Multi-Omics Association Analysis of DOF Transcription Factors Involved in the Drought Resistance of Wheat Induced by Strigolactone
Abstract
1. Introduction
2. Results
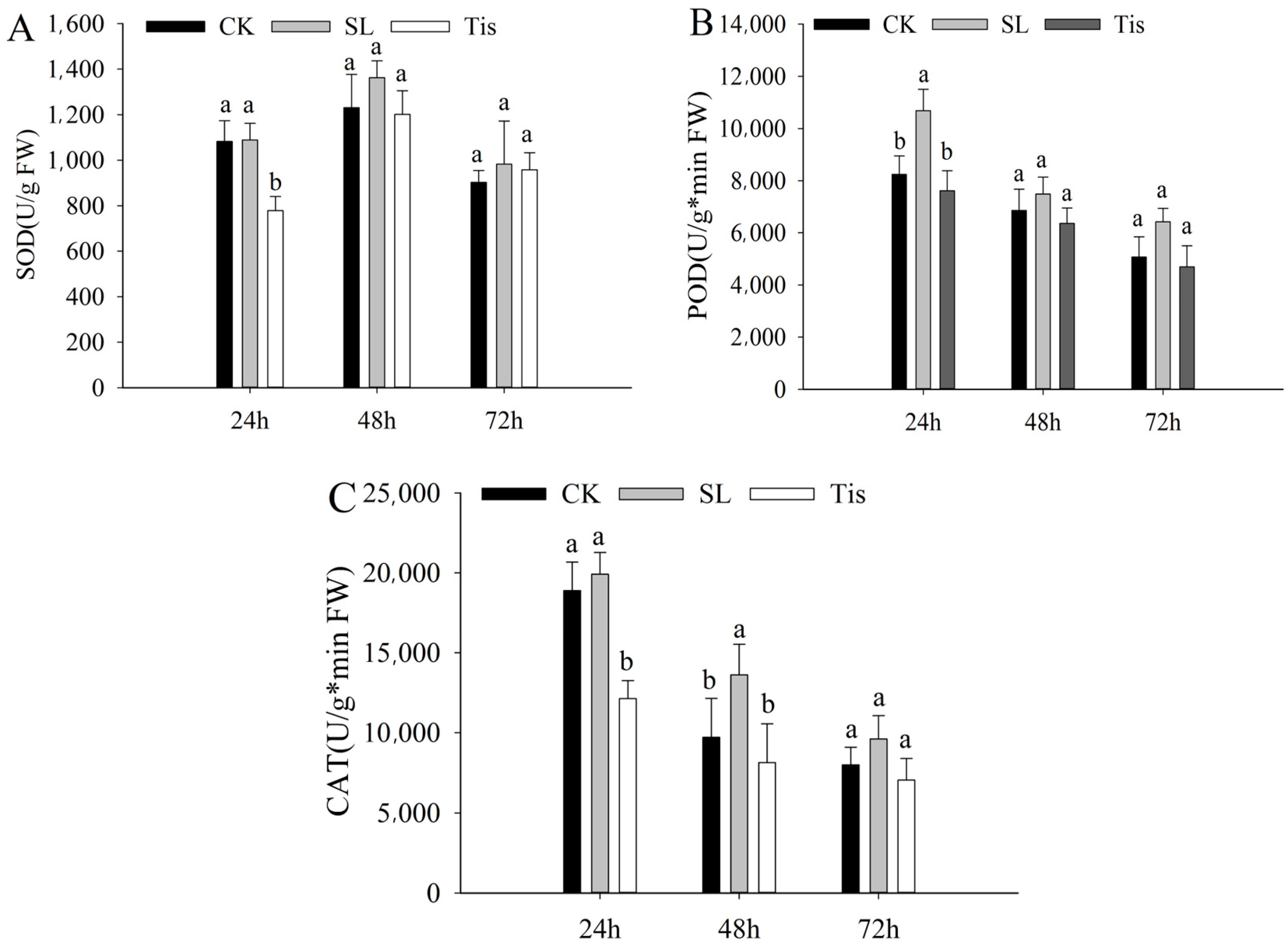
2.1. Morphological and Physiological Changes
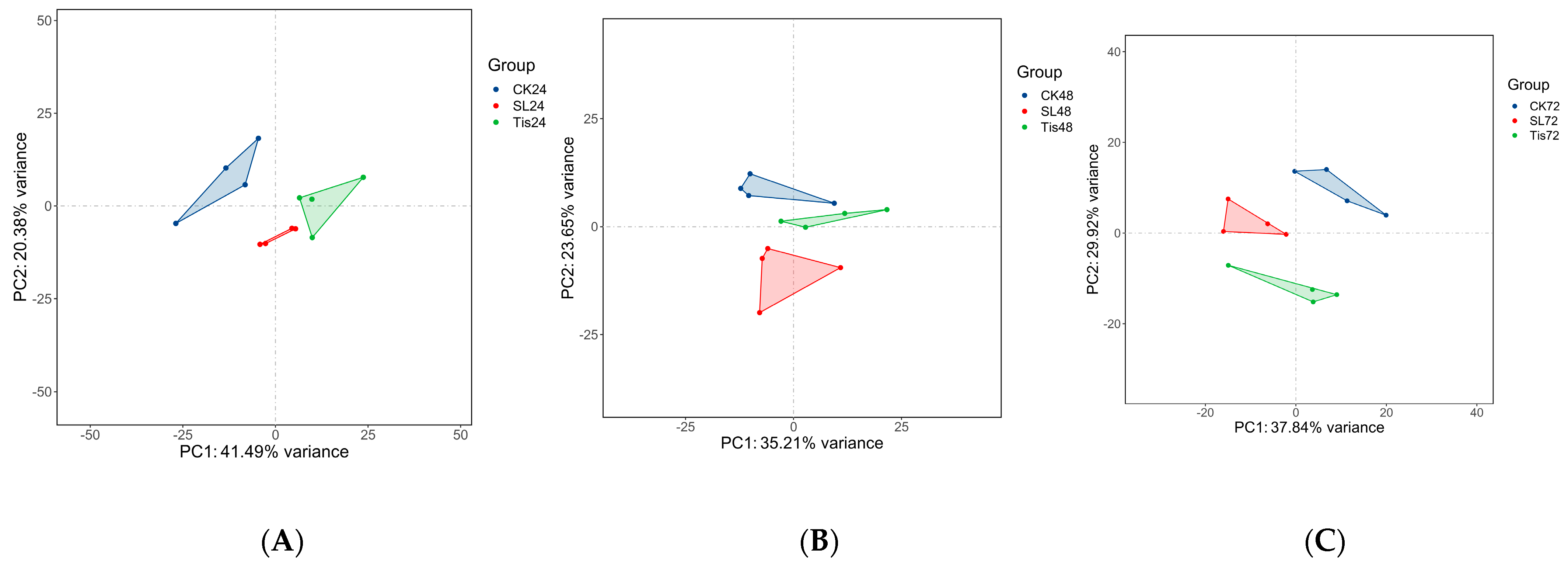
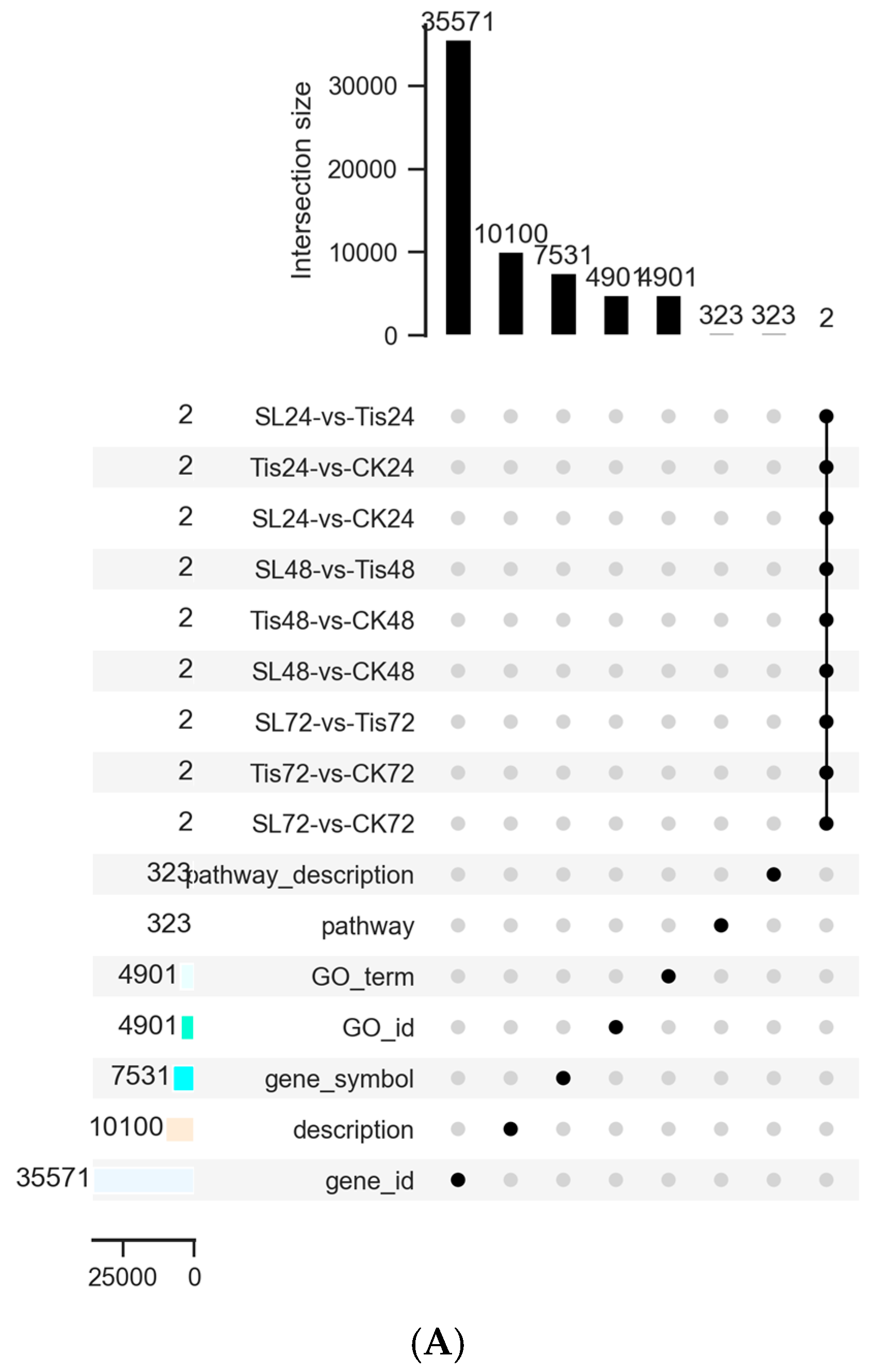
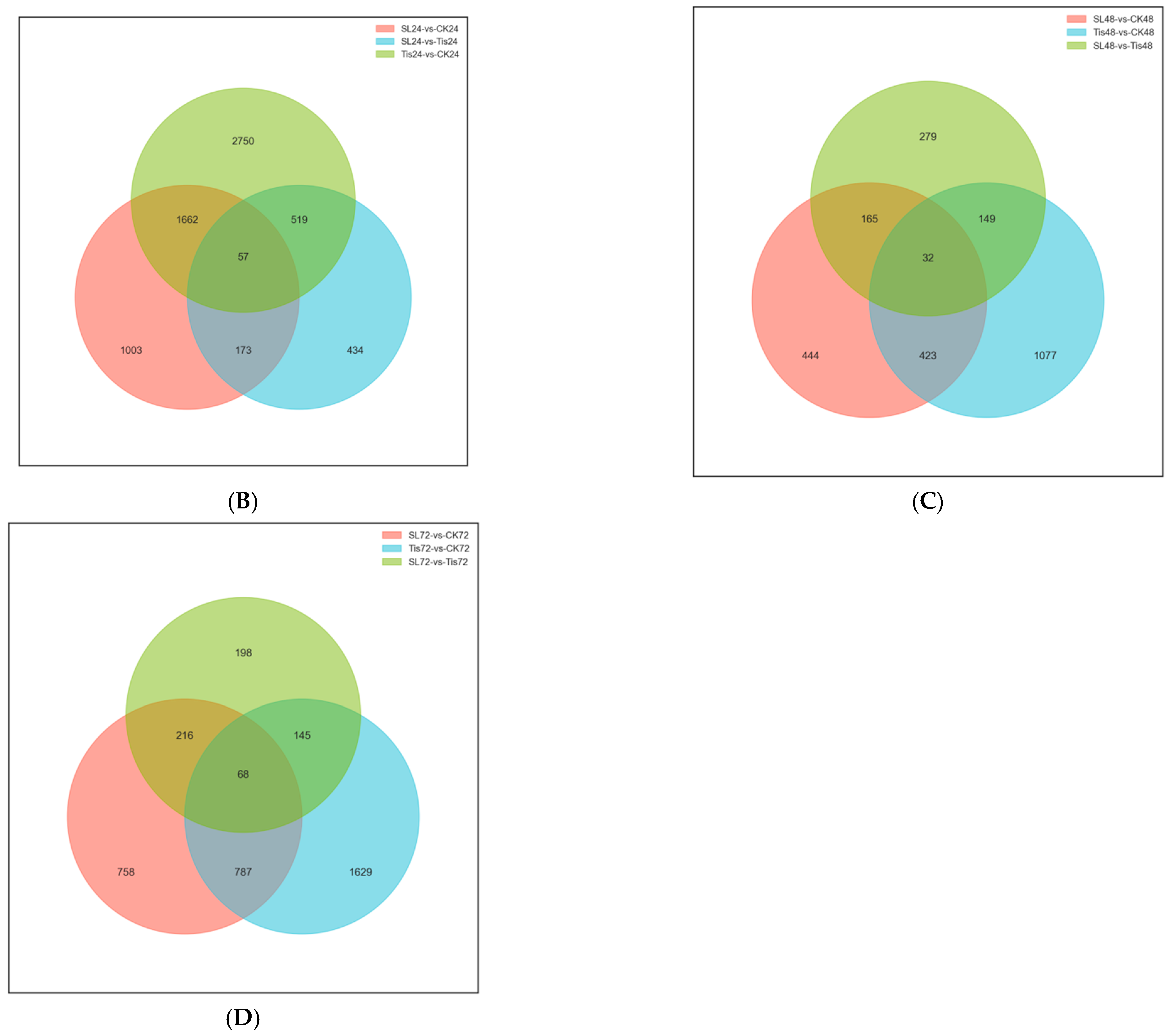
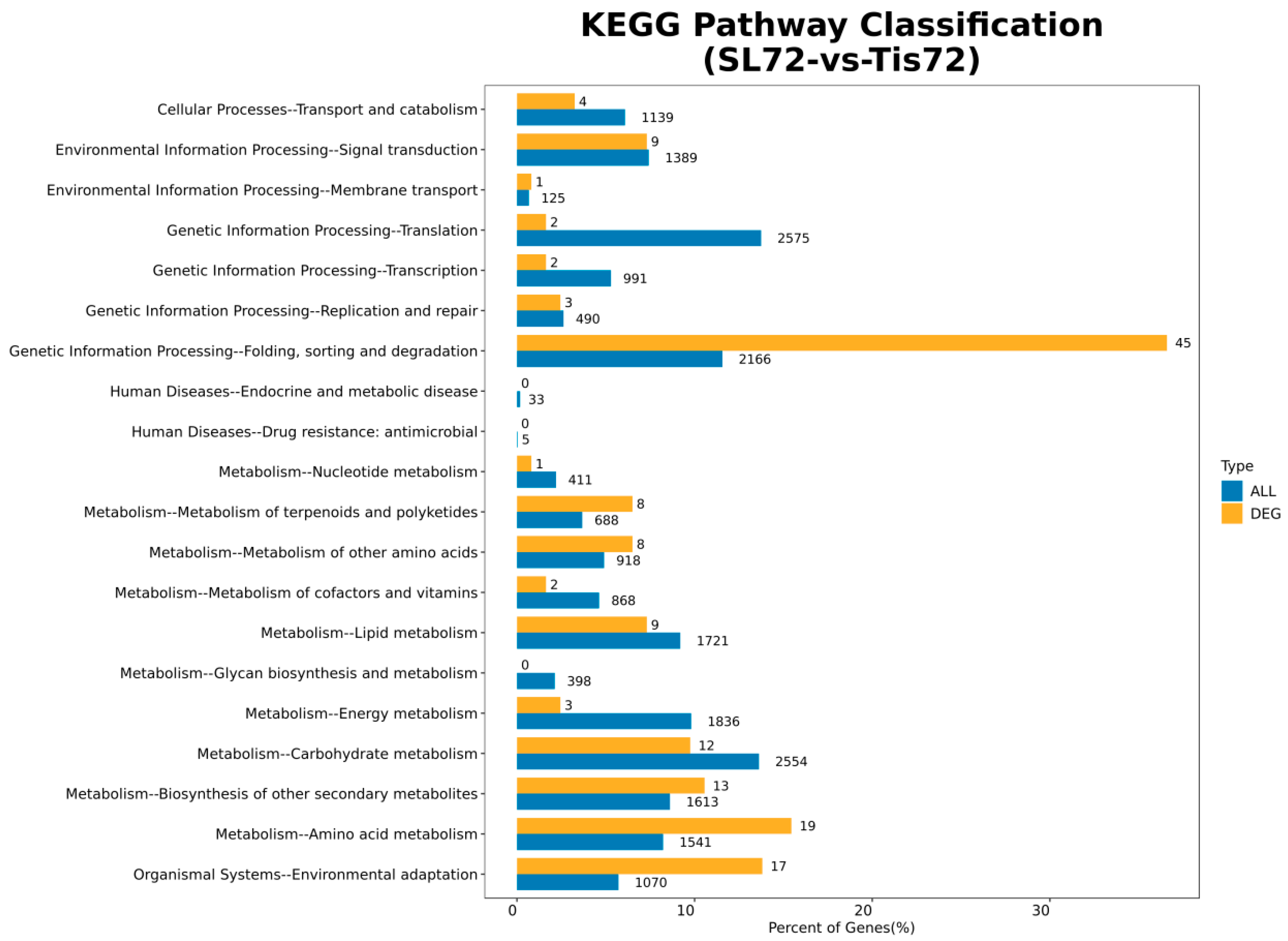
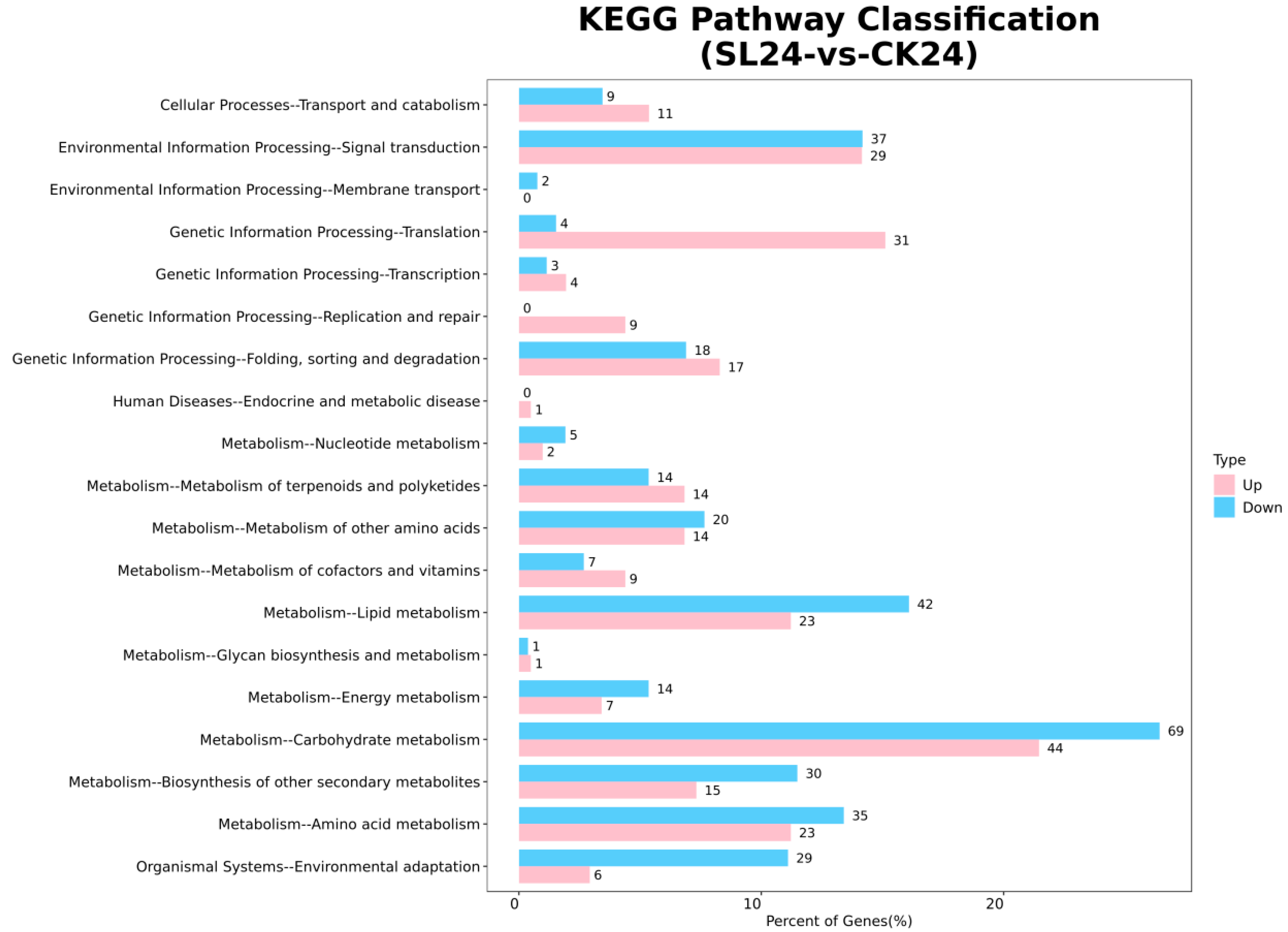
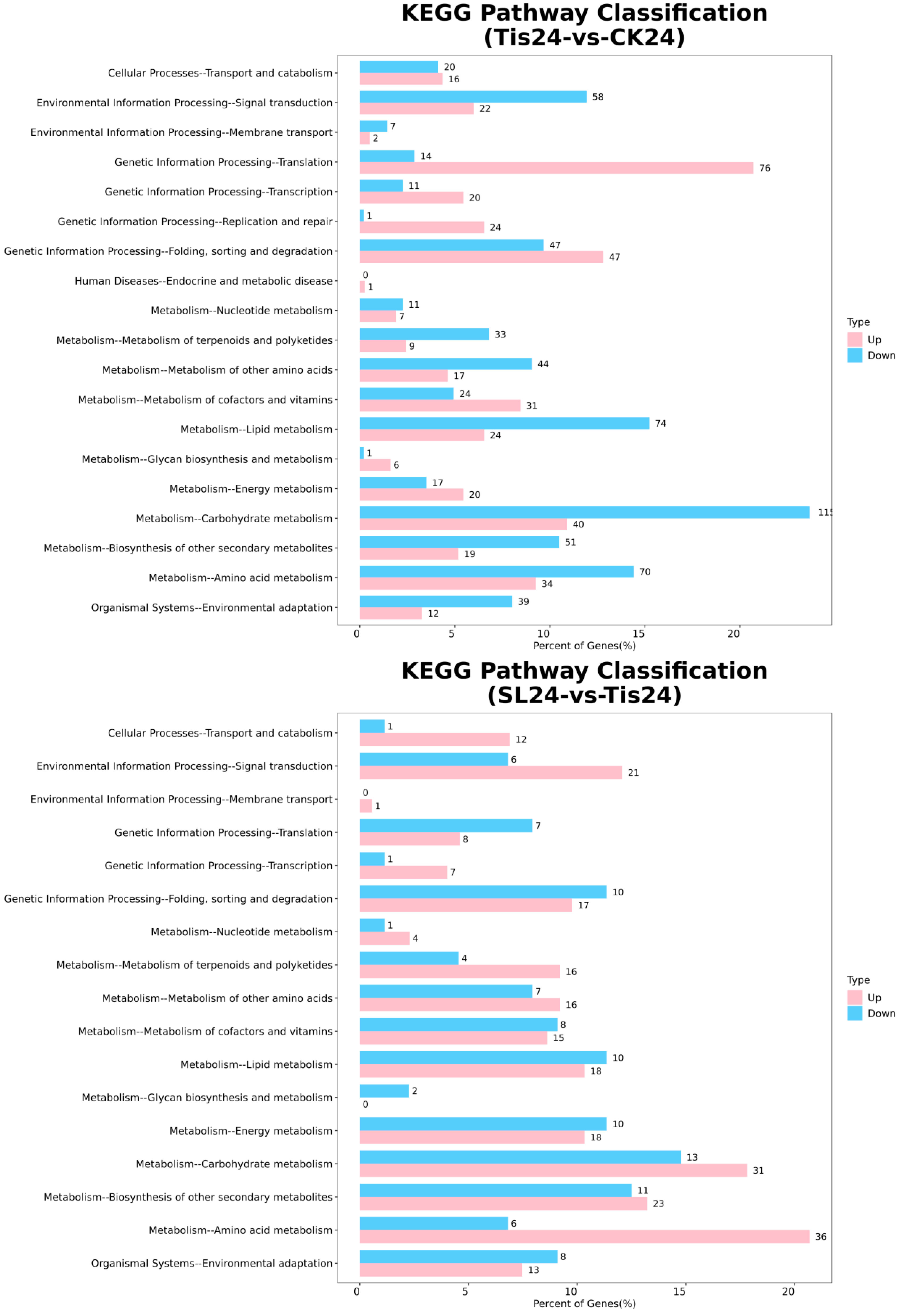
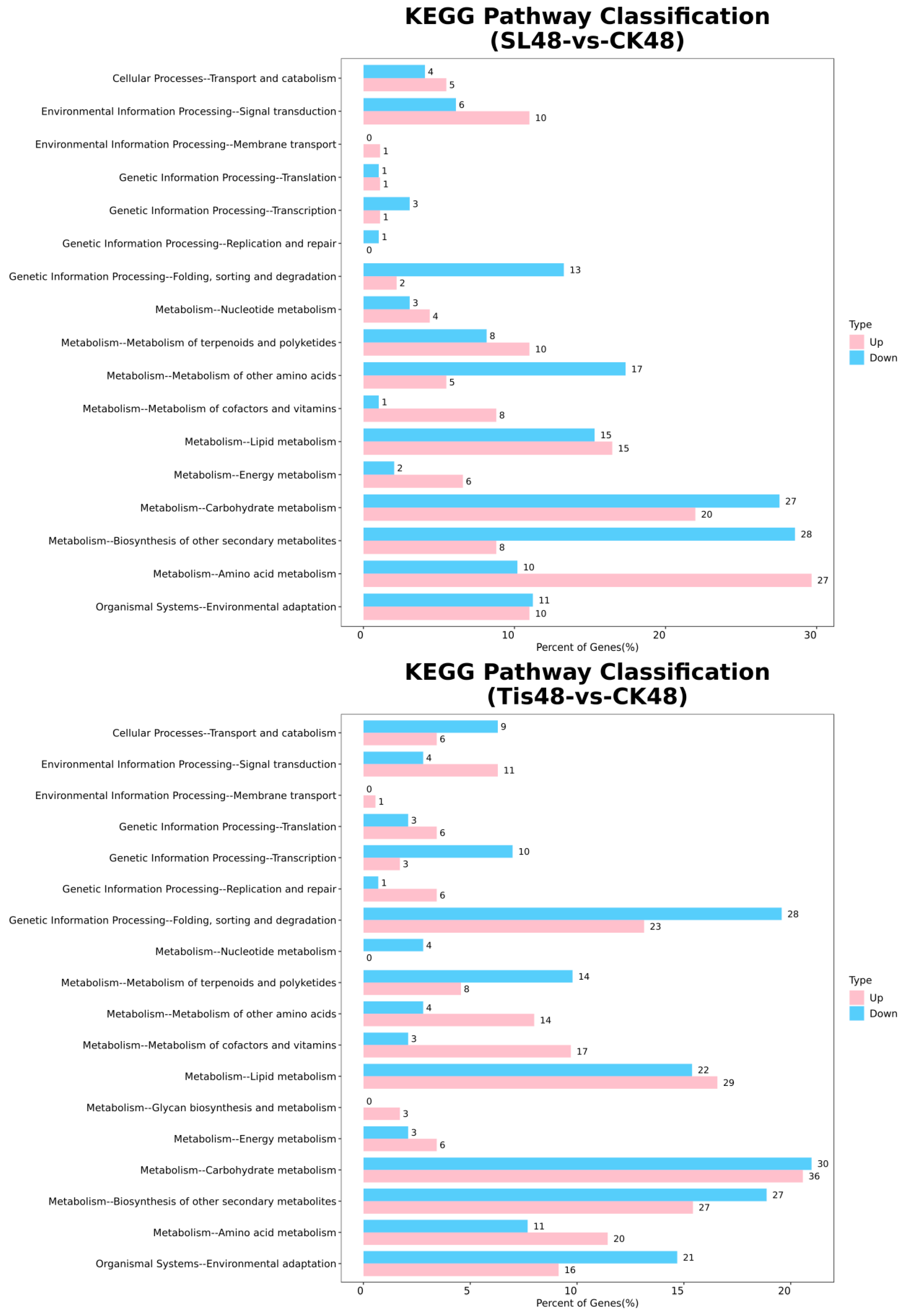
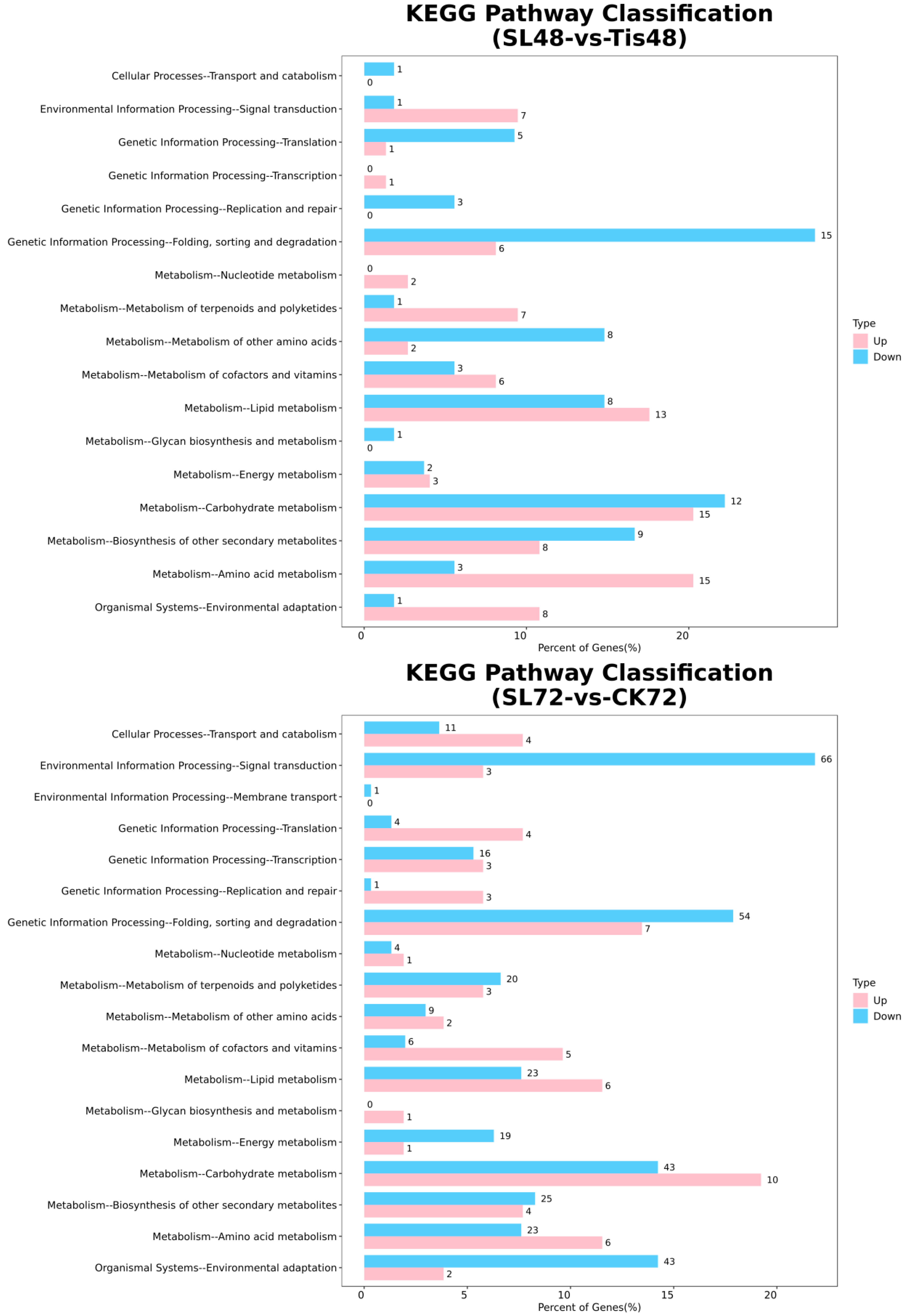
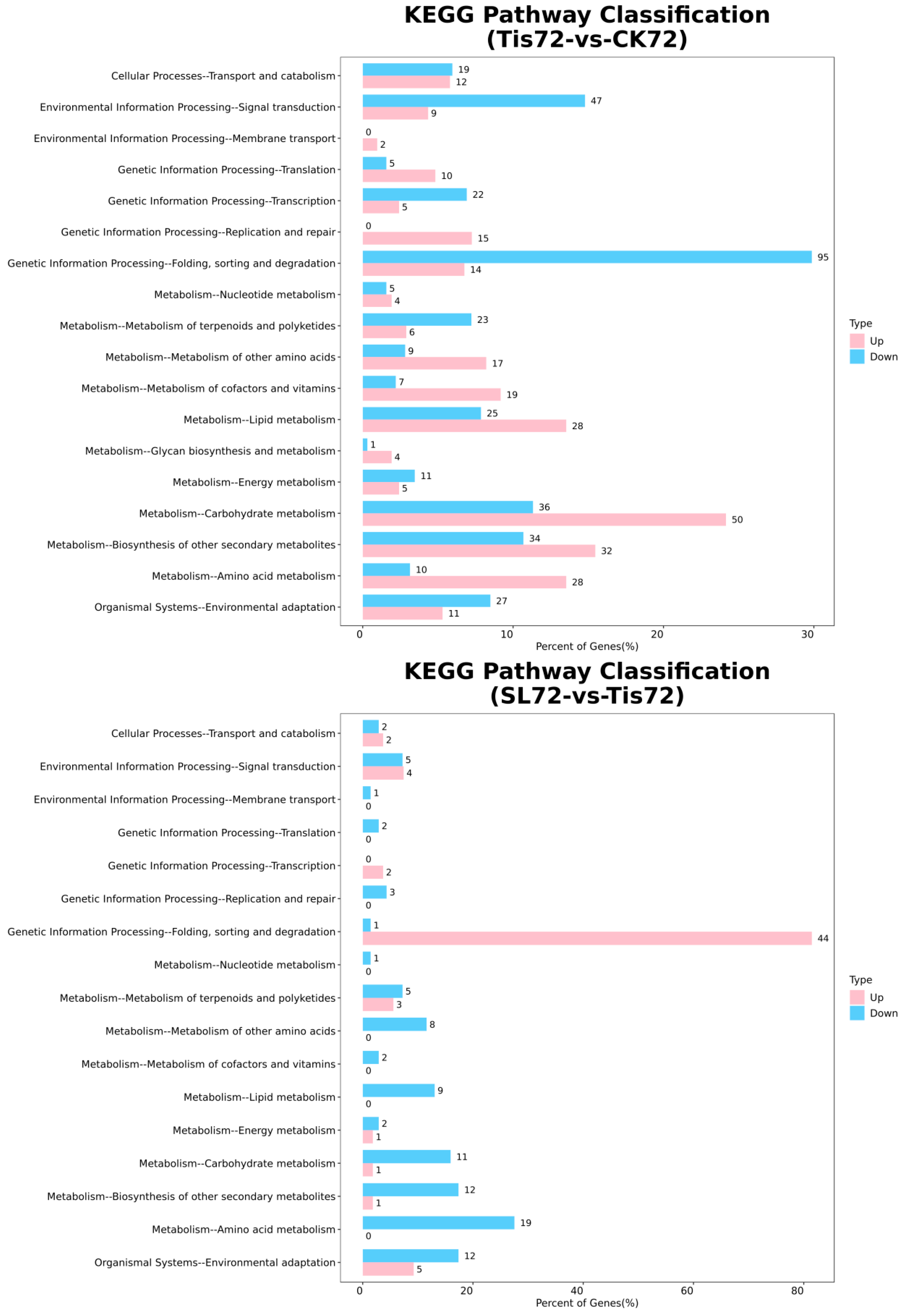
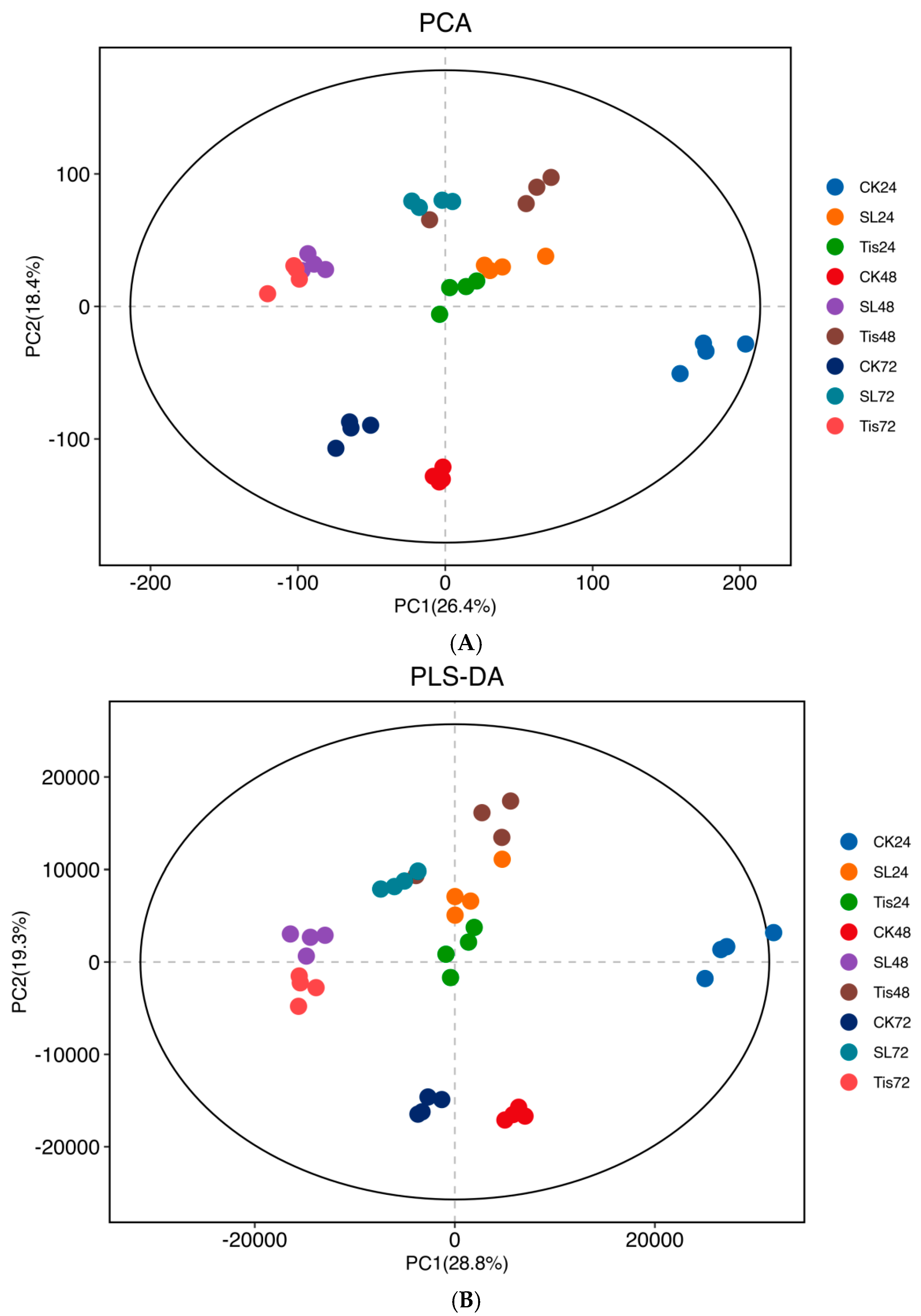
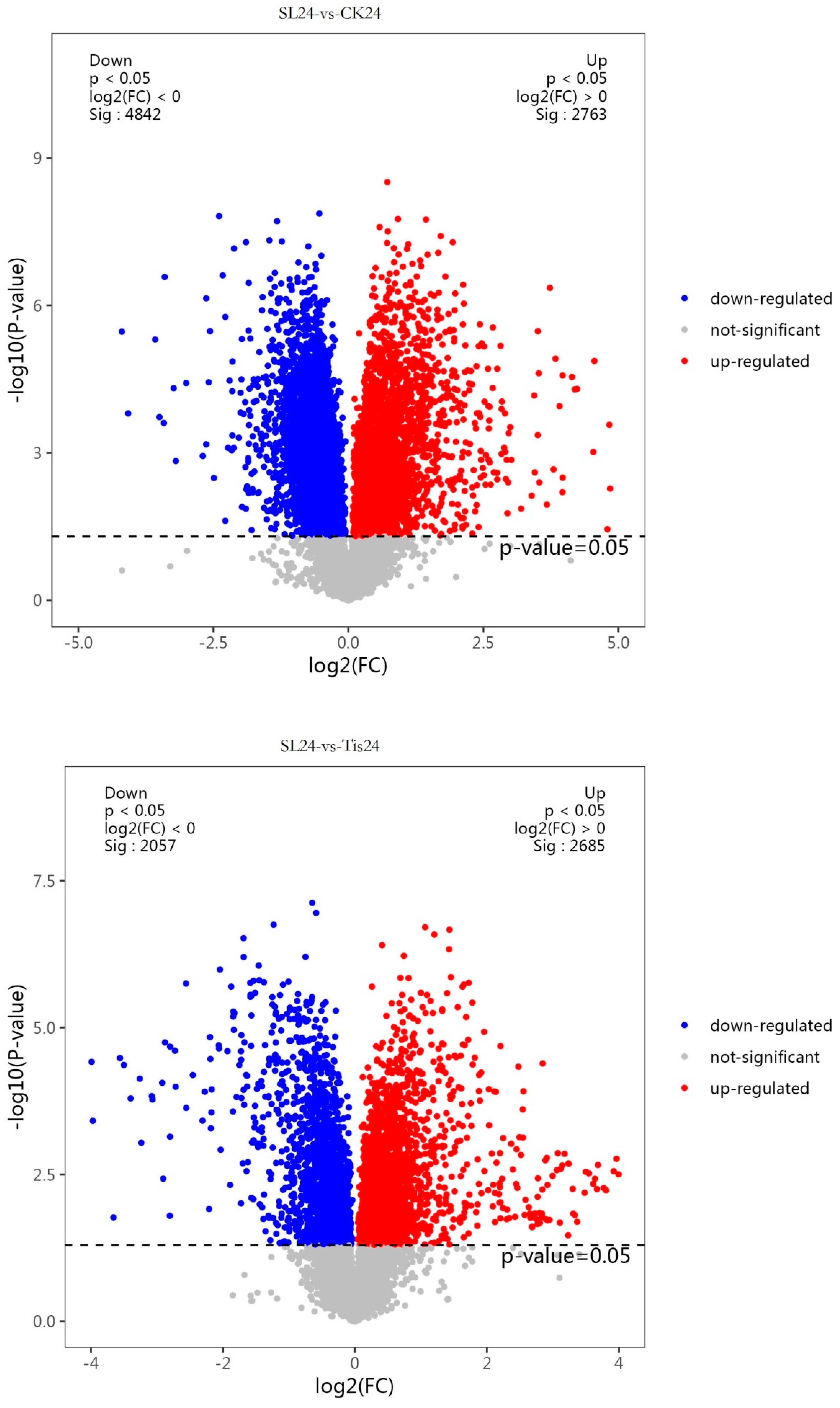
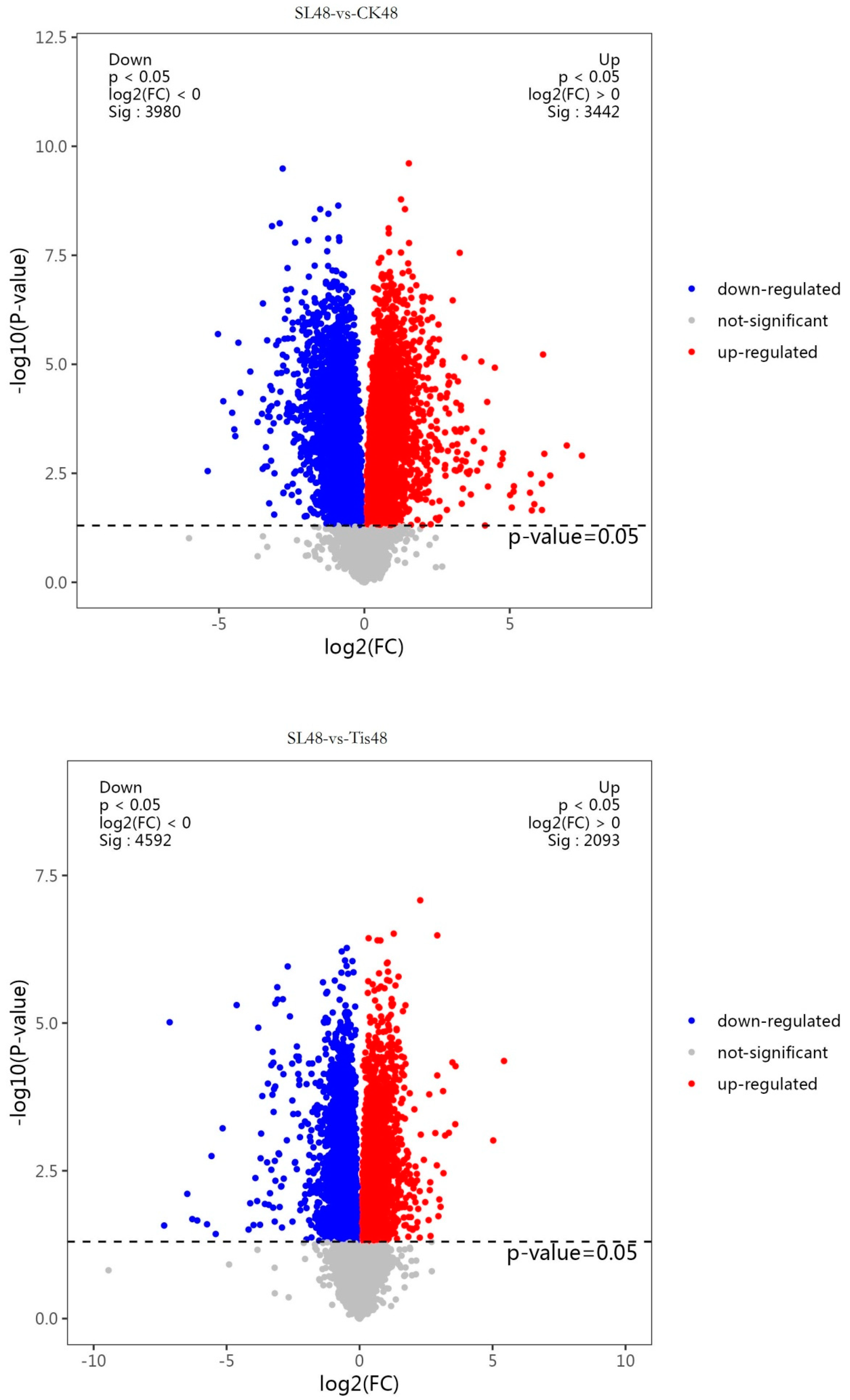
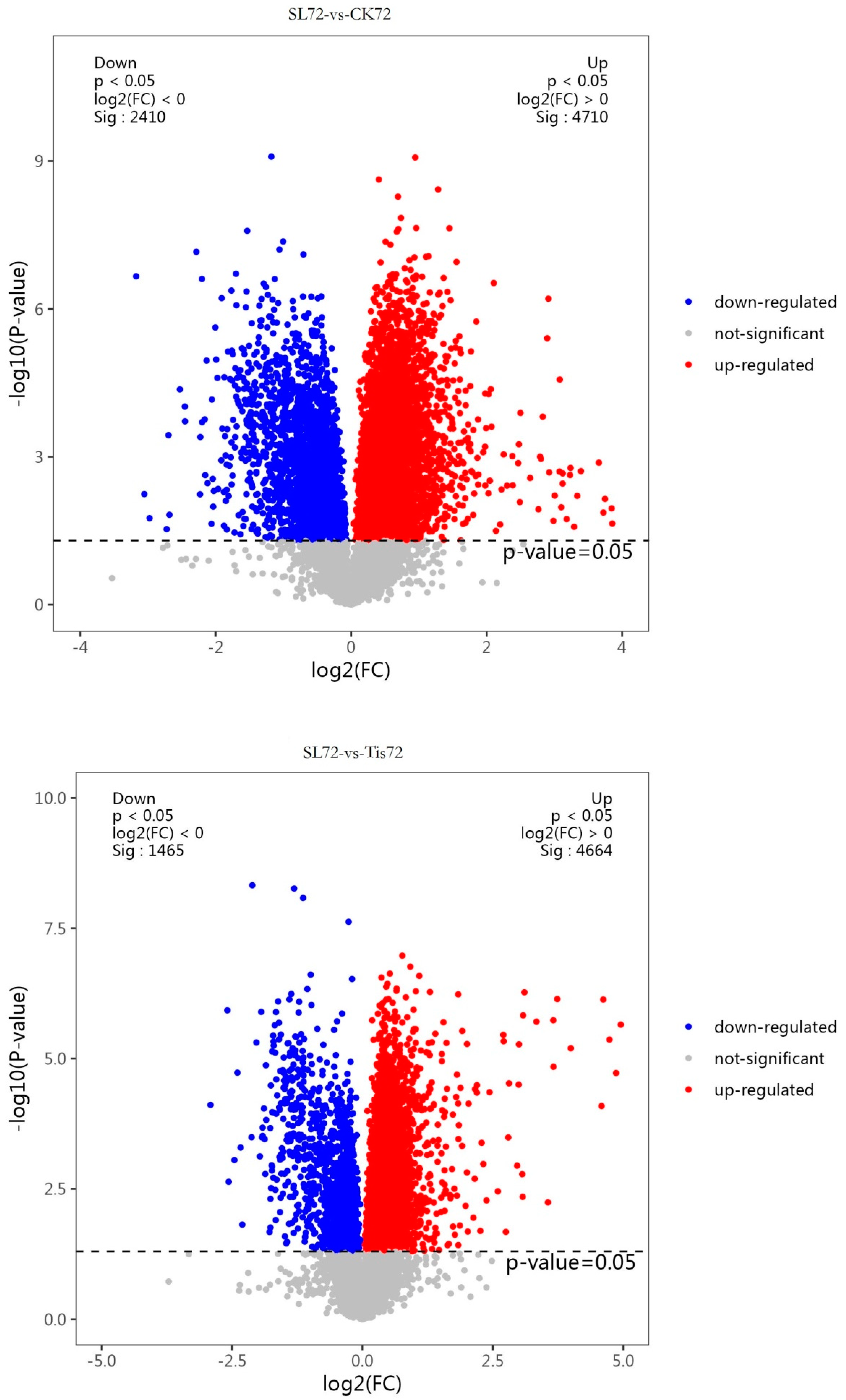
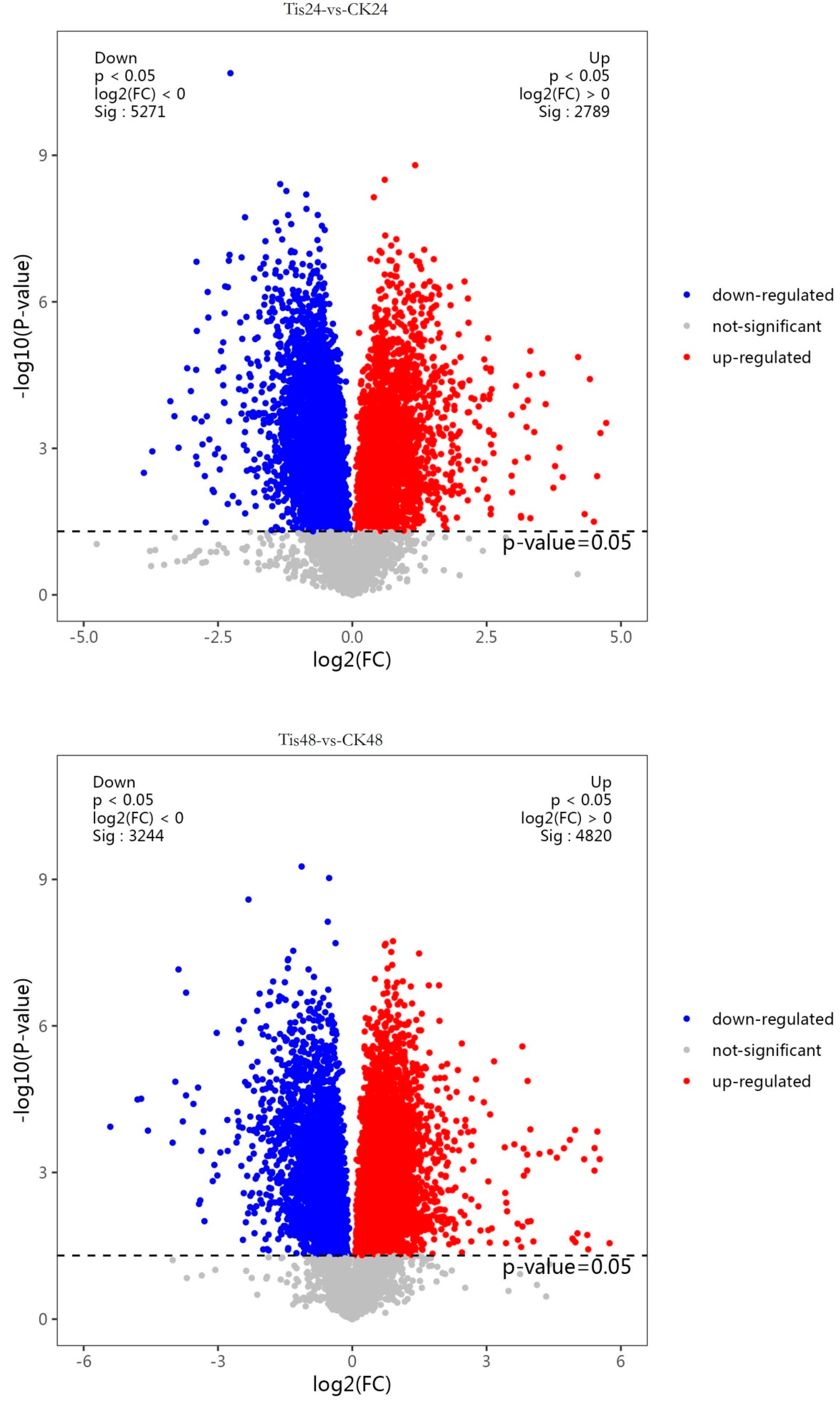
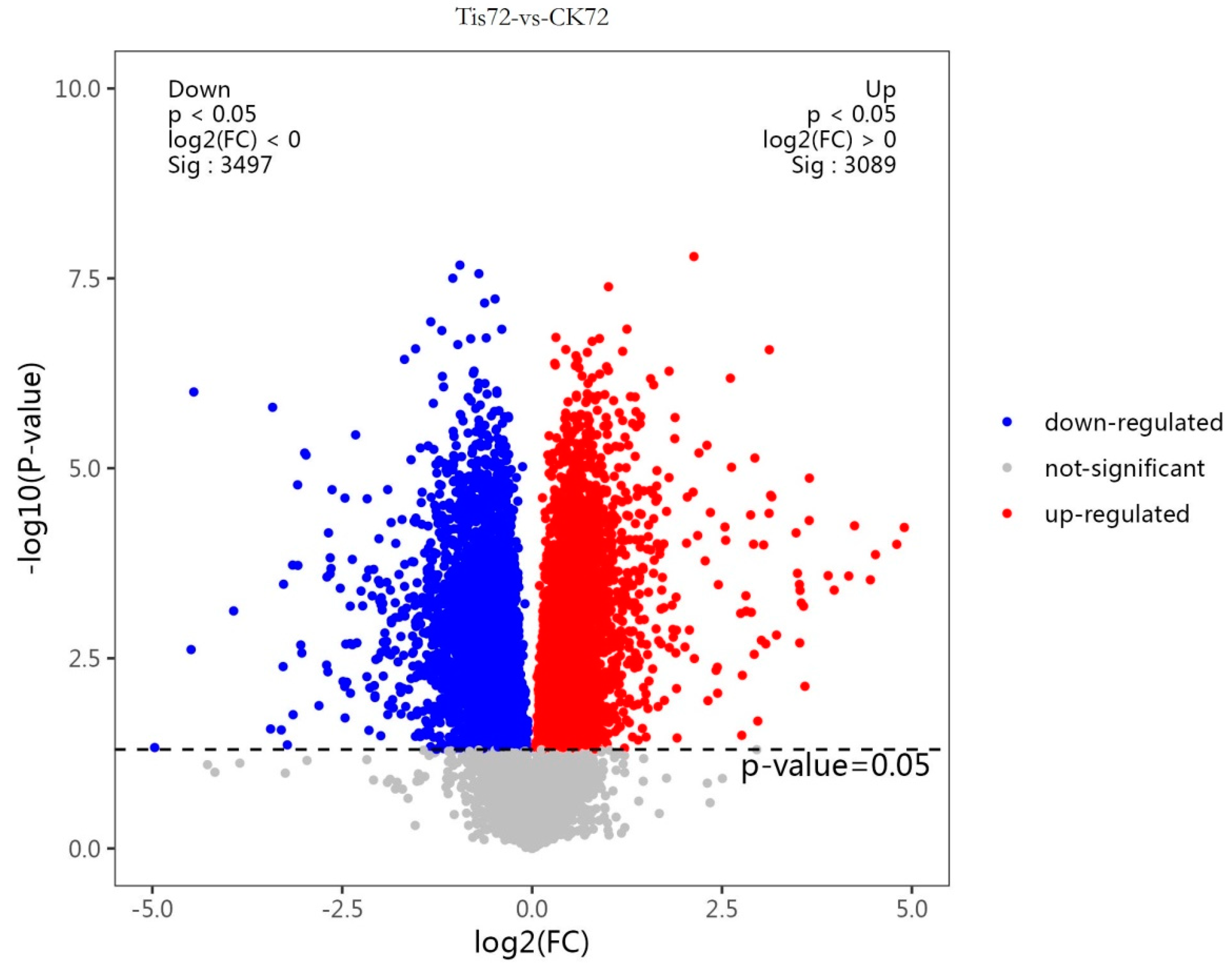
2.2. Transcriptome Analysis and Transcription Factor Screening and Localization
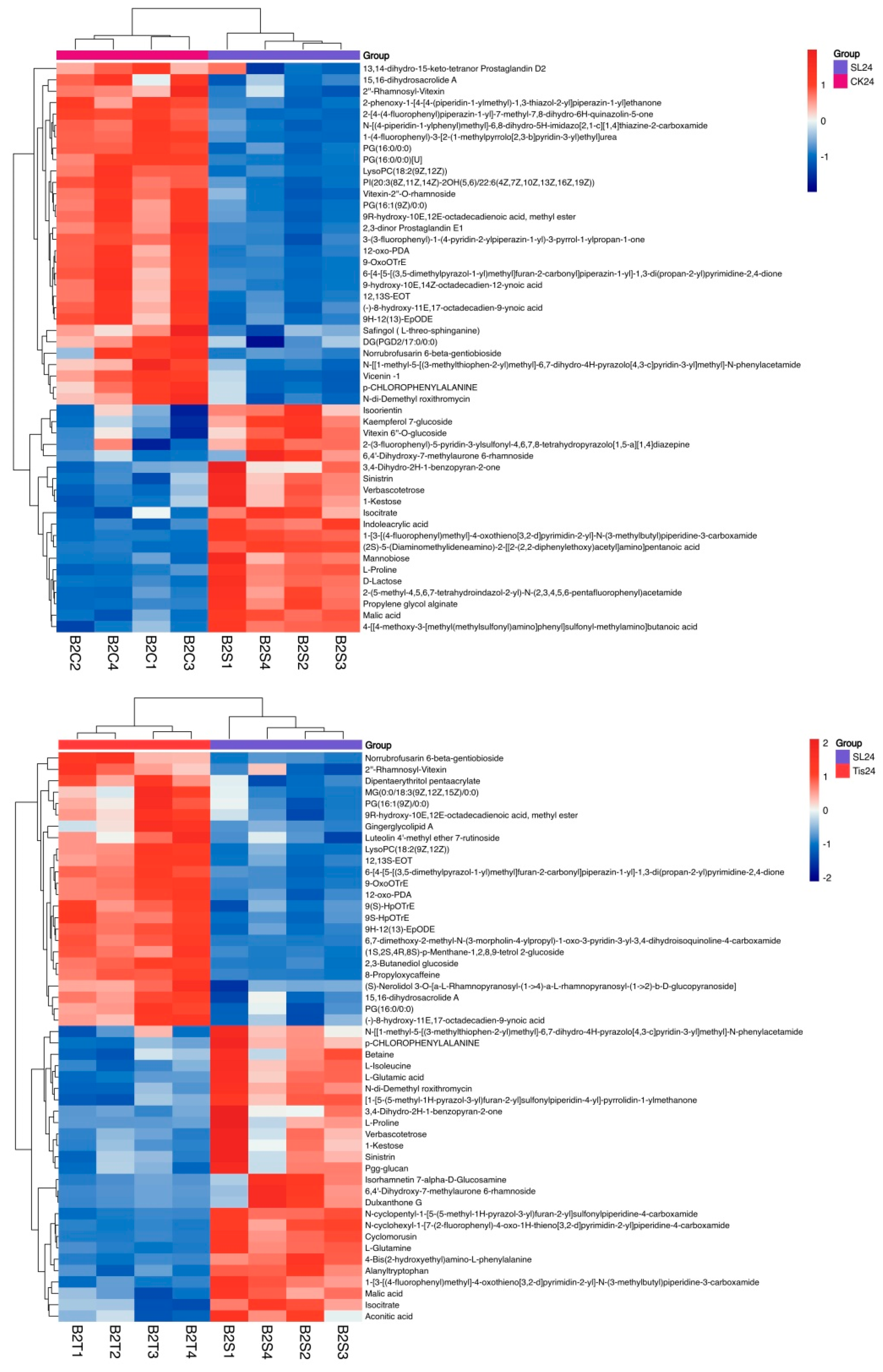
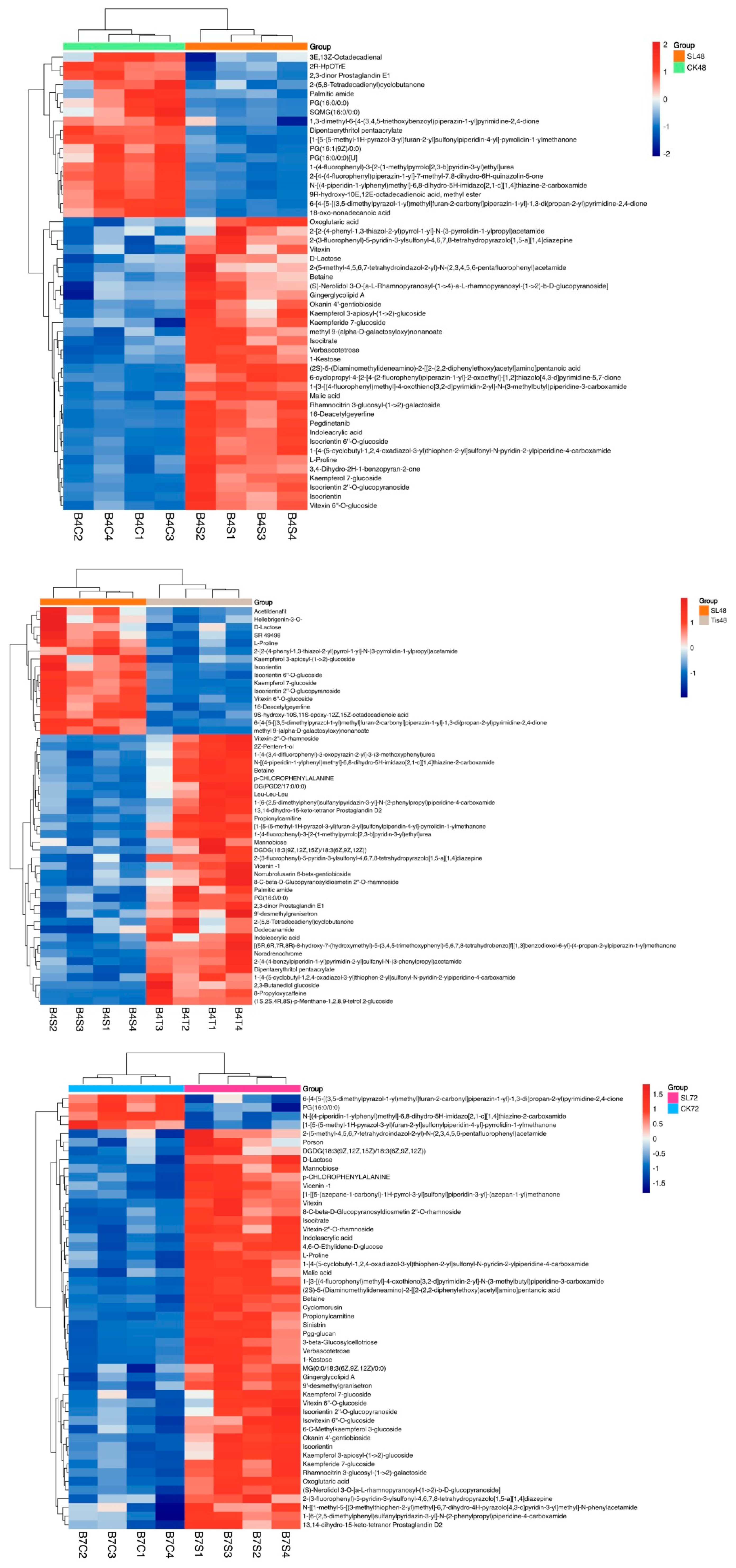
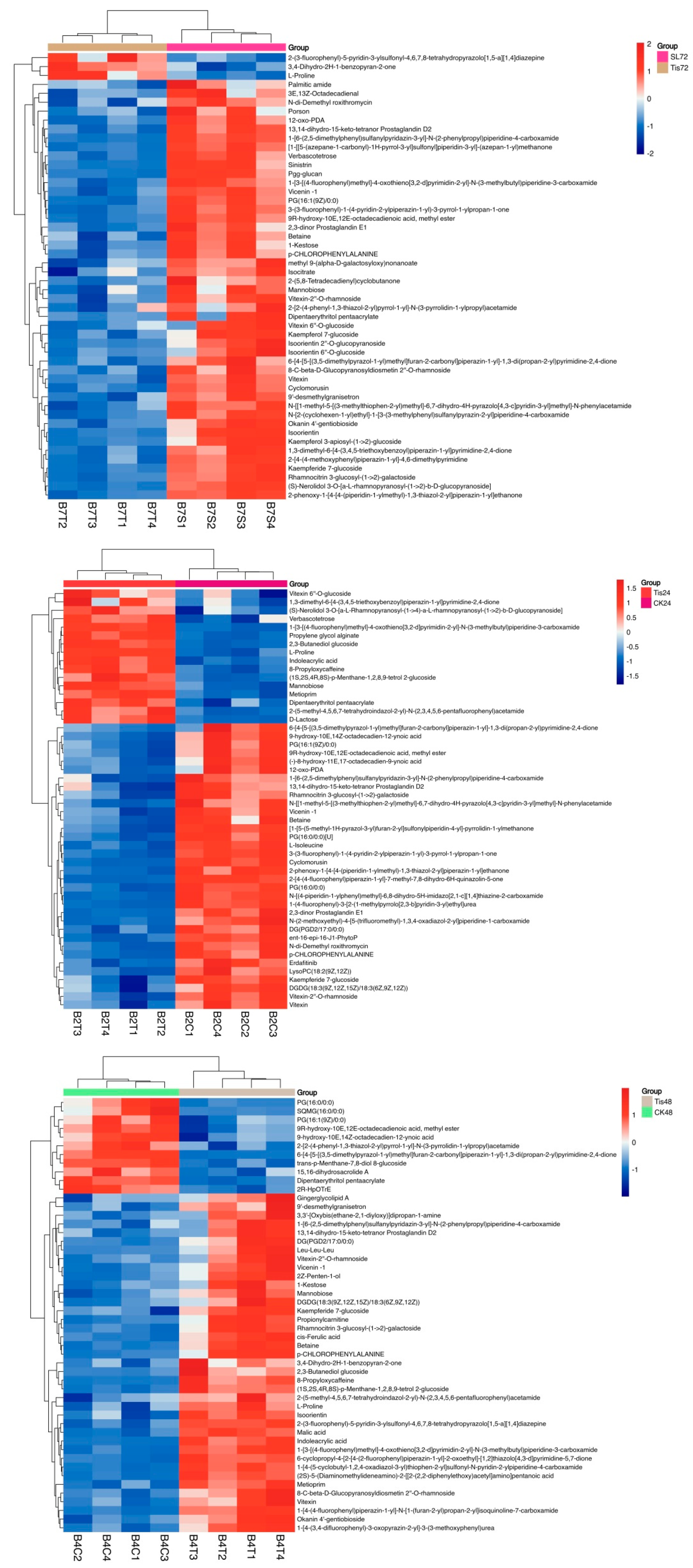
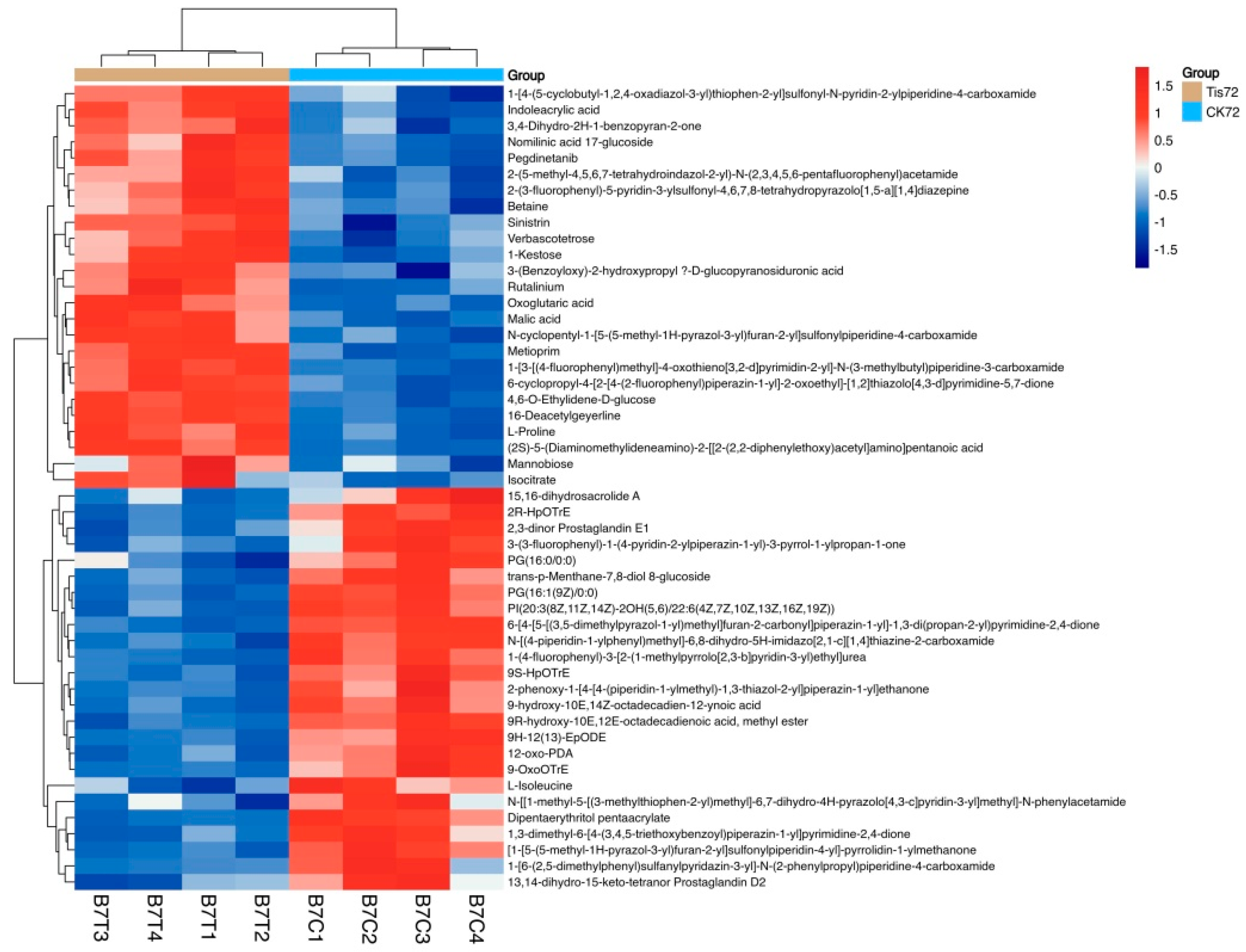
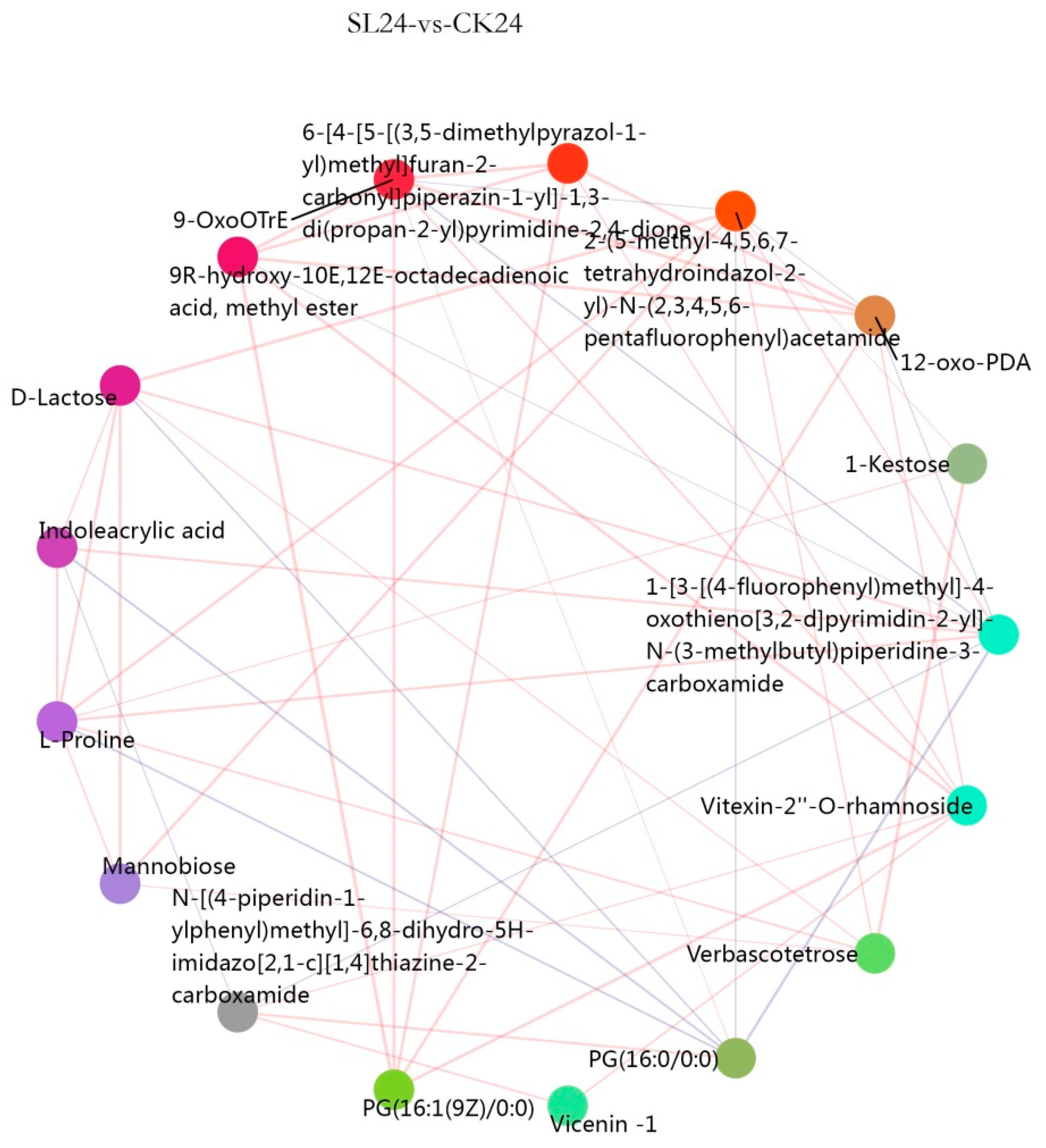
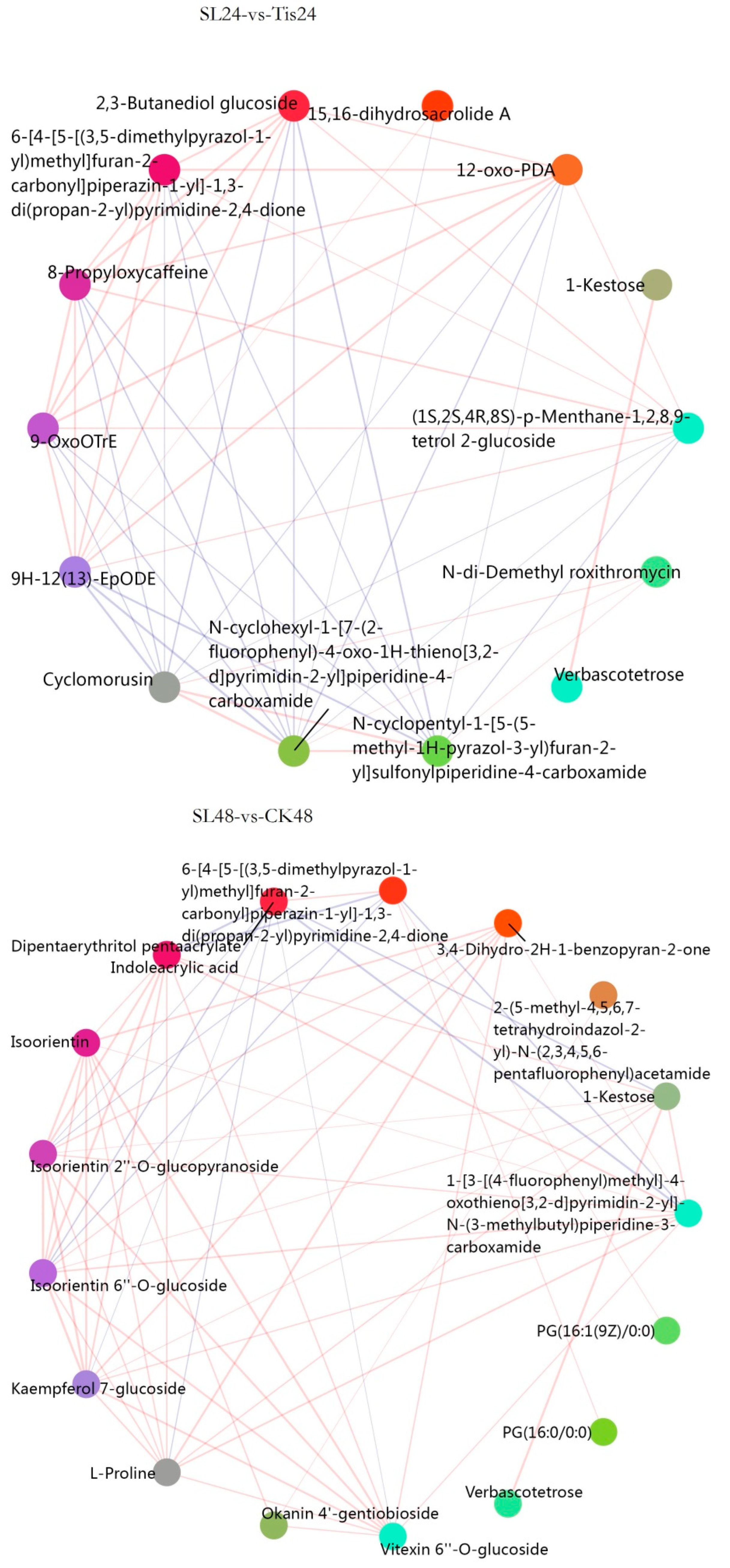
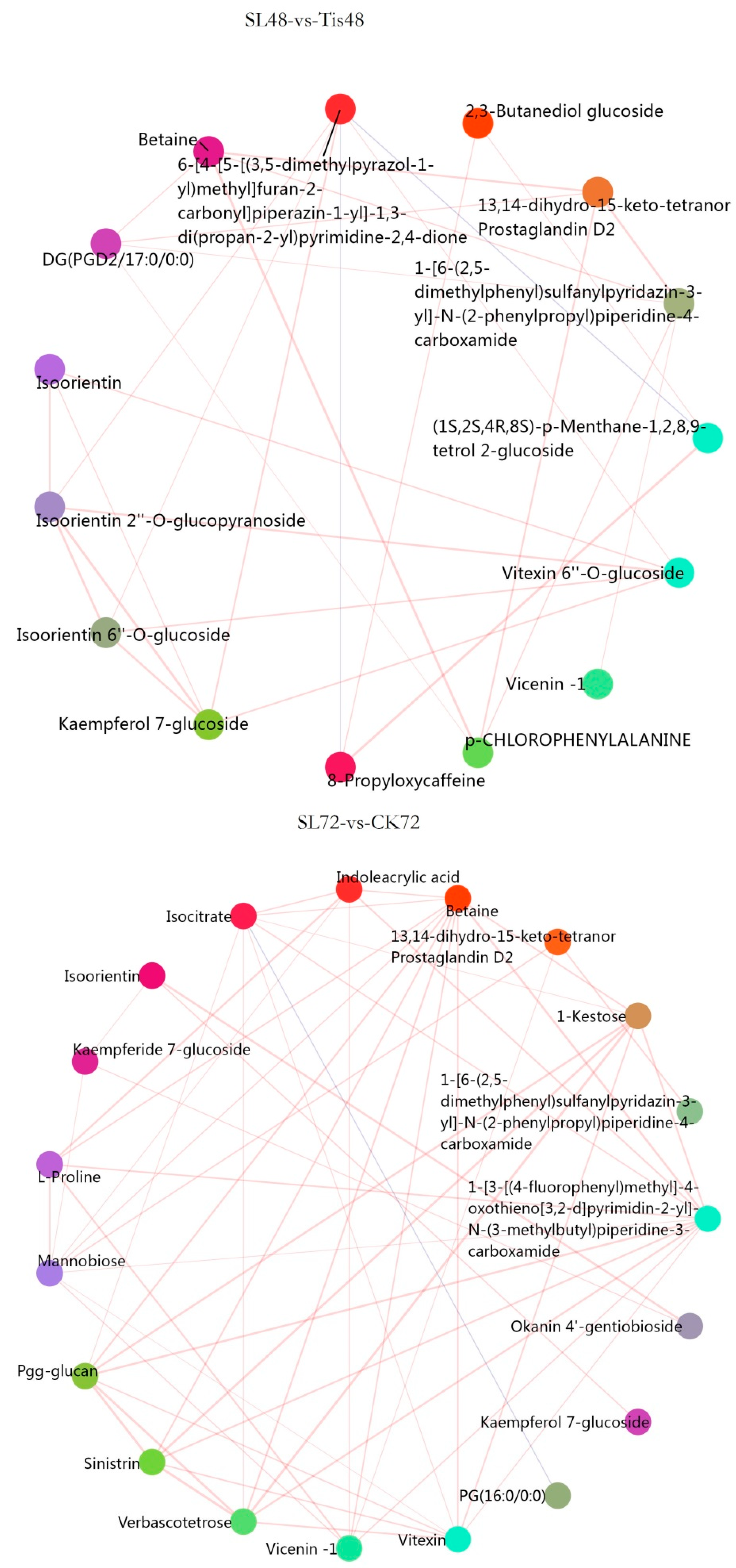
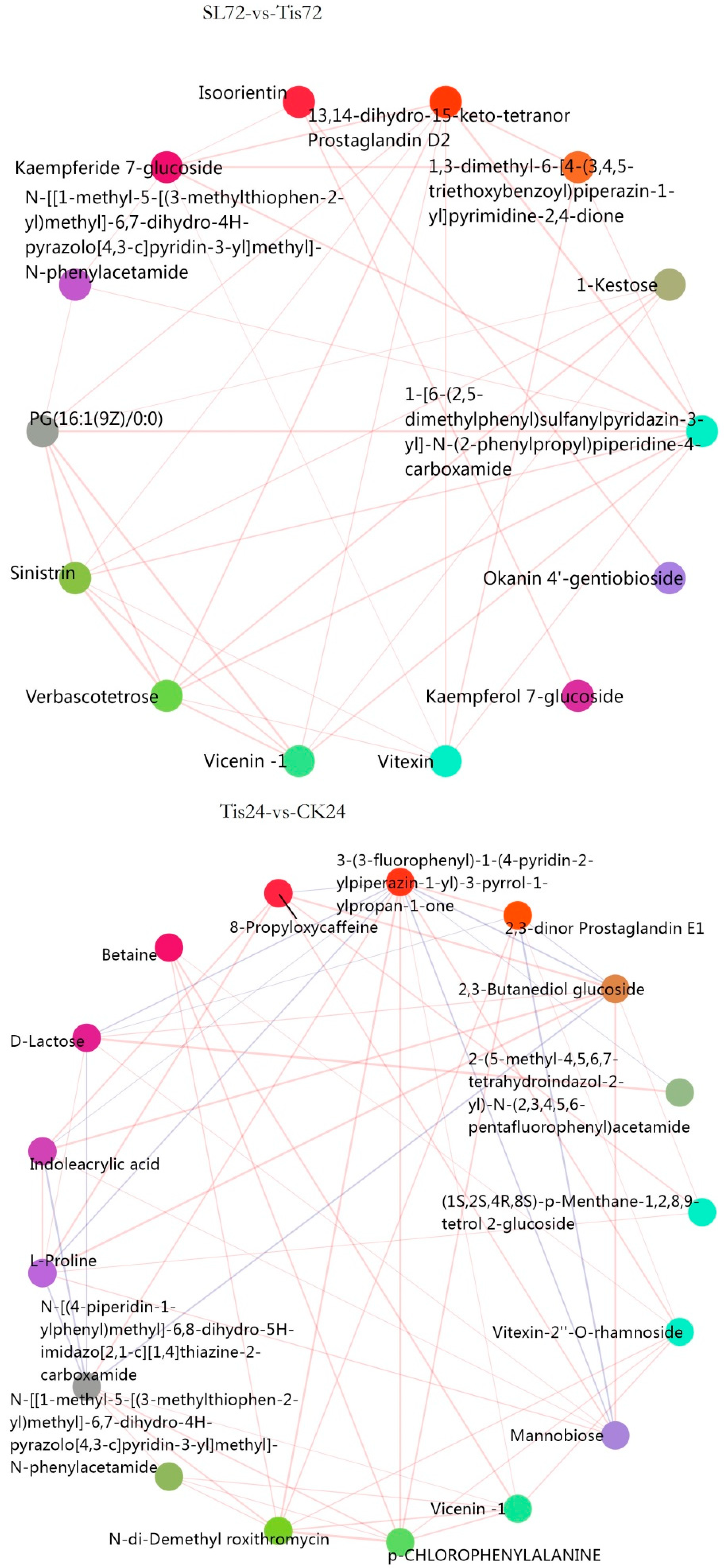
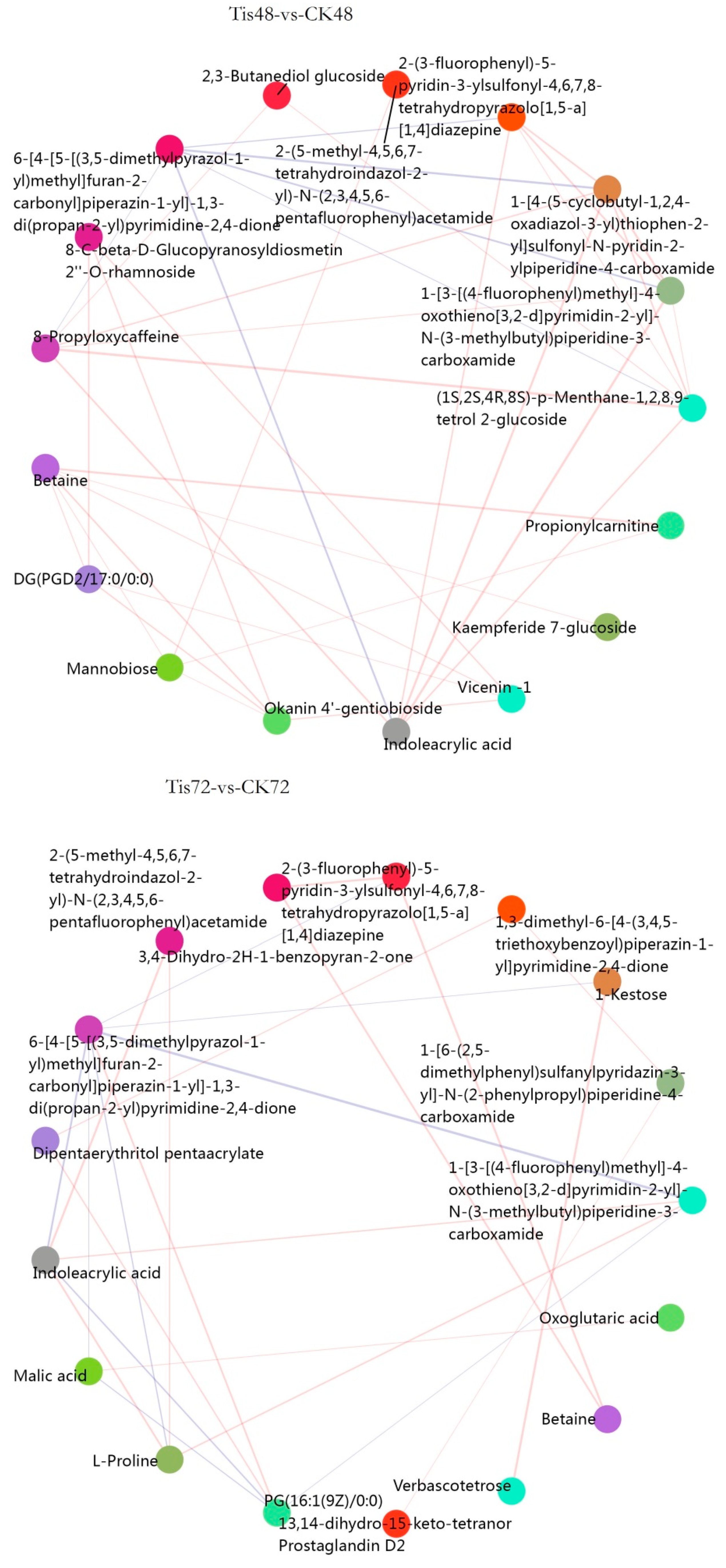
2.3. Metabolome Analysis
3. Discussion
4. Materials and Methods
4.1. Study Settings
4.2. Methods
4.2.1. Metabolome Screening and Analysis
4.2.2. Screening and Analysis of DOF Transcription Factors
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Langridge, P.; Reynolds, M. Breeding for drought and heat tolerance in wheat. Theor. Appl. Genet. 2021, 134, 1753–1769. [Google Scholar] [CrossRef]
- Pequeno, D.N.L.; Reynolds, M.; Kai, S.; Moleromilan, A.; Robertson, R.D.; Lopes, M.S.; Wei, X.; Kropff, M.; Asseng, S. Climate impact and adaptation to heat and drought stress of regional and global wheat production. Environ. Res. Lett. 2021, 16, 054070. [Google Scholar] [CrossRef]
- Zampieri, M.; Ceglar, A.; Dentener, F.; Toreti, A. Wheat yield loss attributable to heat waves, drought and water excess at the global, national and subnational scales. Environ. Res. Lett. 2017, 12, 064008. [Google Scholar] [CrossRef]
