N4-Methylcytosine Supports the Growth of Escherichia coli Uracil Auxotrophs
Abstract
1. Introduction
2. Results and Discussion
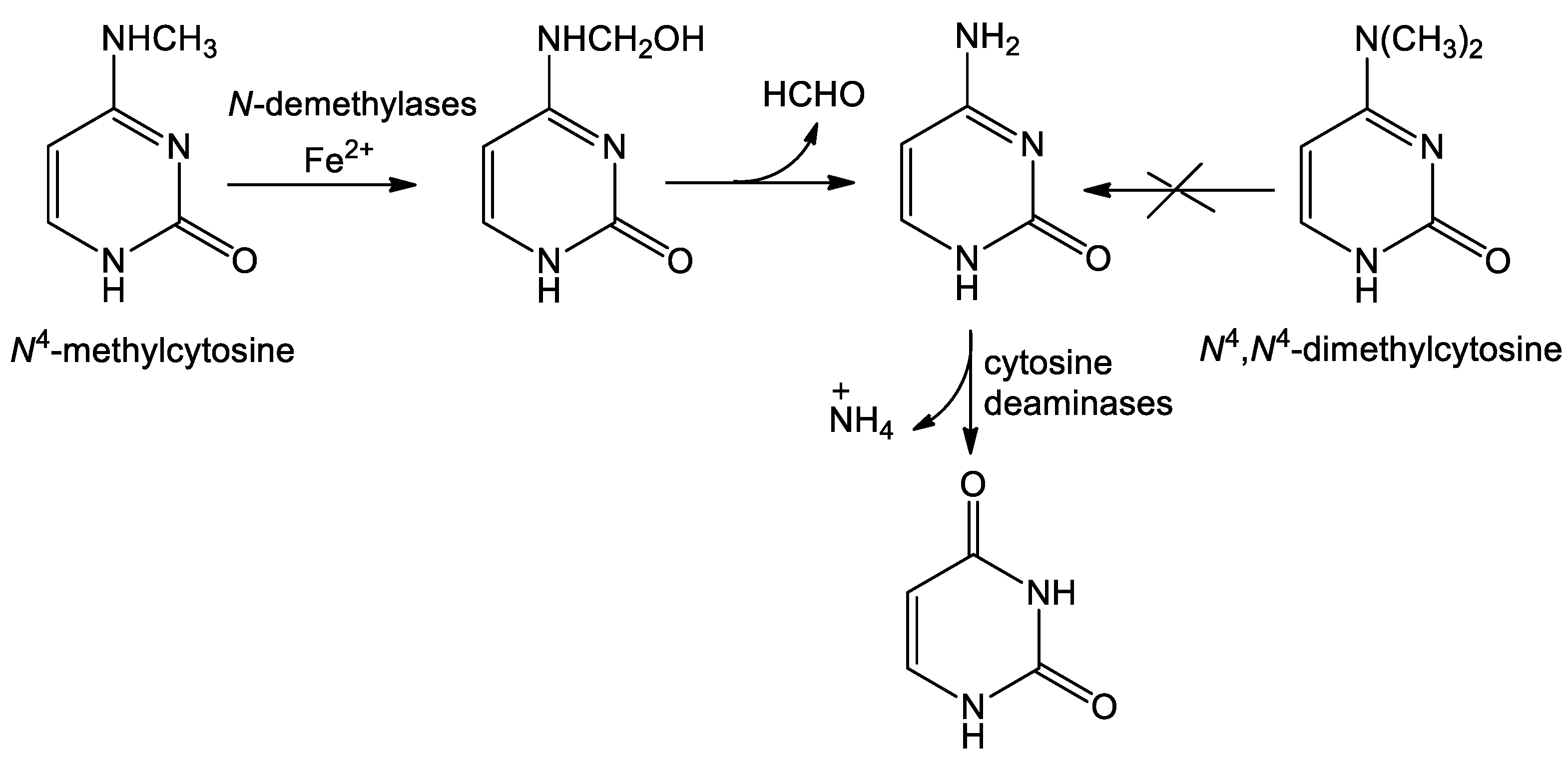
2.1. N4-Methylhcytosine but Not N4,N4-Dimethylcytosine Supports Growth of the Uracil Auxotrophs
2.1.1. Growth of Uracil Auxotrophs in M9 Solid Medium
2.1.2. Growth of the E. coli ∆pyrF::Km in M9 Agar Medium After the Serial Dilutions
2.1.3. Determination of the E. coli ∆pyrF::Km Generation Time
3. Materials and Methods
3.1. Synthesis of Methylated Cytosines
- General information
3.1.1. Synthesis of N4-Methylcytosine
- (i)
- 4-methylthiouracil
- (ii)
- N4-methylcytosine
3.1.2. Synthesis of N4,N4-Dimethylcytosine
3.2. Bacterial Strains and Growth Media
3.3. Growth of Bacteria in Minimal Media
3.4. Determination of Growth Rate of E. coli ∆pyrF::Km Strain
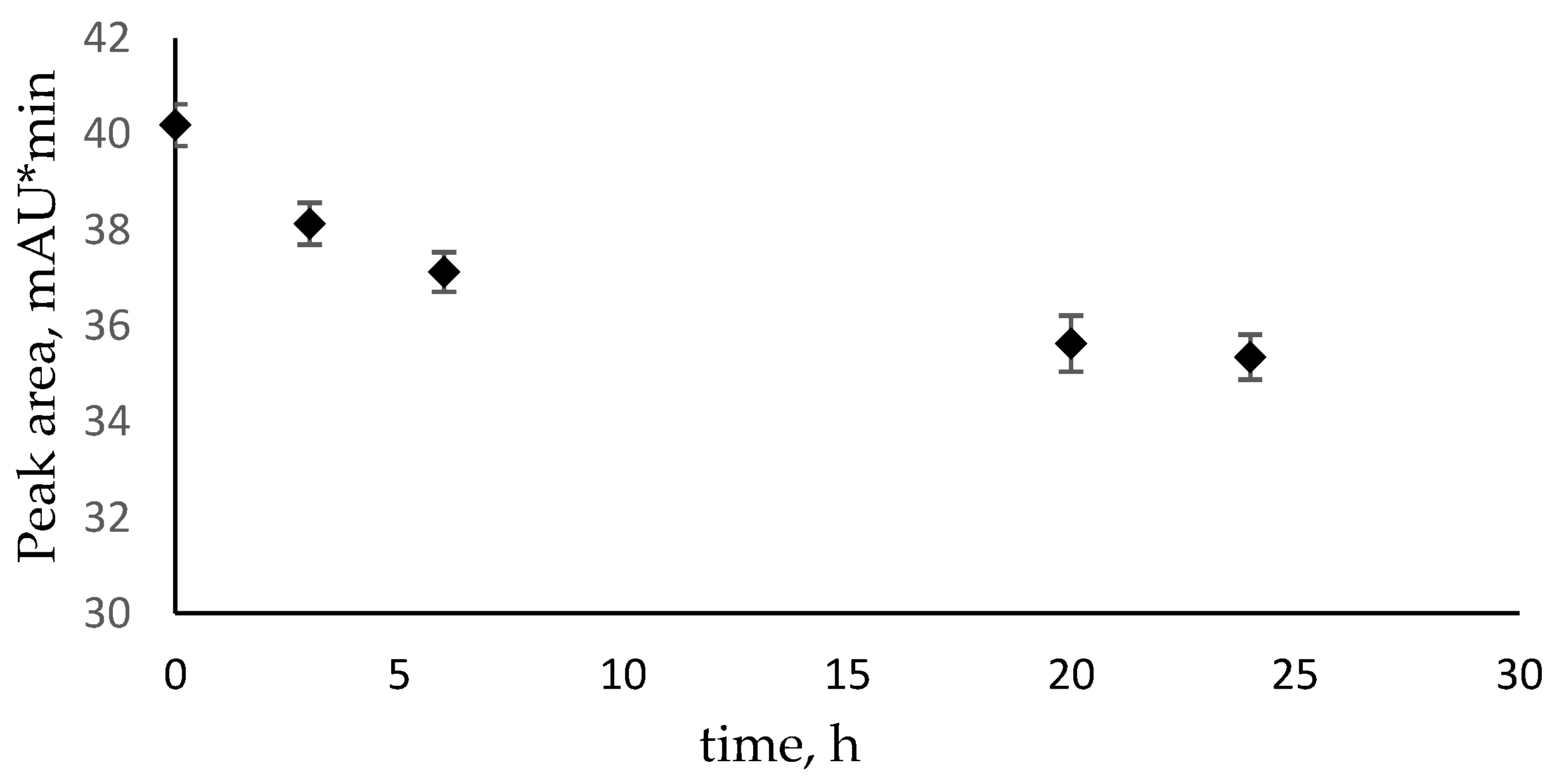
3.5. Determination of N4-Methylcytosine and N4,N4-Dimethylcytosine Concentration and Chemical Stability Using HPLC
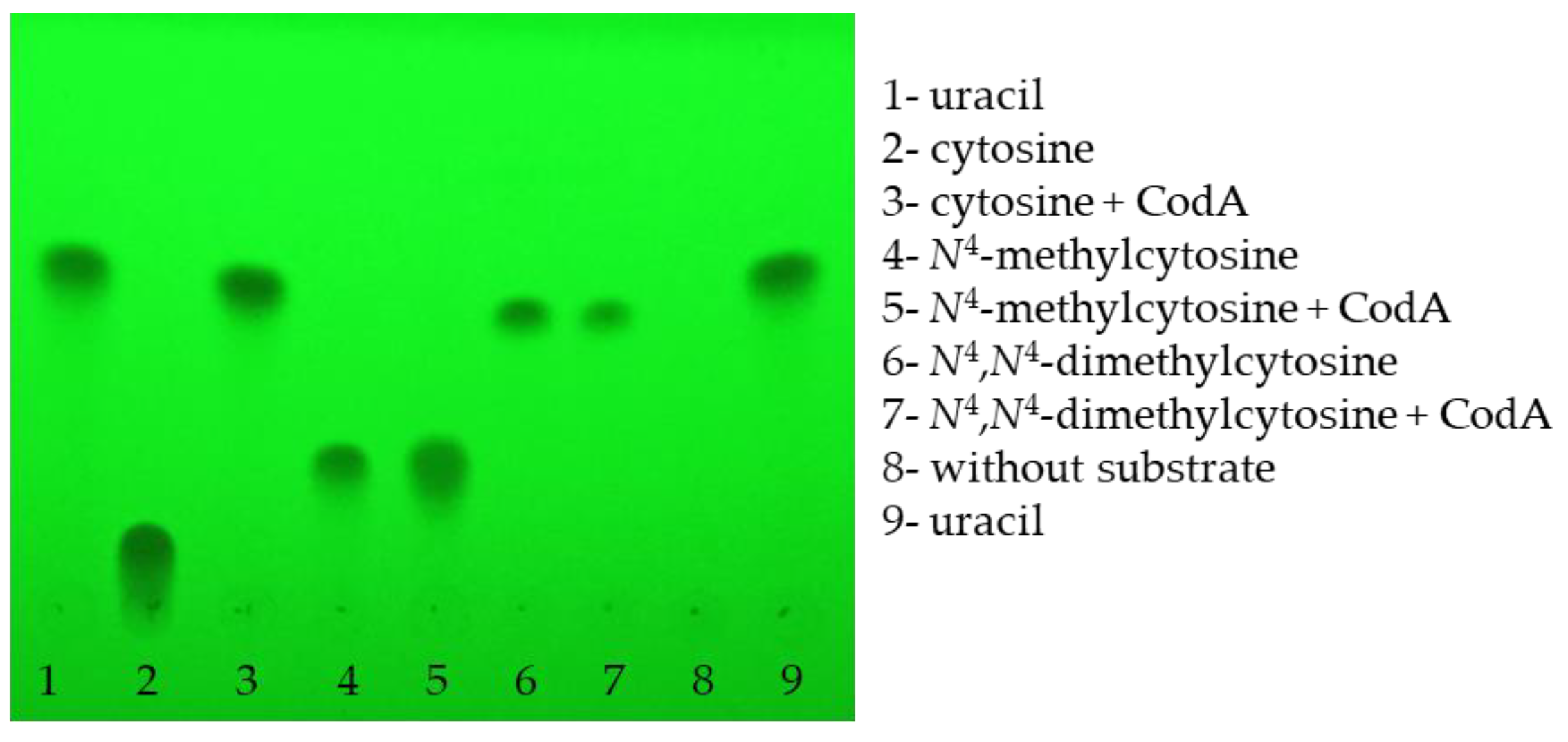
3.6. Purification of E. coli CodA Protein and Determination of Its Enzymatic Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cappannini, A.; Ray, A.; Purta, E.; Mukherjee, S.; Boccaletto, P.; Moafinejad, S.N.; Lechner, A.; Barchet, C.; Klaholz, B.P.; Stefaniak, F.; et al. MODOMICS: A database of RNA modifications and related information. 2023 update. Nucleic Acids Res. 2024, 52, D239–D244. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021, 22, 375–392. [Google Scholar] [CrossRef] [PubMed]
