation Parameters Determined by Orthogonal Experiments
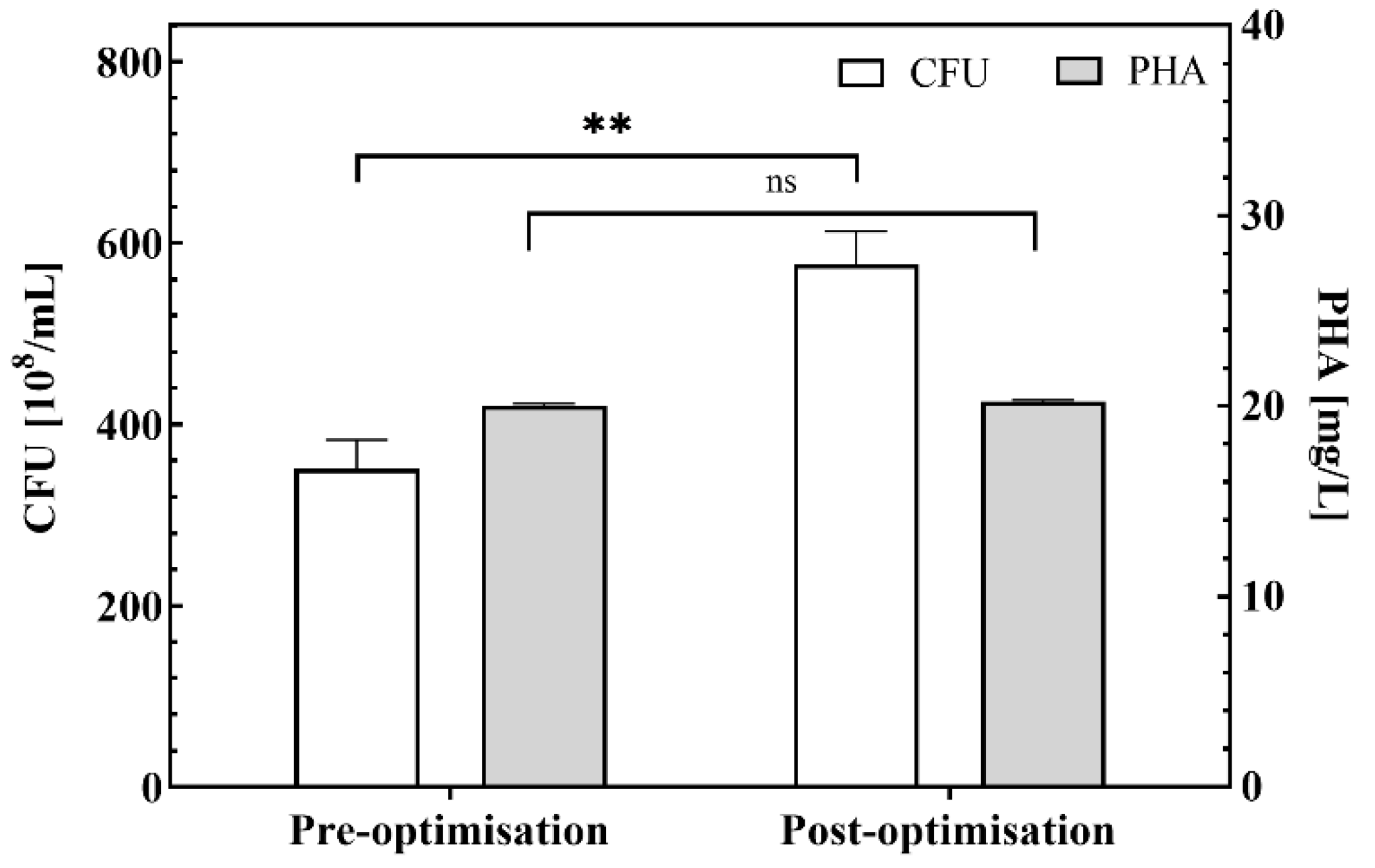
2.3. Validation of Optimization of Fermentation Parameter Combinations
2.4. Effects of Metabolic Engineering of Nitrogen Metabolism on Cell Growth and PHA Production
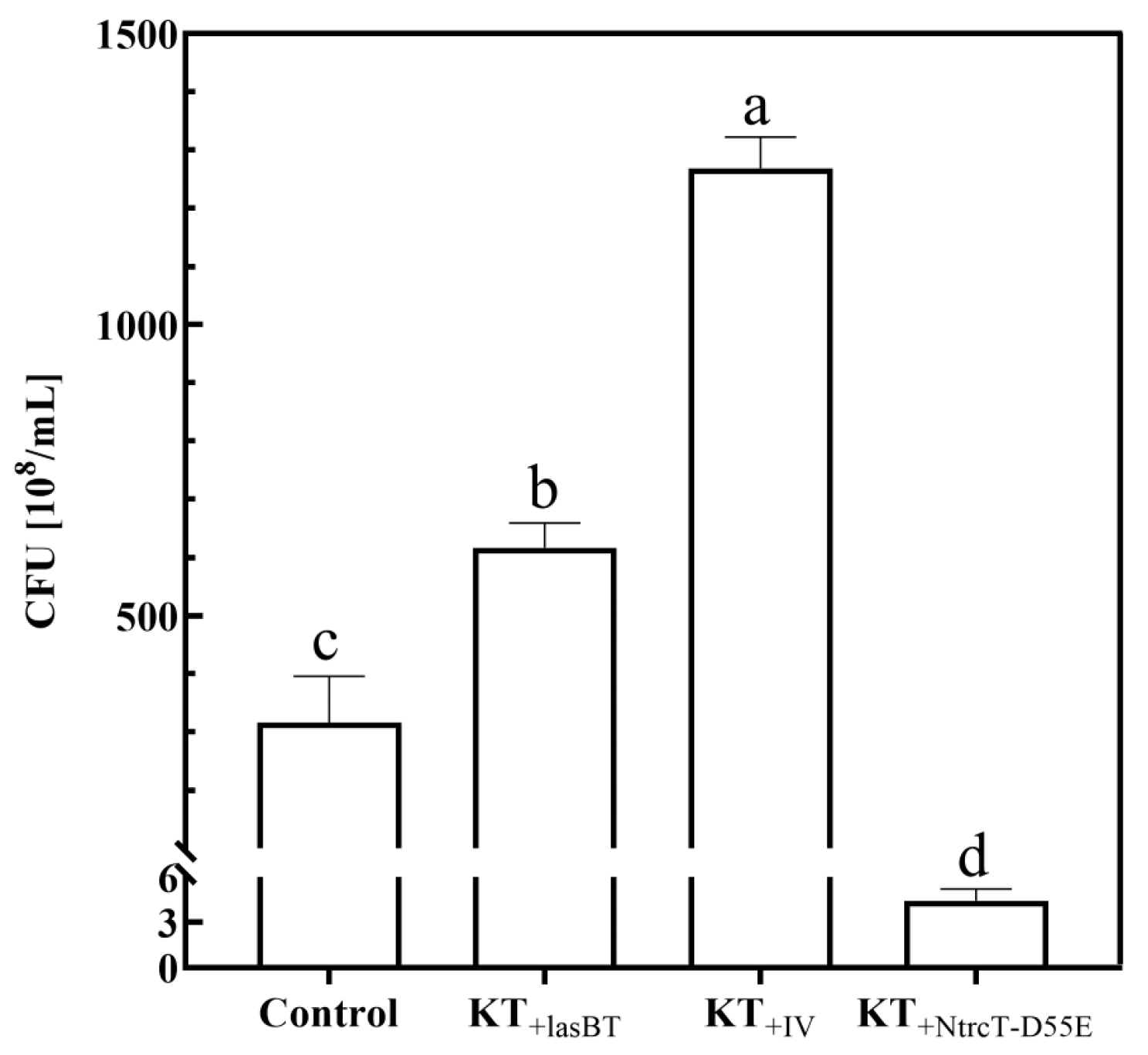
2.4.1. Cell Growth
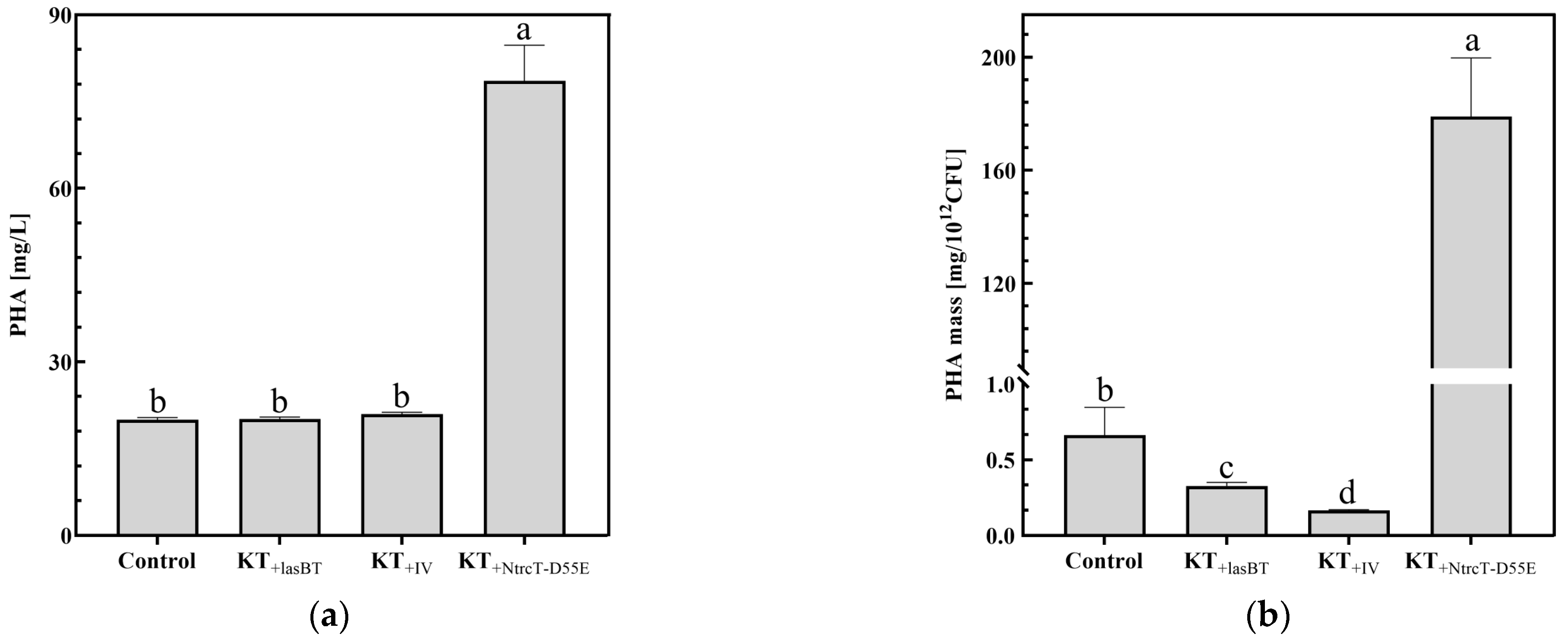
2.4.2. PHA Production
2.4.3. PHA Monomer Composition Analysis
3. Discussion
4. Materials and Methods
4.1. Experimental Materials
4.1.1. Crab Shell Preparation
4.1.2. Culture Media
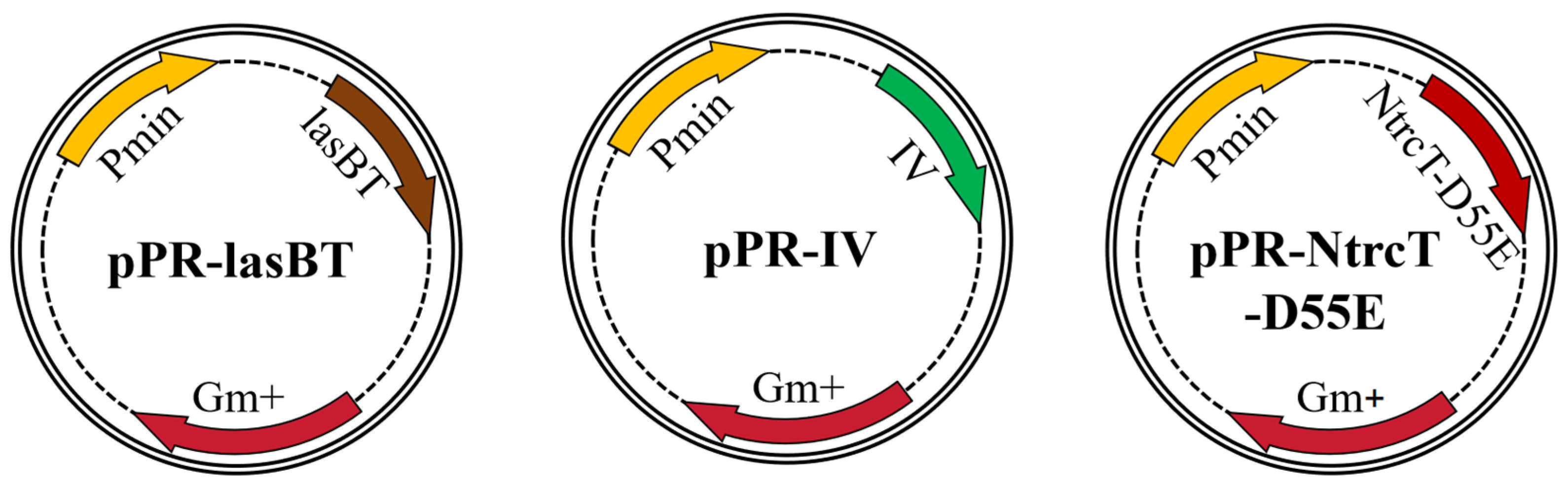
4.1.3. Plasmids and Strains Construction
4.2. Inoculation and Fermentation
4.3. Single Factor of Fermentation
4.4. Orthogonal Experiments to Optimize Fermentation Parameter Combinations
4.5. Measurement of PHA Content
4.6. Data Processing
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lipp, M.; Bessy, C.; Cannavan, A.; Dupouy, E.; Fattori, V.; Kopko, C.; Lejeune, J.; Mukherjee, K.; Ferreira, J.P.; Schulz, D.; et al. Food and Agriculture Organization of the United Nations (FAO). In Encyclopedia of Food Safety (Second Edition); Smithers, G.W., Ed.; Academic Press: Oxford, UK, 2024; pp. 752–760. ISBN 978-0-12-822520-2. [Google Scholar]
- Nekvapil, F.; Ganea, I.V.; Ciorita, A.; Hirian, R.; Ogresta, L.; Glamuzina, B.; Roba, C.; Pinzaru, S.C. Wasted Biomaterials from Crustaceans as a Compliant Natural Product Regarding Microbiological, Antibacterial Properties and Heavy Metal Content for Reuse in Blue Bioeconomy: A Preliminary Study. J. Mater. 2021, 14, 4558. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.C.; Zhao, D.; Xue, C.; Mao, X. An Efficient Method for Chitin Production from Crab Shells by a Natural Deep Eutectic Solvent. Mar. Life Sci. Technol. 2022, 4, 384–388. [Google Scholar] [CrossRef]