- Duddek, P.; Carminati, A.; Koebernick, N.; Ohmann, L.; Lovric, G.; Delzon, S.; Rodriguez-Dominguez, C.M.; King, A.; Ahmed, M.A. The impact of drought-induced root and root hair shrinkage on root–soil contact. Plant Physiol. 2022, 189, 1232–1236. [Google Scholar] [CrossRef]
- Daszkowska-Golec, A.; Szarejko, I. Open or Close the Gate—Stomata Action Under the Control of Phytohormones in Drought Stress Conditions. Front. Plant Sci. 2013, 4, 138. [Google Scholar] [CrossRef]
- Shokat, S.; Großkinsky, D.K.; Roitsch, T.; Liu, F.L. Activities of leaf and spike carbohydrate-metabolic and antioxidant enzymes are linked with yield performance in three spring wheat genotypes grown under well-watered and drought conditions. BMC Plant Biol. 2020, 20, 400. [Google Scholar] [CrossRef]
- Živanović, B.; Milić Komić, S.; Tosti, T.; Vidović, M.; Prokić, L.; Veljović Jovanović, S. Leaf Soluble Sugars and Free Amino Acids as Important Components of Abscisic Acid—Mediated Drought Response in Tomato. Plants 2020, 9, 1147. [Google Scholar] [CrossRef]
- Sharma, P.; Dubey, R.S. Drought Induces Oxidative Stress and Enhances the Activities of Antioxidant Enzymes in Growing Rice Seedlings. Plant Growth Regul. 2005, 46, 209–221. [Google Scholar] [CrossRef]
- De Roo, L.; Salomón, R.L.; Oleksyn, J.; Steppe, K. Woody Tissue photosynthesis delays drought stress in Populus tremula trees and maintains starch reserves in branch xylem Tissues. New Phytol. 2020, 228, 70–81. [Google Scholar] [CrossRef]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Jogawat, A.; Yadav, B.; Chhaya; Lakra, N.; Singh, A.K.; Narayan, O.P. Crosstalk between phytohormones and secondary metabolites in the drought stress tolerance of crop plants: A review. Physiol. Plant 2021, 172, 1106–1132. [Google Scholar] [CrossRef]
- Ramachandran, P.; Wang, G.; Augstein, F.; Vries, J.D.; Carlsbecker, A. Continuous root xylem formation and vascular acclimation to water deficit involves endodermal ABA signalling via miR165. Development 2018, 145, dev.159202. [Google Scholar] [CrossRef]