- Arevalo, C.P.; Bolton, M.J.; Le Sage, V.; Ye, N.; Furey, C.; Muramatsu, H.; Alameh, M.G.; Pardi, N.; Drapeau, E.M.; Parkhouse, K.; et al. A multivalent nucleoside-modified mRNA vaccine against all known influenza virus subtypes. Science 2022, 378, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Van Giesen, K.J.D.; Thompson, M.J.; Meng, Q.; Lovelock, S.L. Biocatalytic synthesis of antiviral nucleosides, cyclic dinucleotides, and oligonucleotide therapies. JACS Au 2023, 3, 13–24. [Google Scholar] [CrossRef]
- Traube, F.R.; Carell, T. The chemistries and consequences of DNA and RNA methylation and demethylation. RNA Biol. 2017, 14, 1099–1107. [Google Scholar] [CrossRef]
- Preumont, A.; Snoussi, K.; Stroobant, V.; Collet, J.F.; Van Schaftingen, E. Molecular identification of pseudouridine-metabolizing enzymes. J. Biol. Chem. 2008, 283, 25238–25246. [Google Scholar] [CrossRef]
- Loh, K.D.; Gyaneshwar, P.; Papadimitriou, E.M.; Fong, R.; Kim, K.S.; Parales, R.; Zhou, Z.; Inwood, W.; Kustu, S. A previously underscribed pathway for pyrimidine catabolism. Proc. Natl. Acad. Sci. USA 2006, 103, 5114–5119. [Google Scholar] [CrossRef]
- Vogels, G.D.; Van Der Drift, C. Degradation of purines and pyrimidines by microorganisms. Bacteriol. Rev. 1976, 40, 403–468. [Google Scholar] [CrossRef]
- Soong, C.L.; Ogawa, J.; Shimizu, S. Novel amidohydrolytic reactions in oxidative pyrimidine metabolism: Analysis of the barbiturase reaction and discovery of a novel enzyme, ureidomalonase. Biochem. Biophys. Res. Commun. 2001, 286, 222–226. [Google Scholar] [CrossRef]
