Overexpression of TCP5 or Its Dominant Repressor Form, TCP5-SRDX, Causes Male Infertility in Arabidopsis
Abstract
1. Introduction
2. Results
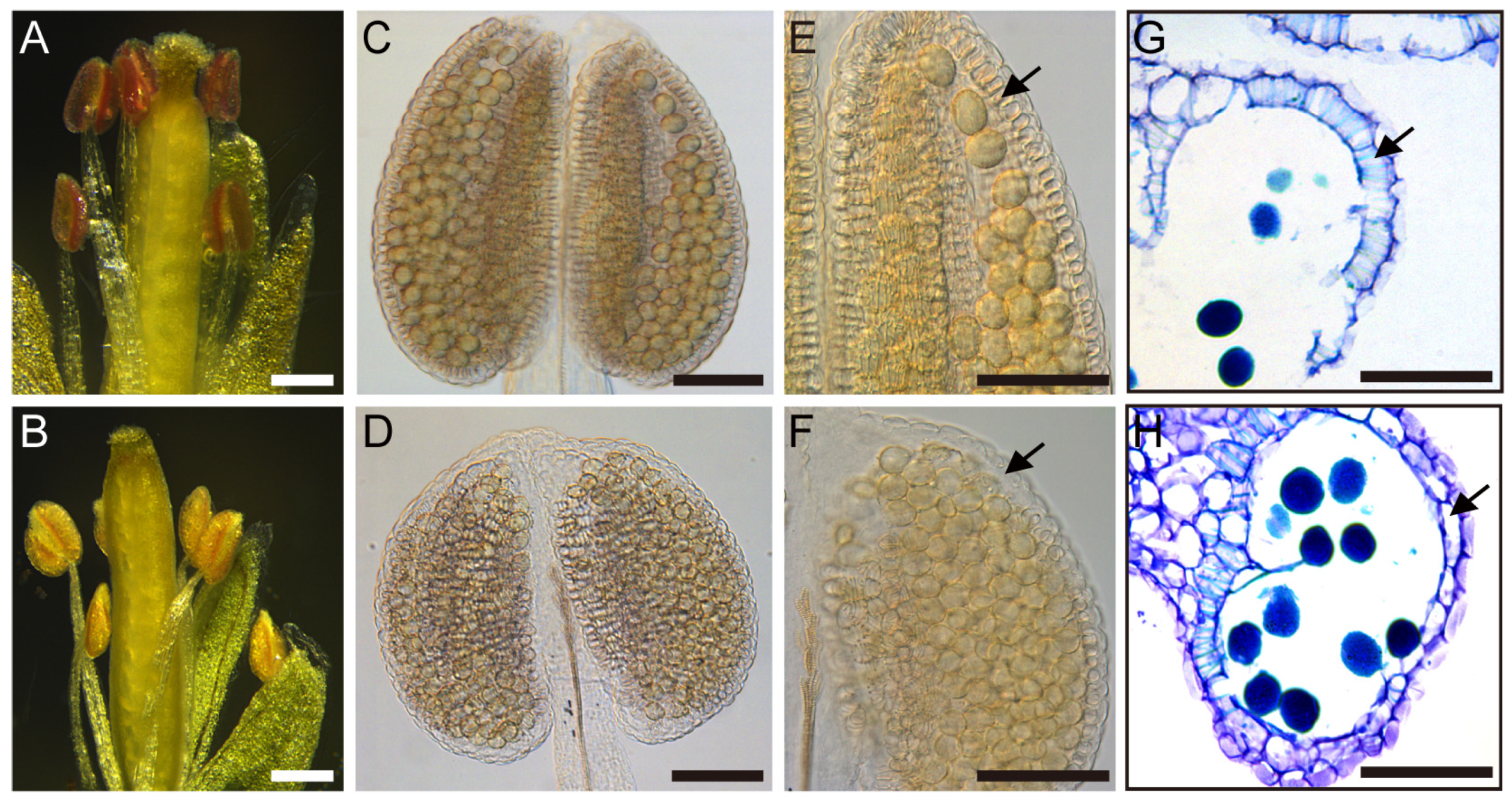
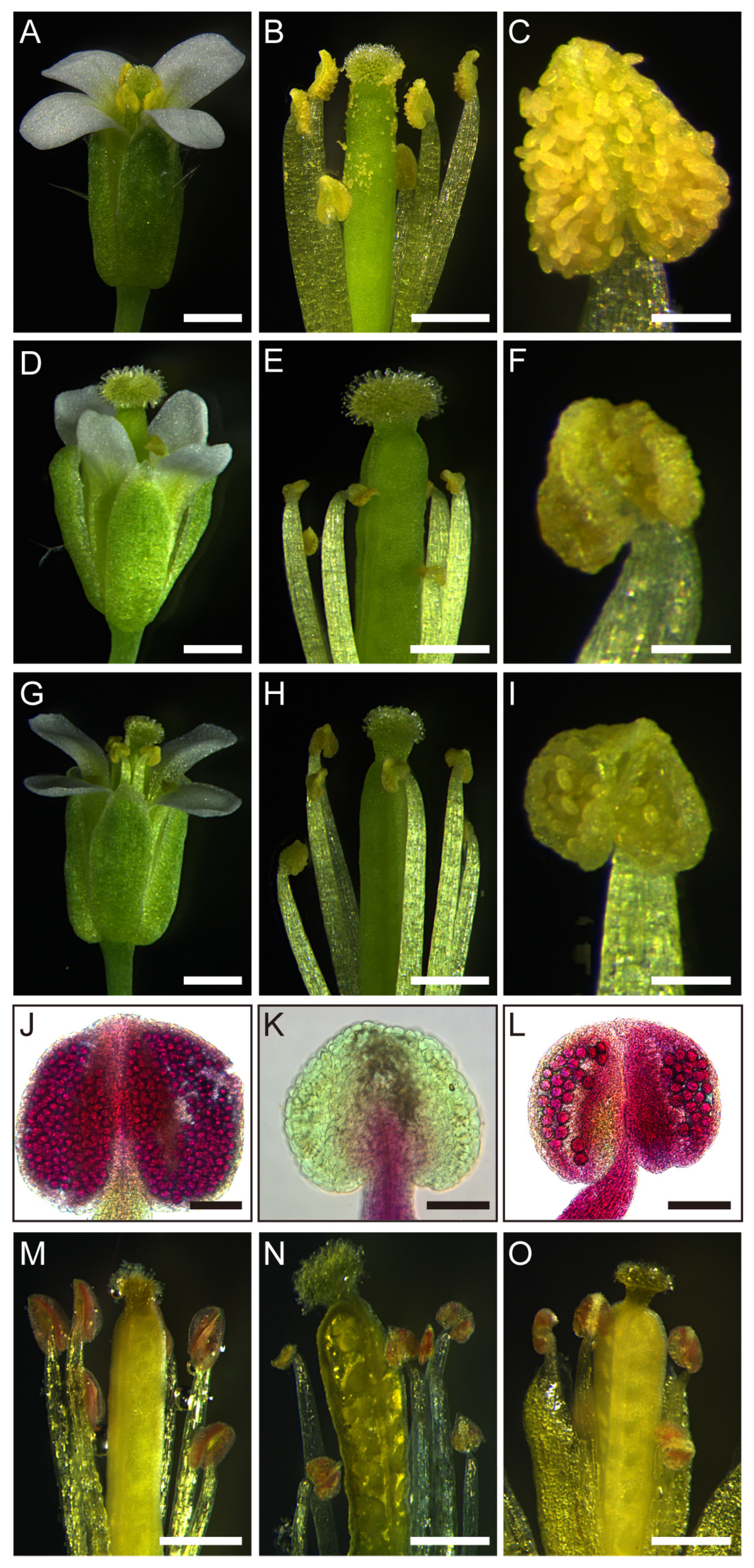
2.1. Overexpression of TCP5 Resulted in Male Sterility