- Sugiyanti, D.; Darmadji, P.; Anggrahini, S.; Anwar, C.; Santoso, U. Preparation and Characterization of Chitosan from Indonesian Tambak Lorok Shrimp Shell Waste and Crab Shell Waste. J. Pak. J. Nutr. 2018, 17, 446–453. [Google Scholar] [CrossRef]
- Zang, B.; Wang, Y.; Wang, G.; Zang, Q.; Chen, H.; Liu, B. A Comparative Study on the Yields of Natural Astaxanthin Esters Extracted from Shrimp Shells Using Four Different Organic Solvents. Zhejiang Chem. Ind. 2020, 51, 21–24. [Google Scholar]
- Drakonaki, A.; Mathioudaki, E.; Geladas, E.D.; Konsolaki, E.; Vitsaxakis, N.; Chaniotakis, N.; Xie, H.; Tsiotis, G. Production of Polyhydroxybutyrate by Genetically Modified Pseudomonas sp. phDV1: A Comparative Study of Utilizing Wine Industry Waste as a Carbon Source. Microorganisms 2023, 11, 1592. [Google Scholar] [CrossRef]
- Wang, Y.; Wilks, J.C.; Danhorn, T.; Ramos, I.; Croal, L.; Newman, D.K. Phenazine-1-Carboxylic Acid Promotes Bacterial Biofilm Development via Ferrous Iron Acquisition. J. Bacteriol. 2011, 193, 3606–3617. [Google Scholar] [CrossRef] [PubMed]
- Letzel, A.C.; Pidot, S.J.; Hertweck, C. Genome Mining for Ribosomally Synthesized and Post-Translationally Modified Peptides (RiPPs) in Anaerobic Bacteria. BMC Genom. 2014, 15, 983. [Google Scholar] [CrossRef]
- Jyot, J.; Balloy, V.; Jouvion, G.; Verma, A.; Touqui, L.; Huerre, M.; Chignard, M.; Ramphal, R. Type II Secretion System of Pseudomonas Aeruginosa: In Vivo Evidence of a Significant Role in Death Due to Lung Infection. J. Infect. Dis. 2011, 203, 1369–1377. [Google Scholar] [CrossRef]
- Reddy, M.V.; Nikhil, G.N.; Mohan, S.V.; Swamy, Y.V.; Sarma, P.N. Pseudomonas Otitidis as a Potential Biocatalyst for Polyhydroxyalkanoates (PHA) Synthesis Using Synthetic Wastewater and Acidogenic Effluents. Bioresour. Technol. 2012, 123, 471–479. [Google Scholar] [CrossRef]