- Kimura, S.; Suzuki, T. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S RRNA. Nucleic Acids Res. 2010, 38, 1341–1352. [Google Scholar] [CrossRef]
- Zou, M.; Mu, Y.; Chai, X.; Ouyang, M.; Yu, L.J.; Zhang, L.; Meurer, J.; Chi, W. The critical function of the plastid rRNA methyltransferase, CMAL, in ribosome biogenesis and plant development. Nucleic Acids Res. 2020, 48, 3195–3210. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, W.; Netzband, R.; Bonenfant, G.; Biegel, J.M.; Miller, C.; Fuchs, G.; Henderson, E.; Arra, M.; Canki, M.; Fabris, D.; et al. Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 2018, 46, 5776–5791. [Google Scholar] [CrossRef] [PubMed]
- Fluke, K.A.; Dai, N.; Wolf, E.J.; Fuchs, R.T.; Ho, P.S.; Talbott, V.; Elkins, L.; Tsai, Y.-L.; Schiltz, J.; Febvre, H.P.; et al. A novel N4,N4-dimethylcytidine in the archaeal ribosome enhances hyperthermophily. Proc. Natl. Acad. Sci. USA 2024, 121, e2405999121. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Romero, M.A.; Cota, I.; Casadesús, J. DNA methylation in bacteria: From the methyl group to the methylome. Curr. Opin. Microbiol. 2015, 25, 9–16. [Google Scholar] [CrossRef]
- Kumar, S.; Karmakar, B.C.; Nagarajan, D.; Mukhopadhyay, A.K.; Morgan, R.D.; Rao, D.N. N4-cytosine DNA methylation regulates transcription and pathogenesis in Helicobacter pylori. Nucleic Acids Res. 2018, 46, 3429–3445. [Google Scholar] [CrossRef]
- Rodriguez, F.; Yushenova, I.A.; DiCorpo, D.; Arkhipova, I.R. Bacterial N4-methylcytosine as an epigenetic mark in eukaryotic DNA. Nat. Commun. 2022, 13, 1072. [Google Scholar] [CrossRef]