2.2. TCP5 Overexpression Reduced Lignin Accumulation in the Anther Endothecium
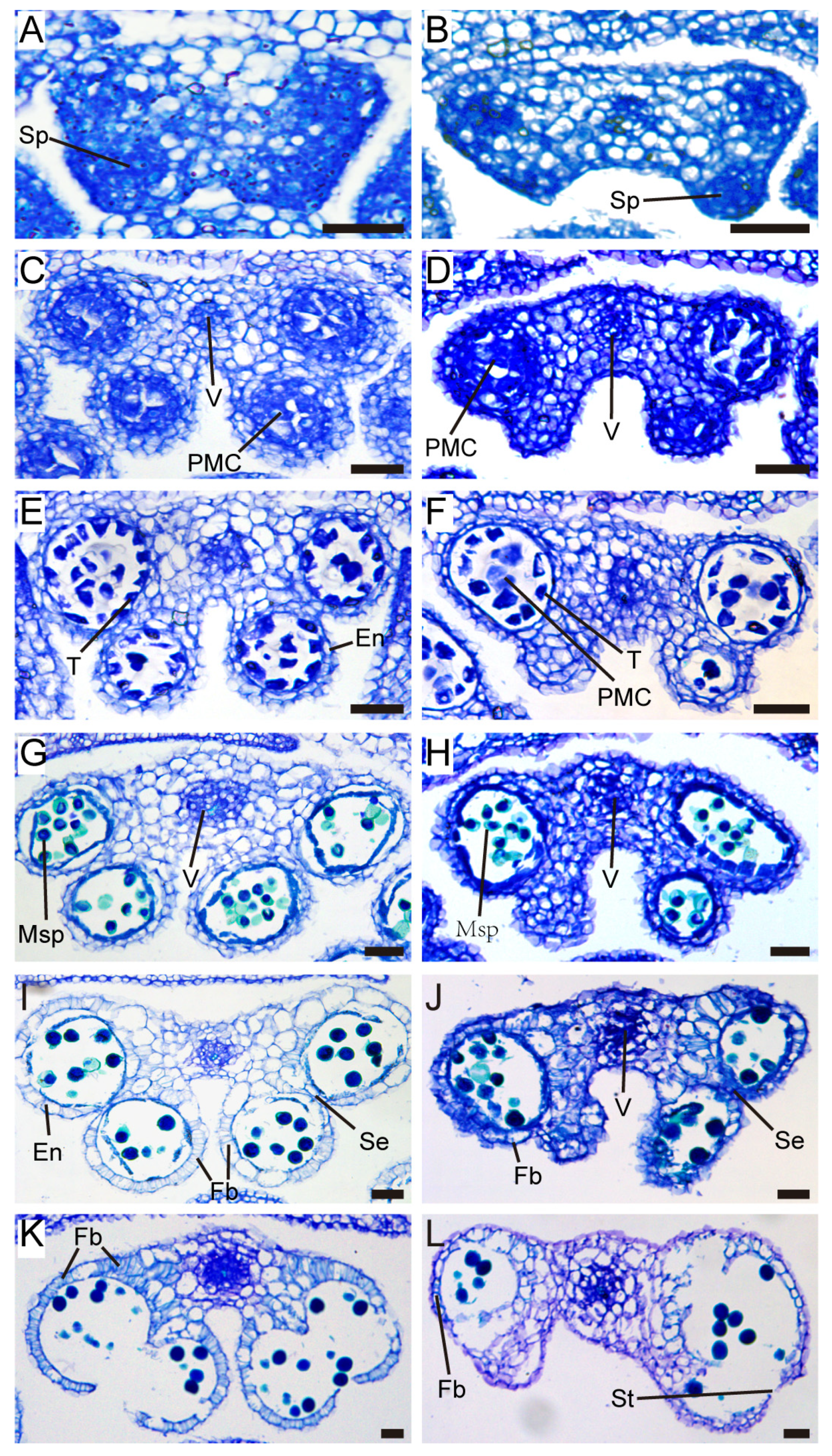