- Fraudentali, I.; Ghuge, S.; Carucci, A.; Tavladoraki, P.T.; Angelini, R.; Rodrigues-Pousada, R.A.; Cona, A. Developmental, hormone- and stress-modulated expression profiles of four members of the Arabidopsis copper-amine oxidase gene family. Plant Physiol. Biochem. PPB 2020, 147, 141–160. [Google Scholar] [CrossRef]
- Khan, A.; Pan, X.; Najeeb, U.; Tan, D.K.Y.; Fahad, S.; Zahoor, R.; Luo, H. Coping with drought: Stress and adaptive mechanisms, and management through cultural and molecular alternatives in cotton as vital constituents for plant stress resilience and fitness. Biol. Res. 2018, 51, 1–17. [Google Scholar] [CrossRef]
- Naeem, M.; Shahzad, K.; Saqib, S.; Shahzad, A.; Nasrullah; Younas, M.; Afridi, M.I. The Solanum melongena COP1LIKE manipulates fruit ripening and flowering time in tomato (Solanum lycopersicum). Plant Growth Regul. 2022, 96, 369–382. [Google Scholar] [CrossRef]
- Müller, M.; Munné-Bosch, S. Hormonal impact on photosynthesis and photoprotection in plants. Plant Physiol. 2021, 185, 1500–1522. [Google Scholar] [CrossRef]
- Verma, S.; Negi, N.P.; Pareek, S.; Mudgal, G.; Kumar, D. Auxin response factors in plant adaptation to drought and salinity stress. Physiol. Plant 2022, 174, e13714. [Google Scholar] [CrossRef]
- Nishiyama, R.; Watanabe, Y.; Fujita, Y.; Le, D.T.; Kojima, M.; Werner, T.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Kakimoto, T. Analysis of Cytokinin Mutants and Regulation of Cytokinin Metabolic Genes Reveals Important Regulatory Roles of Cytokinins in Drought, Salt and Abscisic Acid Responses, and Abscisic Acid Biosynthesis. Plant Cell 2011, 23, 2169–2183. [Google Scholar] [CrossRef] [PubMed]
- Farhangi-Abriz, S.; Torabian, S. Biochar Increased Plant Growth-Promoting Hormones and Helped to Alleviates Salt Stress in Common Bean Seedlings. J. Plant Growth Regul. 2018, 37, 591–601. [Google Scholar] [CrossRef]
- Ronzan, M.; Piacentini, D.; Fattorini, L.; Federica, D.R.; Caboni, E.; Eiche, E.; Ziegler, J.; Hause, B.; Riemann, M.; Betti, C.; et al. Auxin-jasmonate crosstalk in Oryza sativa L. root system formation after cadmium and/or arsenic exposure. Environ. Exp. Bot. 2019, 165, 59–69. [Google Scholar] [CrossRef]