- Koller, M.; Bona, R.; Chiellini, E.; Fernandes, E.G.; Horvat, P.; Kutschera, C.; Hesse, P.; Braunegg, G. Polyhydroxyalkanoate Production from Whey by Pseudomonas Hydrogenovora. Bioresour. Technol. 2008, 99, 4854–4863. [Google Scholar] [CrossRef]
- Wongsirichot, P.; Gonzalez-Miquel, M.; Winterburn, J. Integrated Biorefining Approach for the Production of Polyhydroxyalkanoates from Enzymatically Hydrolyzed Rapeseed Meal under Nitrogen-Limited Conditions. ACS Sustain. Chem. Eng. 2020, 8, 8362–8372. [Google Scholar] [CrossRef]
- Wang, Q.; Nomura, C.T. Monitoring Differences in Gene Expression Levels and Polyhydroxyalkanoate (PHA) Production in Pseudomonas Putida KT2440 Grown on Different Carbon Sources. J. Biosci. Bioeng. 2010, 110, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Torrego-Solana, N.; Martin-Arjol, I.; Bassas-Galia, M.; Diaz, P.; Manresa, A. Hydroxy-Fatty Acid Production in a Pseudomonas Aeruginosa 42A2 PHA Synthase Mutant Generated by Directed Mutagenesis. Appl. Microb. Biotechnol. 2012, 93, 2551–2561. [Google Scholar] [CrossRef]
- Hori, K.; Ichinohe, R.; Unno, H.; Marsudi, S. Simultaneous Syntheses of Polyhydroxyalkanoates and Rhamnolipids by Pseudomonas aeruginosa IFO3924 at Various Temperatures and from Various Fatty Acids. Biochem. Eng. J. 2011, 53, 196–202. [Google Scholar] [CrossRef]
- Chen, J.Y.; Song, G.; Chen, G.Q. A Lower Specificity PhaC2 Synthase from Pseudomonas stutzeri Catalyses the Production of Copolyesters Consisting of Short-Chain-Length and Medium-Chain-Length 3-Hydroxyalkanoates. Antonie Van Leeuwenhoek 2006, 89, 157–167. [Google Scholar] [CrossRef]
- Martinez-Garcia, E.; de Lorenzo, V. Engineering Multiple Genomic Deletions in Gram-Negative Bacteria: Analysis of the Multi-Resistant Antibiotic Profile of Pseudomonas putida KT2440. Environ. Microbiol. 2011, 13, 2702–2716. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Cui, J.; Bilal, M.; Hu, H.; Wang, W.; Zhang, X. Pseudomonas spp. as Cell Factories (MCFs) for Value-Added Products: From Rational Design to Industrial Applications. Crit. Rev. Biotechnol. 2020, 40, 1232–1249. [Google Scholar] [CrossRef]