- Aučynaitė, A.; Rutkienė, R.; Gasparavičiūtė, R.; Meškys, R.; Urbonavičius, J. A gene encoding a DUF523 domain protein is involved in the conversion of 2-thiouracil into uracil. Environ. Microbiol. Rep. 2018, 10, 49–56. [Google Scholar] [CrossRef]
- Hall, R.S.; Fedorov, A.A.; Xu, C.; Fedorov, E.V.; Almo, S.C.; Raushel, F.M. Three-dimensional structure and catalytic mechanism of cytosine deaminase. Biochemistry 2011, 50, 5077–5085. [Google Scholar] [CrossRef][Green Version]
- Urbeliene, N.; Tiškus, M.; Tamulaitiene, G.; Gasparavičiute, R.; Lapinskaite, R.; Jauniškis, V.; Sudžius, J.; Meškiene, R.; Tauraite, D.; Skrodenyte, E.; et al. Cytidine deaminases catalyze the conversion of N(S,O)4-substituted pyrimidine nucleosides. Sci. Adv. 2023, 9, eade4361. [Google Scholar] [CrossRef]
- Cohen, R.M.; Wolfenden, R. Cytidine deaminase from Escherichia coli. Purification, properties and inhibition by the potential transition state analog 3,4,5,6-tetrahydrouridine. J. Biol. Chem. 1971, 246, 7561–7565. [Google Scholar] [CrossRef]
- Casadesús, J.; Sánchez-Romero, M.A. DNA methylation in prokaryotes. In DNA Methyltransferases—Role and Function; Advances in Experimental Medicine and Biology; Jeltsch, A., Jurkowska, R.Z., Eds.; Springer: Cham, Switzerland, 2022; Volume 1389, pp. 21–43. [Google Scholar]
- Meng, W.Y.; Wang, Z.X.; Zhang, Y.; Hou, Y.; Xue, J.H. Epigenetic marks or not? The discovery of novel DNA modifications in eukaryotes. J. Biol. Chem. 2024, 300, 106791. [Google Scholar] [CrossRef] [PubMed]