- Rasheed, A.; Hassan, M.U.; Aamer, M.; Batool, M.; Sheng, F.; WU, Z.; LI, H. A critical review on the improvement of drought stress tolerance in rice (Oryza sativa L.). Not. Bot. Horti Agrobot. Cluj-Napoca 2020, 48, 1756–1788. [Google Scholar] [CrossRef]
- Vishal, B.; Kumar, P.P. Regulation of Seed Germination and Abiotic Stresses by Gibberellins and Abscisic Acid. Front. Plant Sci. 2018, 9, 838. [Google Scholar] [CrossRef]
- Waterman, P.G.; Mole, S. Extrinsic Factors Influencing Production of Secondary Metabolites in Plants. In Insect-Plant Interactions; CRC press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Ullah, A.; Manghwar, H.; Shaban, M.; Khan, A.H.; Akbar, A.; Ali, U.; Ali, E.; Fahad, S. Phytohormones enhanced drought tolerance in plants: A coping strategy. Environ. Sci. Pollut. Res. 2018, 25, 33103–33118. [Google Scholar] [CrossRef]
- Chhaya; Yadav, B.; Jogawat, A.; Gnanasekaran, P.; Kumari, P.; Lakra, N.; Lal, S.K.; Pawar, J.; Narayan, O. An overview of recent advancement in phytohormones-mediated stress management and drought tolerance in crop plants. Plant Gene 2020, 25, 100264. [Google Scholar] [CrossRef]
- Signorelli, S.; Tarkowski, Ł.P.; Wim, V.D.E.; Bassham, D.C. Linking Autophagy to Abiotic and Biotic Stress Responses. Trends Plant Sci. 2019, 24, 413–430. [Google Scholar] [CrossRef]
- Bandurska, H. Salicylic Acid: An Update on Biosynthesis and Action in Plant Response to Water Deficit and Performance Under Drought. In SALICYLIC ACID; Hayat, S., Ahmad, A., Alyemeni, M., Eds.; Springer: Dordrecht, The Netherlands, 2013. [Google Scholar] [CrossRef]
- Ghorbel, M.; Brini, F.; Sharma, A.; Landi, M. Role of jasmonic acid in plants: The molecular point of view. Plant Cell Rep. 2021, 40, 1471–1494. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, K.S.; Husen, A. Plant response to jasmonates: Current developments and their role in changing environment. Bull. Natl. Res. Cent. 2019, 43, 153. [Google Scholar] [CrossRef]
- Chen, E.; Zhang, X.; Yang, Z.; Zhang, C.; Wang, X.; Ge, X.; Li, F. BR deficiency causes increased sensitivity to drought and yield penalty in cotton. BMC Plant Biol. 2019, 19, 220. [Google Scholar] [CrossRef]
- Houben, M.; Van de Poel, B. 1-Aminocyclopropane-1-Carboxylic Acid Oxidase (ACO): The Enzyme That Makes the Plant Hormone Ethylene. Front. Plant Sci. 2019, 10, 695. [Google Scholar] [CrossRef]