- Mitra, R.; Xu, T.; Xiang, H.; Han, J. Current Developments on Polyhydroxyalkanoates Synthesis by Using Halophiles as a Promising Cell Factory. Microb Cell Fact. 2020, 19, 86. [Google Scholar] [CrossRef]
- Chen, G.Q.; Jiang, X.-R.; Guo, Y. Synthetic Biology of Microbes Synthesizing Polyhydroxyalkanoates (PHA). Synth. Syst. Biotechnol. 2016, 1, 236–242. [Google Scholar] [CrossRef]
- Lim, J.; You, M.; Li, J.; Li, Z. Emerging Bone Tissue Engineering via Polyhydroxyalkanoate (PHA)-Based Scaffolds. Mater. Sci. Eng. C 2017, 79, 917–929. [Google Scholar] [CrossRef]
- Costa, S.G.V.A.O.; Lepine, F.; Milot, S.; Deziel, E.; Nitschke, M.; Contiero, J. Cassava Wastewater as a Substrate for the Simultaneous Production of Rhamnolipids and Polyhydroxyalkanoates by Pseudomonas aeruginosa. J. Ind. Microbiol. Biotechnol. 2009, 36, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Kosseva, M.R.; Rusbandi, E. Trends in the Biomanufacture of Polyhydroxyalkanoates with Focus on Downstream Processing. Int. J. Biol. Macromol. 2018, 107, 762–778. [Google Scholar] [CrossRef] [PubMed]
- Komilus, C.F.; Mohamad-Zuki, N.A.; Aminudin, N.H.; Redhwan, A.I.; Mohd-Khairulnizam, N.A.N. Effects of Crab Shell Waste as Feed on Growth Performance and Colouration of Siamese Fighting Fish (Betta Splendens). Malays. Appl. Biol. 2023, 52, 213–220. [Google Scholar] [CrossRef]
- Titz, M.; Kettl, K.H.; Shahzad, K.; Koller, M.; Schnitzer, H.; Narodoslawsky, M. Process Optimization for Efficient Biomediated PHA Production from Animal-Based Waste Streams. Clean Technol. Environ. Policy 2012, 14, 495–503. [Google Scholar] [CrossRef]