- Toh, J.D.W.; Crossley, S.W.M.; Bruemmer, K.J.; Ge, E.J.; He, D.; Iovan, D.A.; Chang, C.J. Distinct RNA N-demethylation pathways catalyzed by conheme iron ALKBH5 and FTO enzymes enable regulation of formaldehyde release rates. Proc. Natl. Acad. Sci. USA 2020, 117, 25284–25292. [Google Scholar] [CrossRef] [PubMed]
- Najmi, A.A.; Bischoff, R.; Permentier, H.P. N-Dealkylation of amines. Molecules 2022, 27, 3293. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Liu, H.; Dai, W.; Liu, X.; Liu, J.; Yu, B. Specific N-demethylation of verapamil by Cytochrome P450 from Streptomyces griseus ATCC 13273. Eng. Life Sci. 2019, 19, 292–301. [Google Scholar] [CrossRef]
- Ensfelder, T.T.; Kurz, M.Q.; Iwan, K.; Geiger, S.; Matheisl, S.; Müller, M.; Beckmann, R.; Carell, T. ALKBH5-induced demethylation of mono- and dimethylated adenosine. Chem. Comm. 2018, 54, 8591–8593. [Google Scholar] [CrossRef]
- Shen, D.; Wang, B.; Gao, Y.; Zhao, L.; Bi, Y.; Zhang, J.; Wang, N.; Kang, H.; Pang, J.; Liu, Y.; et al. Detailed resume of RNA M6A demethylases. Acta Pharm. Sin. B 2022, 12, 2193–2205. [Google Scholar] [CrossRef]
- Moore, L.R.; Caspi, R.; Boyd, D.; Berkmen, M.; Mackie, A.; Paley, S.; Karp, P.D. Revisiting the Y-ome of Escherichia coli. Nucleic Acids Res. 2024, 52, 12201–12207. [Google Scholar] [CrossRef]
- Karp, P.D.; Paley, S.; Caspi, R.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Moore, L.R.; Subhraveti, P.; Gama-Castro, S.; Tierrafria, V.H.; et al. The EcoCyc database (2023). EcoSal Plus 2023, 11, esp-0002. [Google Scholar] [CrossRef]