- Barbier, F.; Fichtner, F.; Beveridge, C. The strigolactone pathway plays a crucial role in integrating metabolic and nutritional signals in plants. Nat. Plants 2023, 9, 1191–1200. [Google Scholar] [CrossRef]
- Yu, C.; Chen, W.; Wang, Z.; Lou, H. Comparative proteomic analysis of tomato (Solanum lycopersicum L.) shoots reveals crosstalk between strigolactone and auxin. Genomics 2021, 113, 3163–3173. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.N.; Min, Z.; Wu, J.R.; Liu, B.C.; Xu, X.L.; Fang, Y.L.; Ju, Y.L. Physiological and transcriptomic analysis of Cabernet Sauvginon (ViTis vinifera L.) reveals the alleviating effect of exogenous strigolactones on the response of grapevine to drought stress. Plant Physiol. Biochem. 2021, 167, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Kapulnik, Y.; Koltai, H. Strigolactone Involvement in Root Development, Response to Abiotic Stress, and Interactions with the Biotic Soil Environment. Plant Physiol. 2014, 166, 560–569. [Google Scholar] [CrossRef]
- Liu, J.; He, H.; Vitali, M.; Visentin, I.; Charnikhova, T.; Haider, I.; Schubert, A.; Ruyter-Spira, C.; Bouwmeester, H.J.; Lovisolo, C.; et al. Osmotic stress represses strigolactone biosynthesis in Lotus japonicus roots: Exploring the interaction between strigolactones and ABA under abiotic stress. Planta 2015, 241, 1435–1451. [Google Scholar] [CrossRef]
- Ruiz-Lozano, J.M.; Aroca, R.; Zamarreño, Á.M.; Molina, S.; Andreo-Jiménez, B.; Porcel, R.; García-Mina, J.M.; Ruyter-Spira, C.; López-Ráez, J.A. Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant Cell Environ. 2016, 39, 441–452. [Google Scholar] [CrossRef]
- Song, M.; Zhou, S.; Hu, N.; Li, J.C.; Huang, Y.; Zhang, J.M.; Chen, X.; Du, X.H.; Niu, J.S.; Yang, X.W.; et al. Exogenous strigolactones alleviate drought stress in wheat (Triticum aestivum L.) by promoting cell wall biogenesis to optimize root architecture. Plant Physiol. Biochem. 2023, 204, 108121. [Google Scholar] [CrossRef] [PubMed]