- Darken, M.A.; Berenson, H.; Shirk, R.J.; Sjolander, N.O. Production of Tetracycline by Streptomyces aureofaciens in Synthetic Media. Appl. Microbiol. 1960, 8, 46–51. [Google Scholar] [CrossRef]
- Shanmugaprakash, M.; Vinoth Kumar, V.; Hemalatha, M.; Melbia, V.; Karthik, P. Solid-state Fermentation for the Production of Debittering Enzyme Naringinase Using Aspergillus niger MTCC 1344. Eng. Life Sci. 2011, 11, 322–325. [Google Scholar] [CrossRef]
- Huang, B.; Li, D.-G.; Huang, Y.; Liu, C. Effects of Spaceflight and Simulated Microgravity on Microbial Growth and Secondary Metabolism. Mil. Med. Res. 2018, 5, 18. [Google Scholar] [CrossRef]
- Piedade, P.J.; Venkat, V.; Al-Shwafy, K.W.A.; Aregawi, M.A.; Dudek, G.; Zygadło, M.; Lukasik, R.M. Comprehensive Wheat Straw Processing with Deep Eutectic Solvent to Deliver Reducing Sugar. Bioenerg. Res. 2024, 17, 1559–1568. [Google Scholar] [CrossRef]
- Park, Y.; Jeon, J.-M.; Park, J.K.; Yang, Y.-H.; Choi, S.S.; Yoon, J.-J. Optimization of Polyhydroxyalkanoate Production in Halomonas Sp. YLGW01 Using Mixed Volatile Fatty Acids: A Study on Mixture Analysis and Fed-Batch Strategy. Microb. Cell Factories 2023, 22, 171. [Google Scholar] [CrossRef]
- Brown, R.B.; Klaus, D.; Todd, P. Effects of Space Flight, Clinorotation, and Centrifugation on the Substrate Utilization Efficiency of E. Coli. Microgravity Sci. Technol. 2002, 13, 24–29. [Google Scholar] [CrossRef]
- Xun, J.; Zhang, X.; Guo, S.; Lu, H.; Chen, J. Editing out HIV: Application of Gene Editing Technology to Achieve Functional Cure. Retrovirology 2021, 18, 39. [Google Scholar] [CrossRef] [PubMed]
- Hervas, A.B.; Canosa, I.; Little, R.; Dixon, R.; Santero, E. NtrC-Dependent Regulatory Network for Nitrogen Assimilation in Pseudomonas putida. J. Bacteriol. 2009, 191, 6123–6135. [Google Scholar] [CrossRef]