- Carbon, S.; Douglass, E.; Good, B.M.; Unni, D.R.; Harris, N.L.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; Hartline, E.; et al. The gene ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Stanislauskienė, R.; Laurynėnas, A.; Rutkienė, R.; Aučynaitė, A.; Tauraitė, D.; Meškienė, R.; Urbelienė, N.; Kaupinis, A.; Valius, M.; Kaliniene, L.; et al. YqfB protein from Escherichia coli: An atypical amidohydrolase active towards N4-acylcytosine derivatives. Sci. Rep. 2020, 10, 788. [Google Scholar] [CrossRef]
- Serviene, E.; Lukša, J.; Orentaite, I.; Lafontaine, D.L.J.; Urbonavičius, J. Screening the budding yeast genome reveals unique factors affecting K2 toxin susceptibility. PLoS ONE 2012, 7, e50779. [Google Scholar] [CrossRef] [PubMed]
- Van Opijnen, T.; Levin, H.L. Transposon insertion sequencing, a global measure of gene function. Annu. Rev. Genet. 2020, 54, 337–365. [Google Scholar] [CrossRef] [PubMed]
- Augustin, M.M.; Augustin, J.M.; Brock, J.R.; Kutchan, T.M. Enzyme morphinan N-demethylase for more sustainable opiate processing. Nat. Sustain. 2019, 2, 465–474. [Google Scholar] [CrossRef]
- Aučynaite, A.; Rutkiene, R.; Tauraite, D.; Meškys, R.; Urbonavičius, J. Discovery of bacterial deaminases that convert 5-fluoroisocytosine into 5-fluorouracil. Front. Microbiol. 2018, 9, 2375. [Google Scholar] [CrossRef]
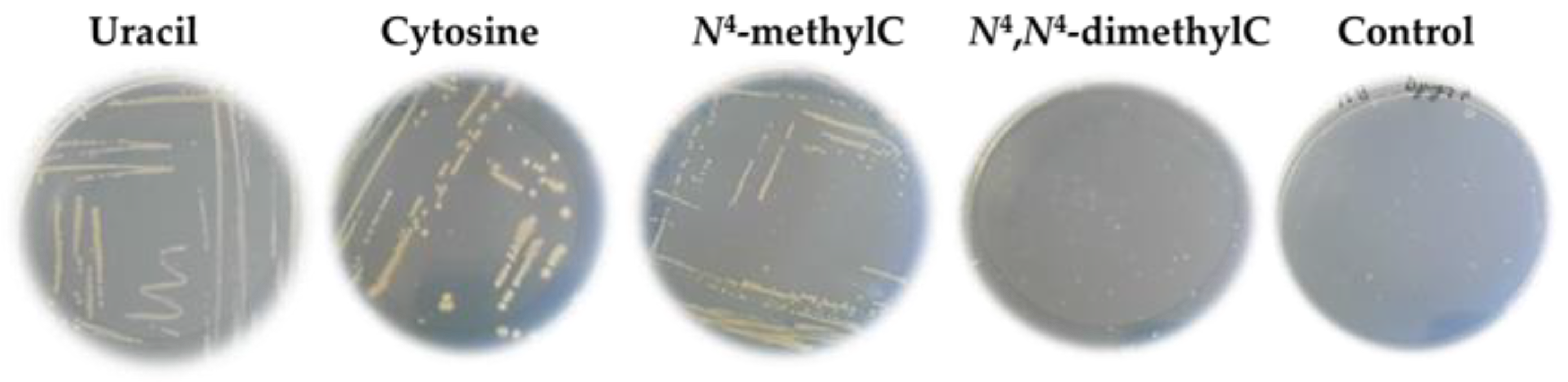
 ; cytosine
; cytosine  ; N4-methylcytosine
; N4-methylcytosine  ; or N4,N4-dimethylcytosine
; or N4,N4-dimethylcytosine  . Error bars show the standard deviation of the three experiments.
. Error bars show the standard deviation of the three experiments.