- Sedaghat, M.; Emam, Y.; Mokhtassi-Bidgoli, A.; Hazrati, S.; Lovisolo, C.; Visentin, I.; Cardinale, F.; Tahmasebi-Sarvestani, Z. The Potential of the Synthetic Strigolactone Analogue GR24 for the Maintenance of Photosynthesis and Yield in Winter Wheat under Drought: Investigations on the Mechanisms of Action and Delivery Modes. Plants 2021, 10, 1223. [Google Scholar] [CrossRef]
- Song, M.; Hu, N.; Zhou, S.; Xie, S.; Yang, J.; Ma, W.; Teng, Z.; Liang, W.; Wang, C.; Bu, M.; et al. Physiological and RNA-Seq Analyses on Exogenous Strigolactones Alleviating Drought by Improving Antioxidation and Photosynthesis in Wheat (Triticum aestivum L.). Antioxidants 2023, 12, 1884. [Google Scholar] [CrossRef] [PubMed]
- Daszkowska-Golec, A.; Mehta, D.; Uhrig, R.G.; Brąszewska, A.; Novak, O.; Fontana, I.M.; Melzer, M.; Płociniczak, T.; Marzec, M. Multi-omics insights into the positive role of strigolactone perception in barley drought response. BMC Plant Biol. 2023, 23, 445. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, F.; Jiao, P.; Liu, J.; Zhang, H.; Liu, S.; Guan, S.; Ma, Y. The Overexpression of Zea mays Strigolactone Receptor Gene D14 Enhances Drought Resistance in Arabidopsis thaliana L. Int. J. Mol. Sci. 2024, 25, 1327. [Google Scholar] [CrossRef]
- Corrales, A.R.; Carrillo, L.; Lasierra, P.; Nebauer, S.G.; Dominguez-Figueroa, J.; Renau-Morata, B.; Pollmann, S.; Granell, A.; Molina, R.V.; Vicente-Carbajosa, J.; et al. Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liang, W.; Gu, P.; Huang, Z. Salt tolerance function of the novel C2H2-type zinc finger protein TaZNF in wheat. Plant Physiol. Biochem. 2016, 106, 129–140. [Google Scholar] [CrossRef]
- Chen, P.; Yan, M.; Li, L.; He, J.Q.; Zhou, S.X.; Li, Z.X.; Niu, C.D.; Bao, C.N.; Zhi, F.; Ma, F.W.; et al. The apple DNA-binding one zinc-finger protein MdDof54 promotes drought resistance. Hortic. Res. 2020, 7, 195. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Chandrasekaran, U.; Liu, A. Genome-wide analysis of the Dof transcription factors in castor bean (Ricinus communis L.). Genes Genom. 2014, 36, 527–537. [Google Scholar] [CrossRef]