- Traidej, M.; Caballero, A.R.; Marquart, M.E.; Thibodeaux, B.A.; O’Callaghan, R.J. Molecular Analysis of Pseudomonas aeruginosa Protease IV Expressed in Pseudomonas putida. Investig. Ophthalmol. Vis. Sci. 2003, 44, 190–196. [Google Scholar] [CrossRef]
- Thibodeaux, B.A.; Caballero, A.R.; Marquart, M.E.; Tommassen, J.; O’Callaghan, R.J. Corneal Virulence of Pseudomonas aeruginosa Elastase B and Alkaline Protease Produced by Pseudomonas putida. Curr. Eye Res. 2007, 32, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Tan, I.K.P.; Foong, C.P.; Tan, H.T.; Lim, H.; Zain, N.-A.A.; Tan, Y.C.; Hoh, C.C.; Sudesh, K. Polyhydroxyalkanoate (PHA) Synthase Genes and PHA-Associated Gene Clusters in Pseudomonas Spp. and Janthinobacterium spp. Isolated from Antarctica. J. Biotechnol. 2020, 313, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual. Anal. Anal. Biochem. 2001, 186, 182–183. [Google Scholar] [CrossRef]
- Cao, L.; Lin, L.; Sui, H.; Wang, H.; Zhang, Z.; Jiao, N.; Zhou, J. Efficient Extracellular Laccase Secretion via Bio-Designed Secretory Apparatuses to Enhance Bacterial Utilization of Recalcitrant Lignin. Green Chem. 2021, 23, 2079–2094. [Google Scholar] [CrossRef]
- Lin, L.; Cheng, Y.; Pu, Y.; Sun, S.; Li, X.; Jin, M.; Pierson, E.A.; Gross, D.C.; Dale, B.E.; Dai, S.Y.; et al. Systems Biology-Guided Biodesign of Consolidated Lignin Conversion. Green Chem. 2016, 18, 5536–5547. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, X.; Yu, W.; Fu, Y.; Ni, L.; Yu, J.; Wang, X.; Song, W.; Wang, C. Enhancing Pseudomonas Cell Growth for the Production of Medium-Chain-Length Polyhydroxyalkanoates from Antarctic Krill Shell Waste. Int. J. Biol. Macromol. 2024, 277, 133364. [Google Scholar] [CrossRef]

| Experiment Number | A | B | C | D | |
|---|---|---|---|---|---|
| Temp (°C) | Time (h) | Solid/Liquid Ratio (%) | Rotation Speed (rpm) | CFU (108/mL) | |