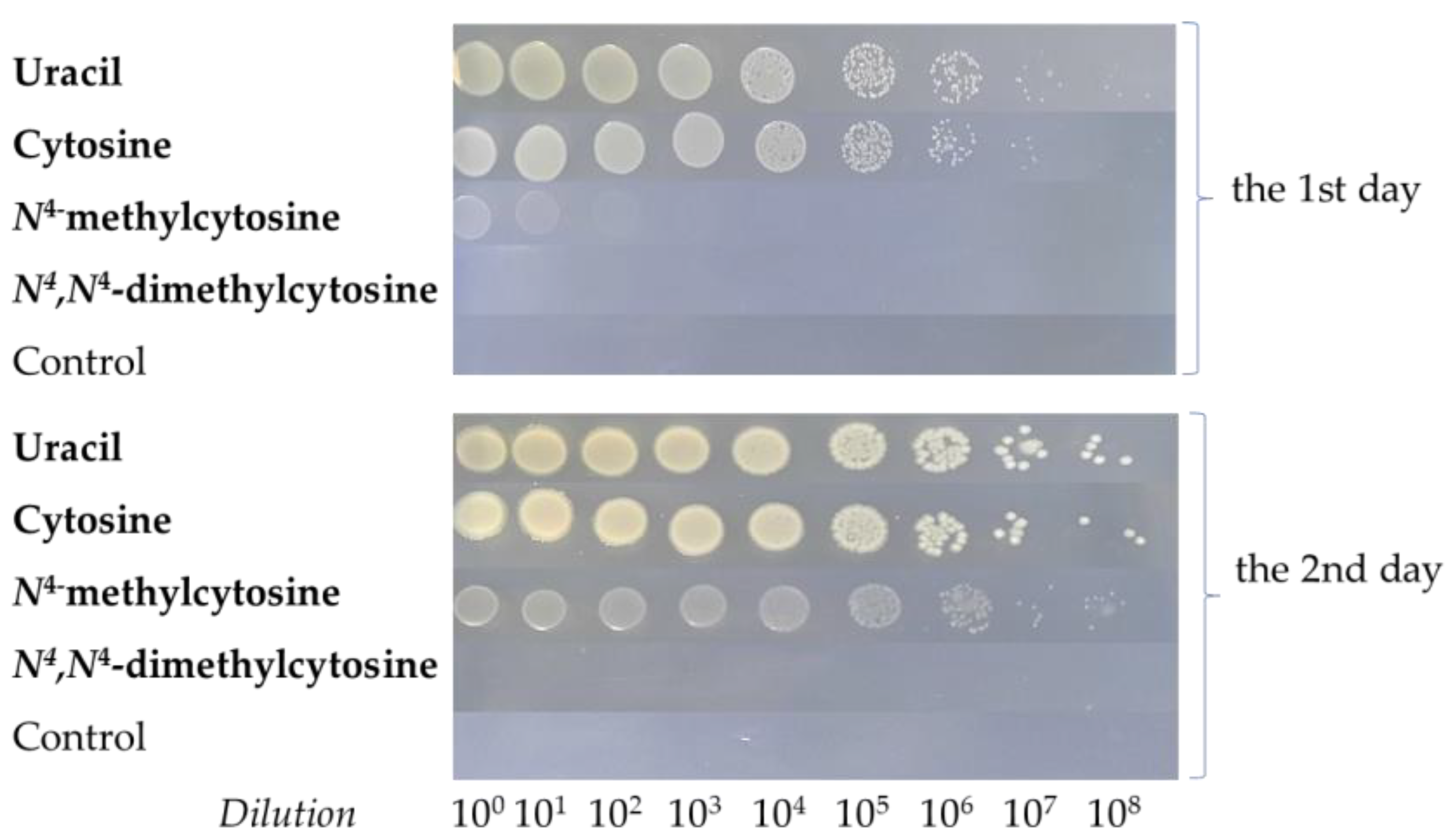
 ; cytosine
; cytosine  ; N4-methylcytosine
; N4-methylcytosine  ; or N4,N4-dimethylcytosine
; or N4,N4-dimethylcytosine  . Error bars show the standard deviation of the three experiments.