- Song, A.; Gao, T.; Li, P.; Chen, S.M.; Guan, Z.Y.; Guan, Z.Y.; Wu, D.; Xin, J.J.; Fan, Q.Q.; Zhao, K.K.; et al. Transcriptome-Wide Identification and Expression Profiling of the DOF Transcription Factor Gene Family in Chrysanthemum morifolium. Front. Plant Sci. 2016, 7, 199. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.; Trapnell, C.; Donaghey, J.; Rinn, J.L.; Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011, 12, R22. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36 (Suppl. S1), D480–D484. [Google Scholar] [CrossRef] [PubMed]

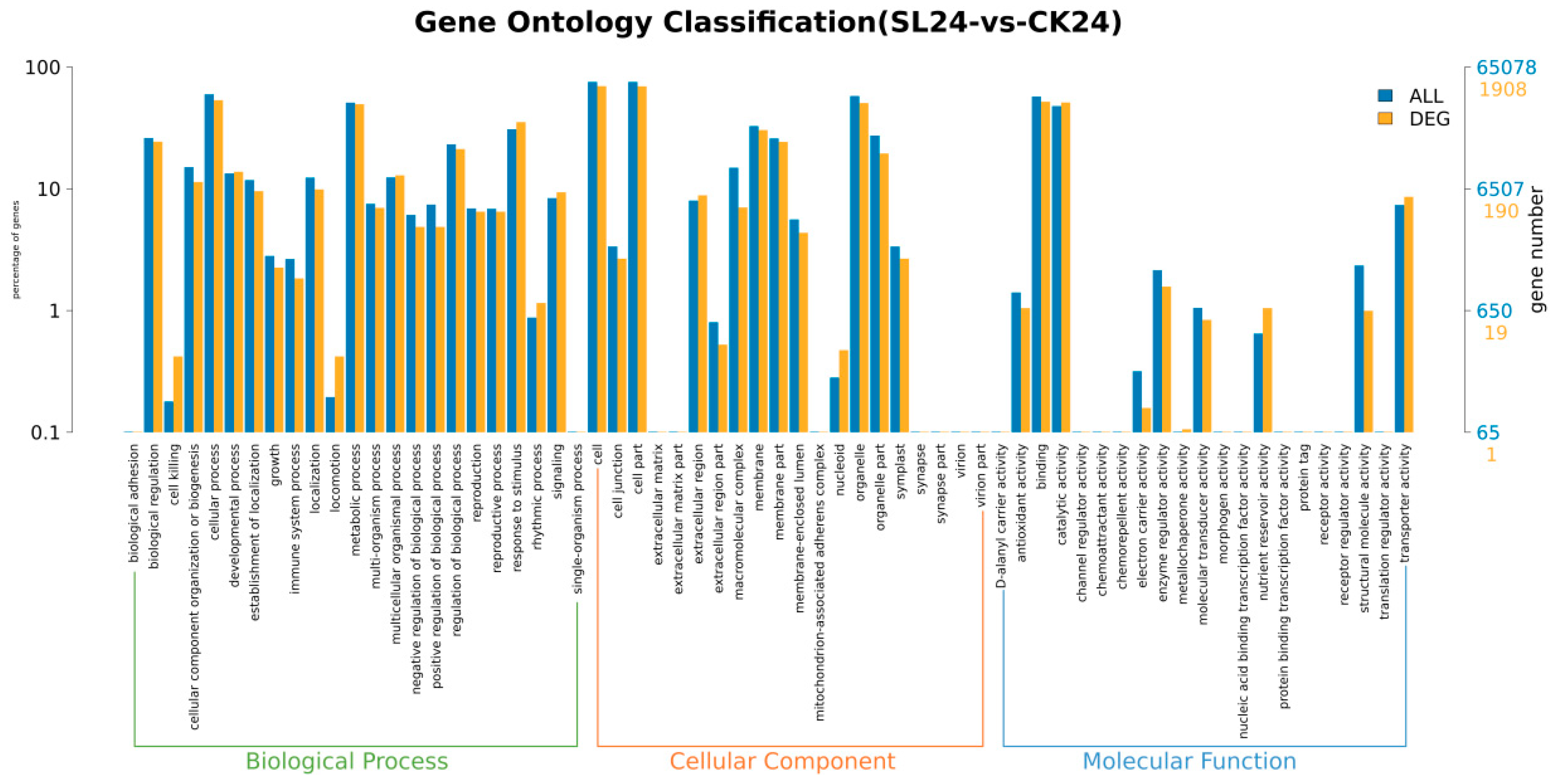
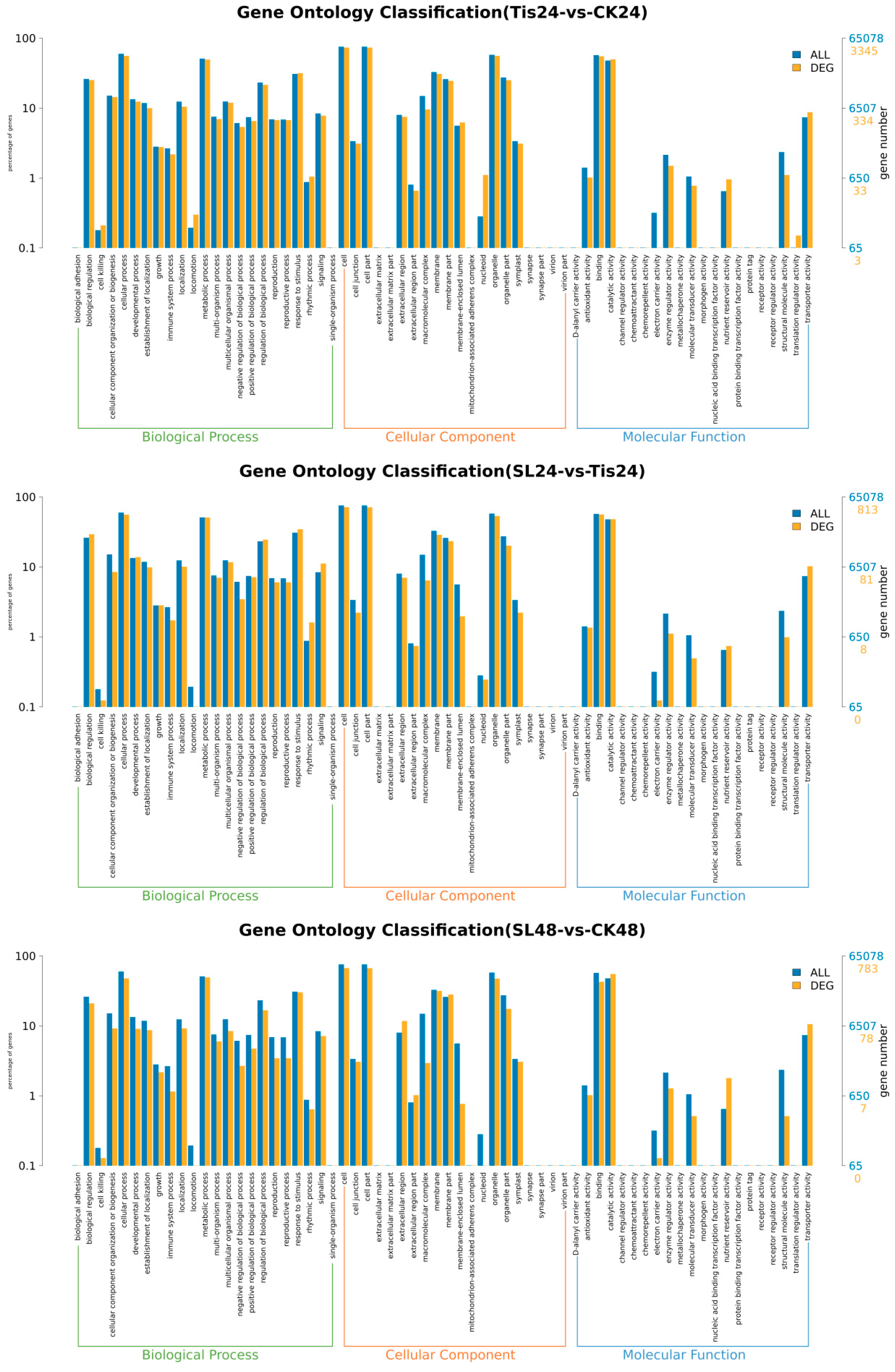
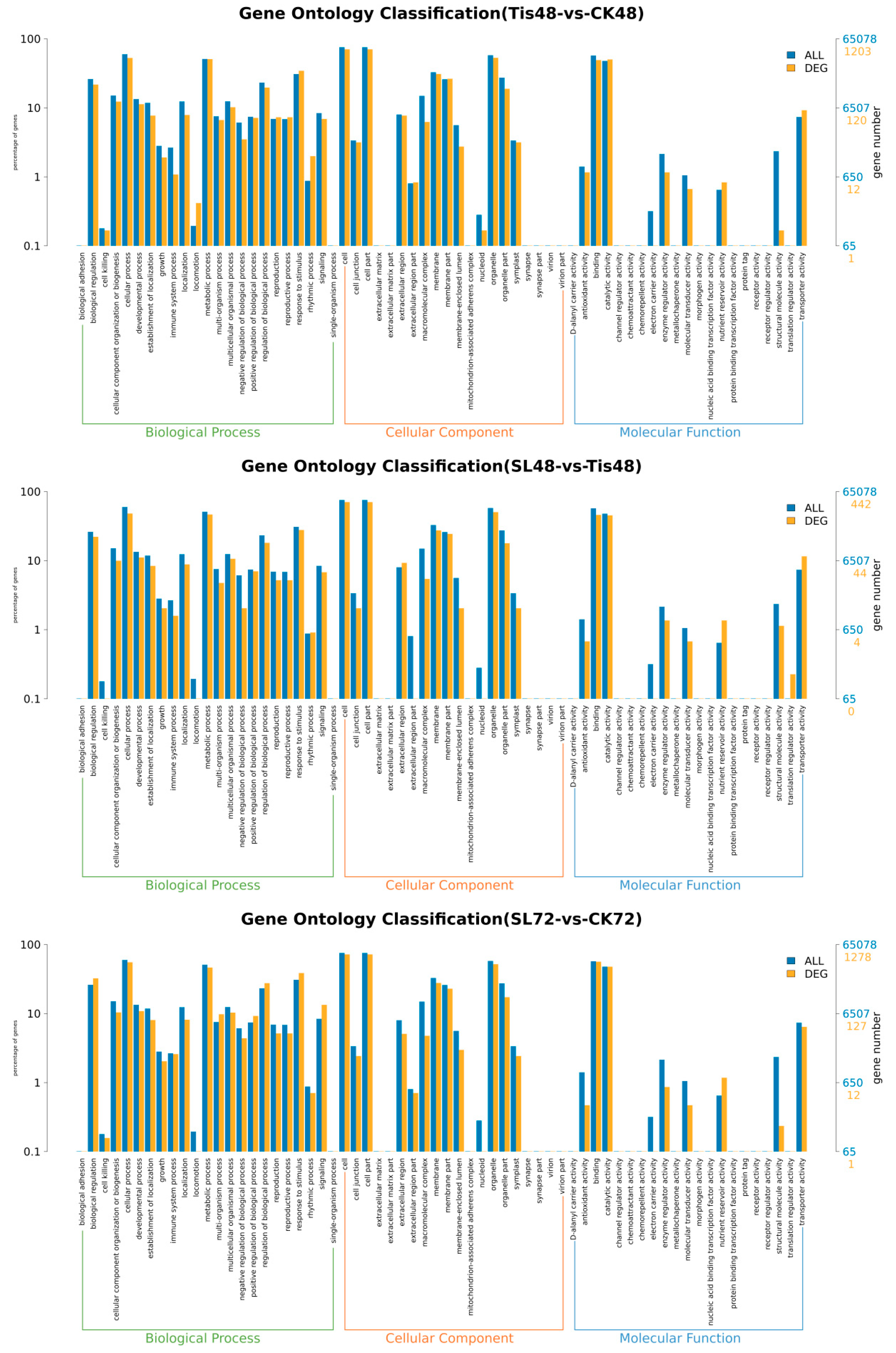
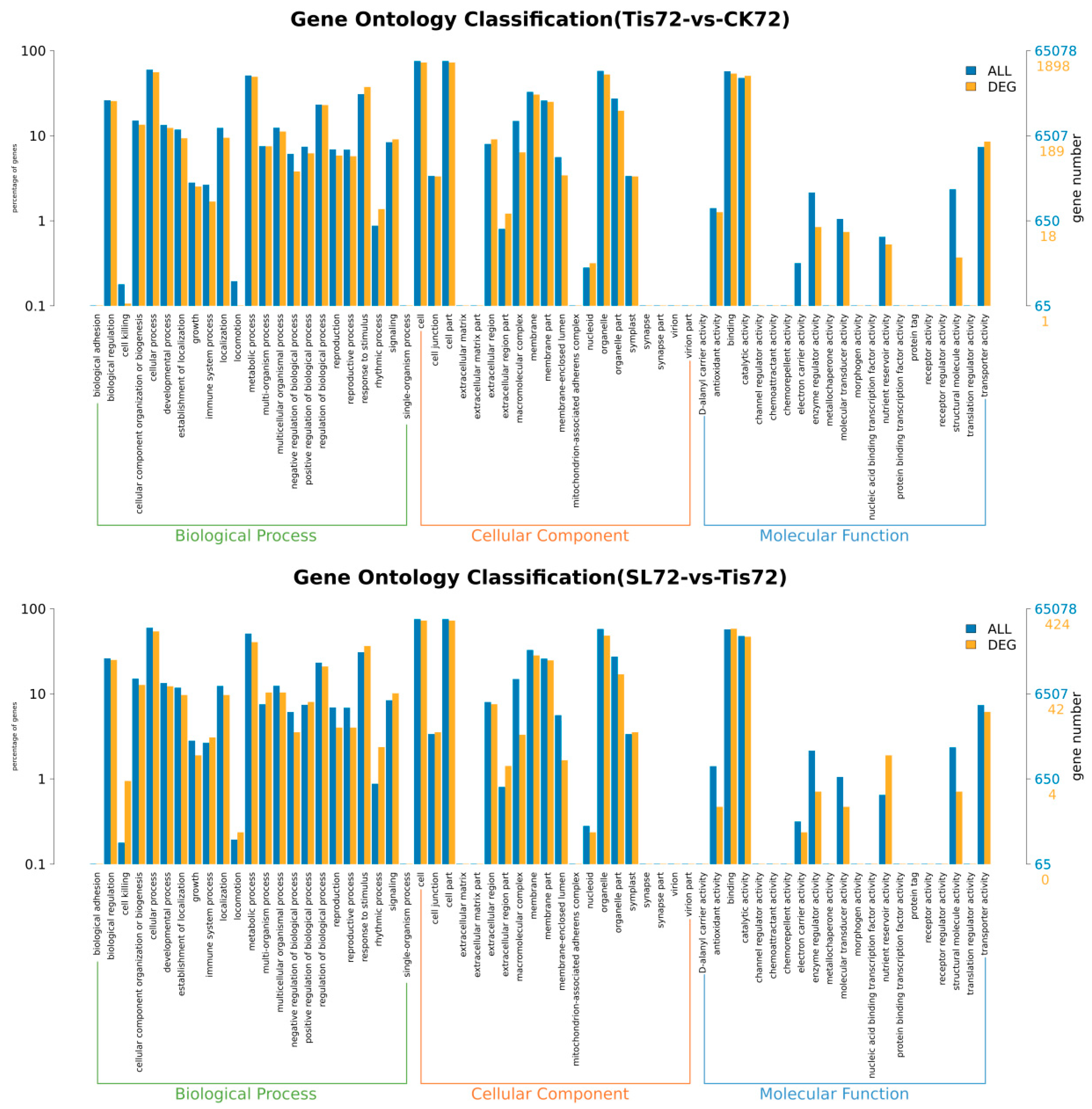
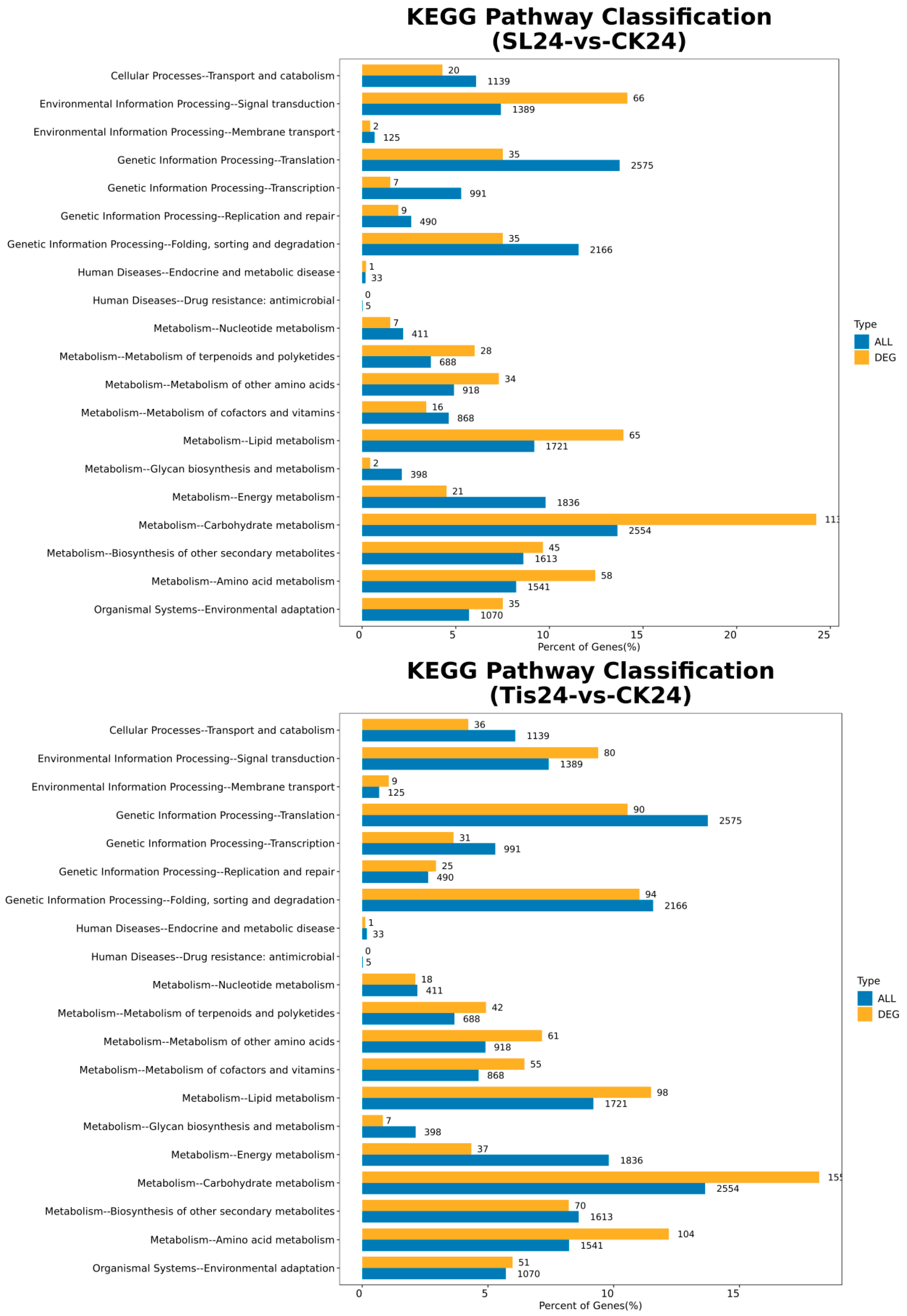
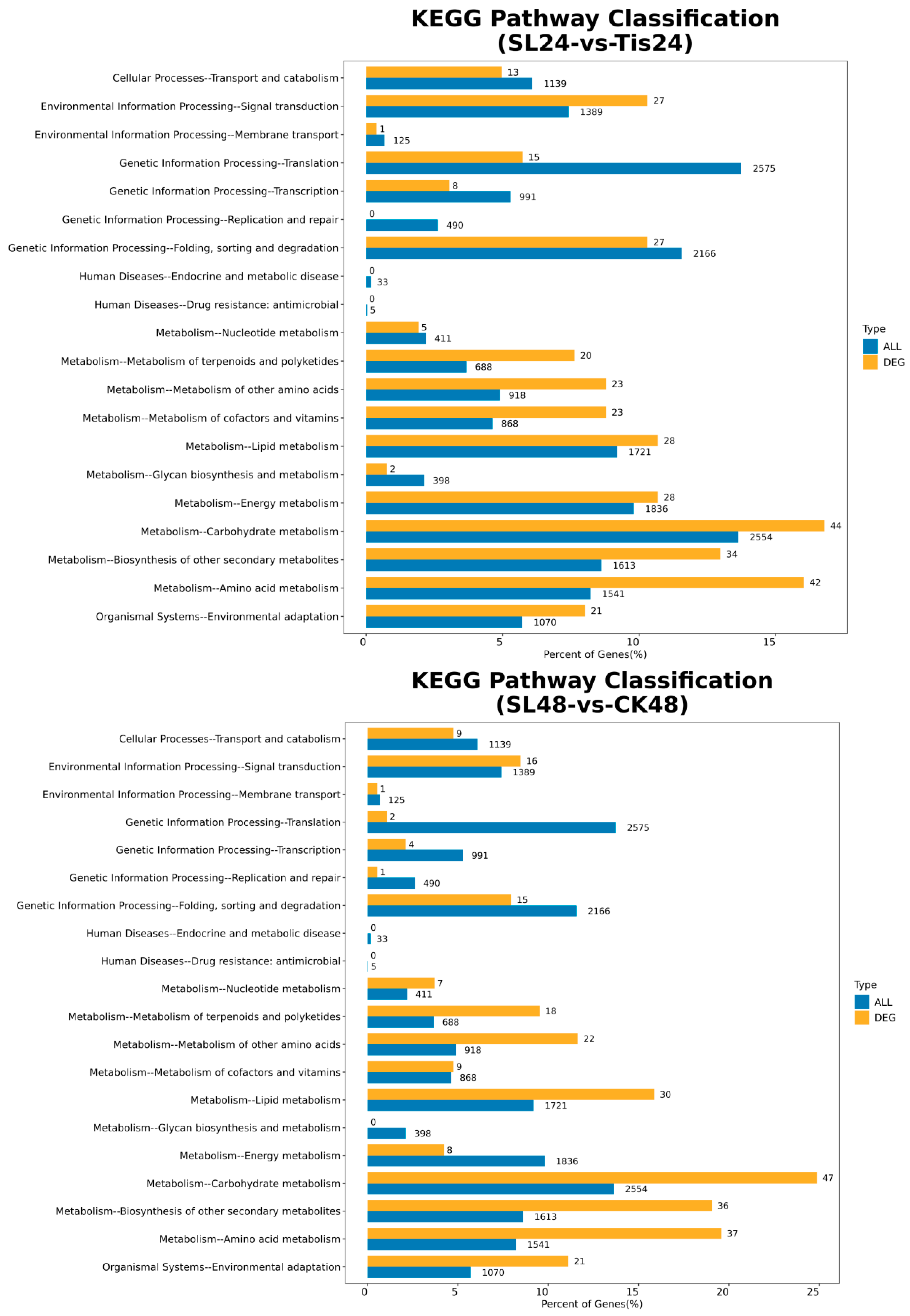
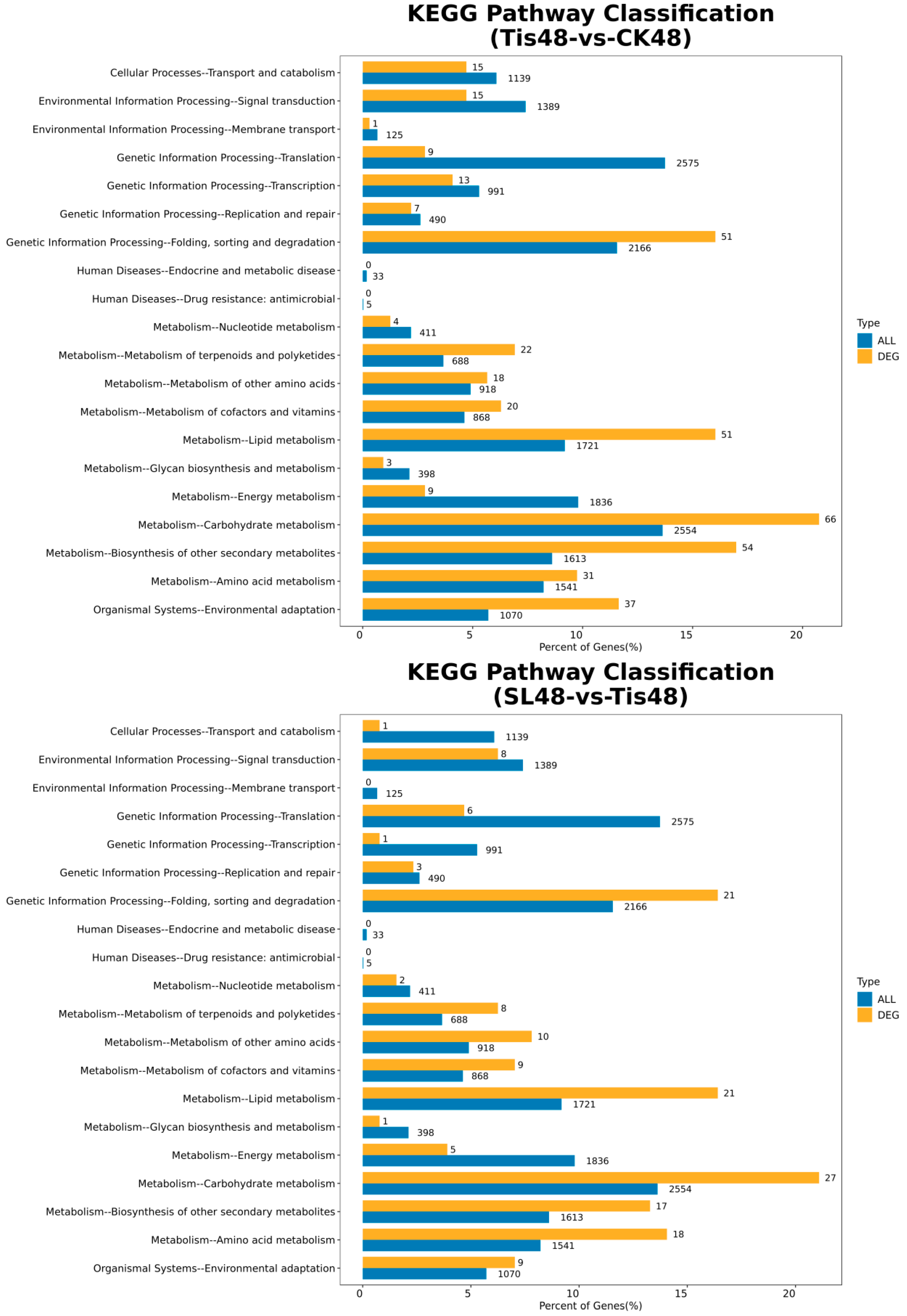
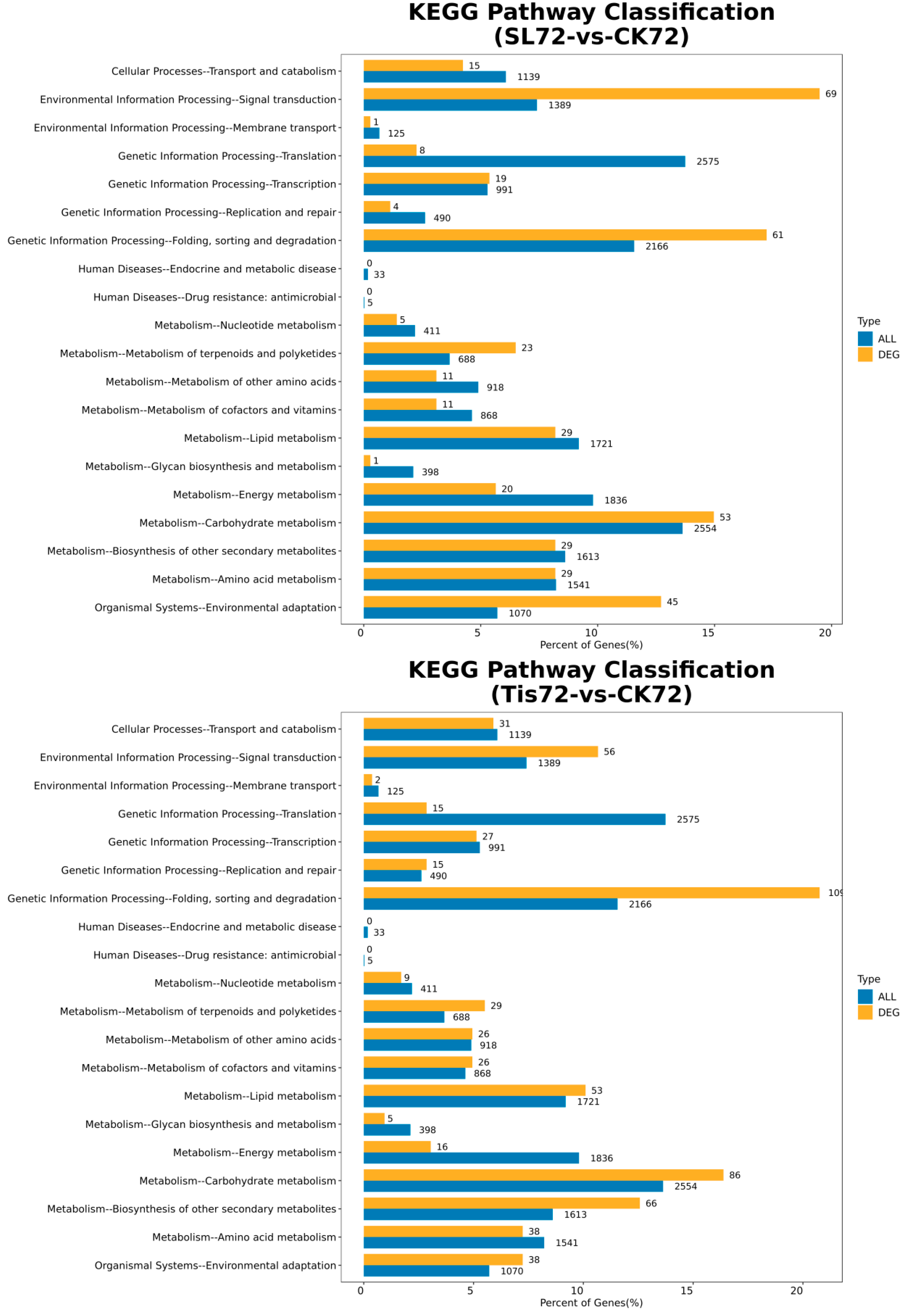
| Geneid | Description | Chromosome | Orientation |
|---|---|---|---|
| 123064986 | dof zinc finger protein DOF3.1-like | 3B | minus |
| 100415886 | dof zinc finger protein DOF1.7 | 2A | minus |
| 123127979 | dof zinc finger protein 2-like | 6A | minus |
| 123190041 | dof zinc finger protein 1-like | 2A | plus |
| 123137649 | dof zinc finger protein 2-like | 6B | plus |
| 123046287 | dof zinc finger protein 1-like | 2B | plus |
| 123104950 | dof zinc finger protein DOF3.6-like | 1B | minus |
| 123187442 | cyclic dof factor 1-like | 2A | minus |
| 123179992 | dof zinc finger protein DOF3.6-like | 1D | minus |
| 123116001 | dof zinc finger protein 3-like | 5B | minus |
| 123145076 | dof zinc finger protein 2-like | 6D | minus |
| 123051520 | cyclic dof factor 1-like | 2D | minus |
| 123186581 | dof zinc finger protein DOF2.2-like | 2A | minus |
| 123188464 | dof zinc finger protein DOF5.8-like | 2A | minus |
| 123044711 | dof zinc finger protein DOF5.8-like | 2B | minus |
| 123052574 | dof zinc finger protein DOF5.8-like | 2D | minus |
| 123093292 | dof zinc finger protein DOF1.8-like | 1B | plus |
| 123040442 | dof zinc finger protein DOF5.7-like | 2B | plus |
| 606390 | dof zinc finger protein 4 | 6A | minus |
| 100037553 | dof zinc finger protein 4 | 6B | minus |
| Time and Treatment | Transcriptome Number | Metabolome Number |
|---|---|---|
| 24 h CK | R2C1 | B2C1 |
| 24 h CK | R2C2 | B2C2 |
| 24 h CK | R2C3 | B2C3 |
| 24 h CK | R2C4 | B2C4 |
| 24 h SL | R2S1 | B2S1 |
| 24 h SL | R2S2 | B2S2 |
| 24 h SL | R2S3 | B2S3 |