| 1 | 1 (27 °C) | 1 (30 h) | 1 (3%) | 1 (150 rpm) | 176 |
| 2 | 1 | 2 (36 h) | 2 (5%) | 2 (200 rpm) | 301 |
| 3 | 1 | 3 (42 h) | 3 (7%) | 3 (250 rpm) | 337 |
| 4 | 2 (30 °C) | 1 | 2 | 3 | 386 |
| 5 | 2 | 2 | 3 | 1 | 399 |
| 6 | 2 | 3 | 1 | 2 | 299 |
| 7 | 3 (33 °C) | 1 | 3 | 2 | 318 |
| 8 | 3 | 2 | 1 | 3 | 187 |
| 9 | 3 | 3 | 2 | 1 | 261 |
| K1 | 814 | 880 | 662 | 836 | |
| K2 | 1084 | 887 | 948 | 918 | |
| K3 | 766 | 897 | 1054 | 910 | |
| k1 | 271.33 | 293.33 | 220.67 | 278.67 | |
| k2 | 361.33 | 295.67 | 316.00 | 306.00 | |
| k3 | 255.33 | 299.00 | 351.33 | 303.33 | |
| R | 106.00 | 5.67 | 130.67 | 27.33 | |
| p-value | p < 0.05 | p > 0.05 | p < 0.01 | p > 0.05 | |
| Priority of factors | C > A > D > B | ||||
| Optimal composition | A2B3C3D2 | ||||
| Monomer | PHA Composition (mol%) | |||||
|---|---|---|---|---|---|---|
| WT | Control | KT+lasBT | KT+IV | KT+NtrcT-D55E | p-Value | |
| 3HHx (C6) | 1.15 ± 0.05 | 1.64 ± 0.35 | 2.58 ± 0.90 | 1.34 ± 0.04 | 2.79 ± 0.02 | 0.077 |
| 3HO (C8) | 21.98 ± 0.18 c | 21.67 ± 0.27 c | 21.61 ± 0.19 c | 23.09 ± 0.16 b | 24.63 ± 0.08 a | 0.000 |
| 3HD (C10) | 40.18 ± 0.08 b | 39.93 ± 0.16 b | 39.55 ± 0.36 b | 39.92 ± 0.04 b | 44.75 ± 0.75 a | 0.000 |
| 3HDD (C12) | 36.69 ± 0.21 a | 36.76 ± 0.09 a | 36.26 ± 0.36 a | 35.66 ± 0.16 a | 27.83 ± 0.81 b | 0.000 |
| Plasmid Vectors/Strains | Plasmid/Strain Characteristics | Source |
|---|---|---|
| Plasmid vector | ||
| pUCP18-Gm | Ampr; Gmr, shuttle vector; lacZ with MCS | Cao et al. [39] |
| pPR-NtrcT-D55E | pUCP18-Gm derivative, Pmin:: NtrcT-D55E | This study |
| pPR-IV | pUCP18-Gm derivative, Pmin:: IV | This study |
| pPR-lasBT | pUCP18-Gm derivative, Pmin:: lasBT | This study |
| Strains | ||
| P. putida KT2440 | Wild type | Lab stock |
| KTG (control strain) | KT2440 carrying pUCP18-Gm | This study |
| KT+NtrcT-D55E | KT2440 carrying pPR-NtrcT-D55E | This study |