. Error bars show the standard deviation of the three experiments.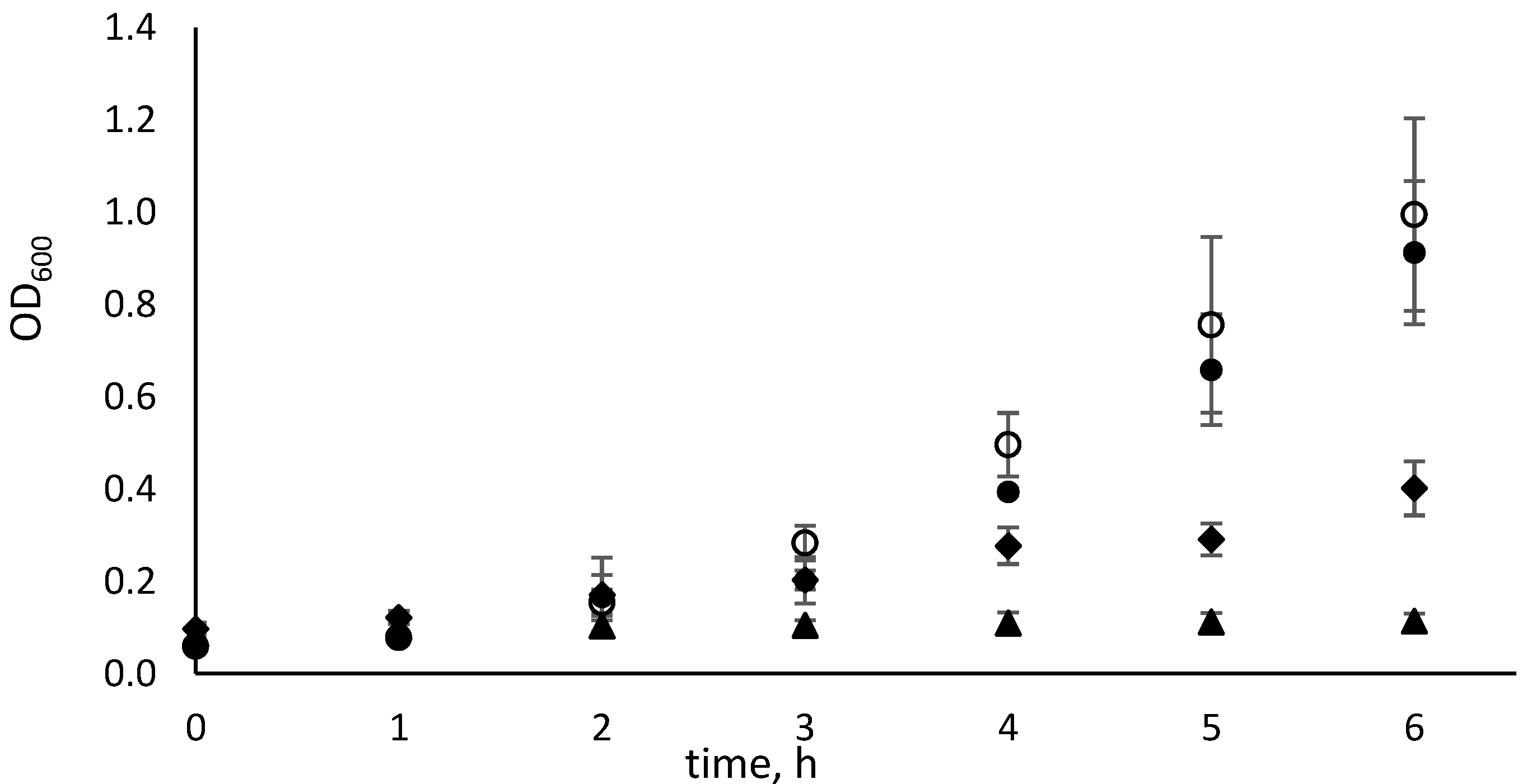
| Uracil | Cytosine | N4-MethylC | N4,N4-DimethylC | |
|---|---|---|---|---|
| Generation time, h | 1.53 ± 0.04 | 1.49 ± 0.10 | 2.99 ± 0.59 | 33.03 ± 11.41 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urbonavičius, J.; Čekytė, A.; Tauraitė, D. N4-Methylcytosine Supports the Growth of Escherichia coli Uracil Auxotrophs. Int. J. Mol. Sci. 2025, 26, 1812. https://doi.org/10.3390/ijms26051812
Urbonavičius J, Čekytė A, Tauraitė D. N4-Methylcytosine Supports the Growth of Escherichia coli Uracil Auxotrophs. International Journal of Molecular Sciences. 2025; 26(5):1812. https://doi.org/10.3390/ijms26051812
Chicago/Turabian StyleUrbonavičius, Jaunius, Aušrinė Čekytė, and Daiva Tauraitė. 2025. "N4-Methylcytosine Supports the Growth of Escherichia coli Uracil Auxotrophs" International Journal of Molecular Sciences 26, no. 5: 1812. https://doi.org/10.3390/ijms26051812
APA StyleUrbonavičius, J., Čekytė, A., & Tauraitė, D. (2025). N4-Methylcytosine Supports the Growth of Escherichia coli Uracil Auxotrophs. International Journal of Molecular Sciences, 26(5), 1812. https://doi.org/10.3390/ijms26051812