| 24 h SL | R2S4 | B2S4 |
| 24 h Tis | R2T1 | B2T1 |
| 24 h Tis | R2T2 | B2T2 |
| 24 h Tis | R2T3 | B2T3 |
| 24 h Tis | R2T4 | B2T4 |
| 48 h CK | R4C1 | B4C1 |
| 48 h CK | R4C2 | B4C2 |
| 48 h CK | R4C3 | B4C3 |
| 48 h CK | R4C4 | B4C4 |
| 48 h SL | R4S1 | B4S1 |
| 48 h SL | R4S2 | B4S2 |
| 48 h SL | R4S3 | B4S3 |
| 48 h SL | R4S4 | B4S4 |
| 48 h Tis | R4T1 | B4T1 |
| 48 h Tis | R4T2 | B4T2 |
| 48 h Tis | R4T3 | B4T3 |
| 48 h Tis | R4T4 | B4T4 |
| 72 h CK | R7C1 | B7C1 |
| 72 h CK | R7C2 | B7C2 |
| 72 h CK | R7C3 | B7C3 |
| 72 h CK | R7C4 | B7C4 |
| 72 h SL | R7S1 | B7S1 |
| 72 h SL | R7S2 | B7S2 |
| 72 h SL | R7S3 | B2C1 |
| 72 h SL | R7S4 | B2C2 |
| 72 h Tis | R7T1 | B2C3 |
| 72 h Tis | R7T2 | B2C4 |
| 72 h Tis | R7T3 | B2S1 |
| 72 h Tis | R7T4 | B2S2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Jin, H.; Du, S.; Fang, B.; Yue, J.; Yang, C.; Wang, H.; Zhang, D.; Wang, J.; Song, H.; et al. Multi-Omics Association Analysis of DOF Transcription Factors Involved in the Drought Resistance of Wheat Induced by Strigolactone. Int. J. Mol. Sci. 2025, 26, 2396. https://doi.org/10.3390/ijms26062396
Wang Y, Jin H, Du S, Fang B, Yue J, Yang C, Wang H, Zhang D, Wang J, Song H, et al. Multi-Omics Association Analysis of DOF Transcription Factors Involved in the Drought Resistance of Wheat Induced by Strigolactone. International Journal of Molecular Sciences. 2025; 26(6):2396. https://doi.org/10.3390/ijms26062396
Chicago/Turabian StyleWang, Yanjing, Haiyang Jin, Simeng Du, Baoting Fang, Junqin Yue, Cheng Yang, Hanfang Wang, Deqi Zhang, Jiarui Wang, Hang Song, and et al. 2025. "Multi-Omics Association Analysis of DOF Transcription Factors Involved in the Drought Resistance of Wheat Induced by Strigolactone" International Journal of Molecular Sciences 26, no. 6: 2396. https://doi.org/10.3390/ijms26062396
APA StyleWang, Y., Jin, H., Du, S., Fang, B., Yue, J., Yang, C., Wang, H., Zhang, D., Wang, J., Song, H., Shao, Y., & Li, X. (2025). Multi-Omics Association Analysis of DOF Transcription Factors Involved in the Drought Resistance of Wheat Induced by Strigolactone. International Journal of Molecular Sciences, 26(6), 2396. https://doi.org/10.3390/ijms26062396