| KT+IV | KT2440 carrying pPR-IV | This study |
| KT+lasBT | KT2440 carrying pPR-lasBT | This study |
| Primer Name | Sequence (5′-3′) |
|---|---|
| D55E F61 | tggtaaagagctcatgagccgaagtg |
| D55E R61 | cacctcagtggtcatcaccttcctc |
| IV F | cggaattccatgcataagagaacgtacctgaat |
| IV R | ggatcctcagggcgcgaagtagcgggagat |
| lasBT F | ccgcggatccaaataaaacgaaaggctcagtcg |
| lasBT R | cccggaattcaaaaggccatccgtcaggat |
| Level | A | B | C | D |
|---|---|---|---|---|
| Temp (°C) | Time (h) | Solid/Liquid Ratio (%) | Rotation Speed (rpm) | |
| 1 | 27 | 30 | 3 | 150 |
| 2 | 30 | 36 | 5 | 200 |
| 3 | 33 | 42 | 7 | 250 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, X.; Wei, H.; Zhou, Y.; Song, W.; Shi, C.; Mu, C.; Wang, C.; Wang, X. Utilization of Crab Shell Waste for Value-Added Bioplastics by Pseudomonas-Based Microbial Cell Factories. Int. J. Mol. Sci. 2025, 26, 2543. https://doi.org/10.3390/ijms26062543
Song X, Wei H, Zhou Y, Song W, Shi C, Mu C, Wang C, Wang X. Utilization of Crab Shell Waste for Value-Added Bioplastics by Pseudomonas-Based Microbial Cell Factories. International Journal of Molecular Sciences. 2025; 26(6):2543. https://doi.org/10.3390/ijms26062543
Chicago/Turabian StyleSong, Xiaofen, Hansheng Wei, Yueyue Zhou, Weiwei Song, Ce Shi, Changkao Mu, Chunlin Wang, and Xiaopeng Wang. 2025. "Utilization of Crab Shell Waste for Value-Added Bioplastics by Pseudomonas-Based Microbial Cell Factories" International Journal of Molecular Sciences 26, no. 6: 2543. https://doi.org/10.3390/ijms26062543
APA StyleSong, X., Wei, H., Zhou, Y., Song, W., Shi, C., Mu, C., Wang, C., & Wang, X. (2025). Utilization of Crab Shell Waste for Value-Added Bioplastics by Pseudomonas-Based Microbial Cell Factories. International Journal of Molecular Sciences, 26(6), 2543. https://doi.org/10.3390/ijms26062543