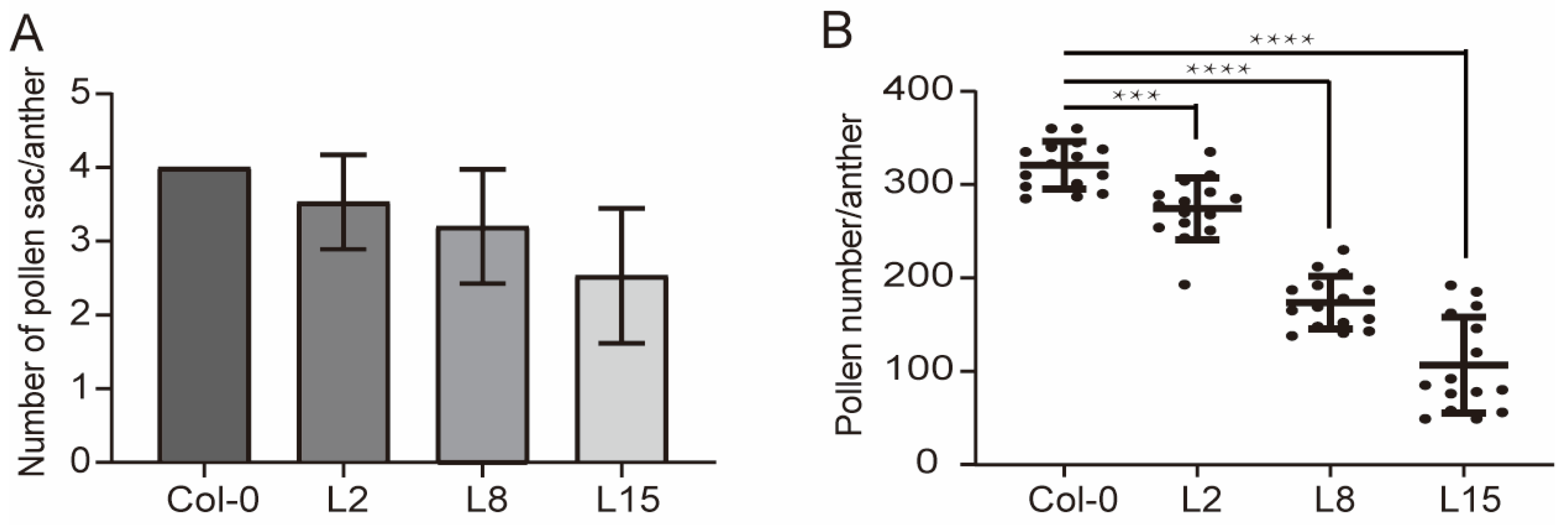
2.3. TCP5 Overexpression Resulted in Fewer Pollen Sacs and Pollen
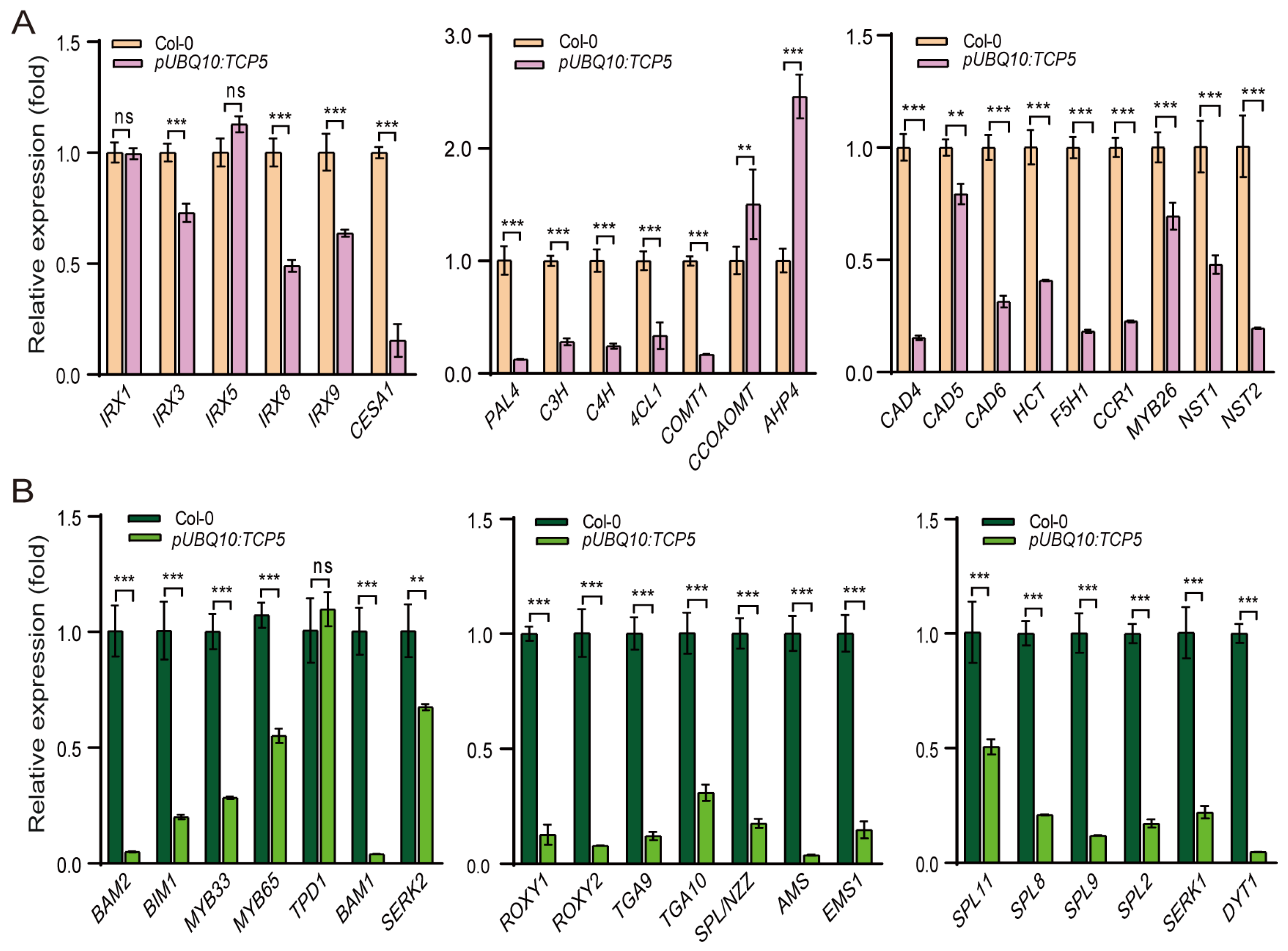
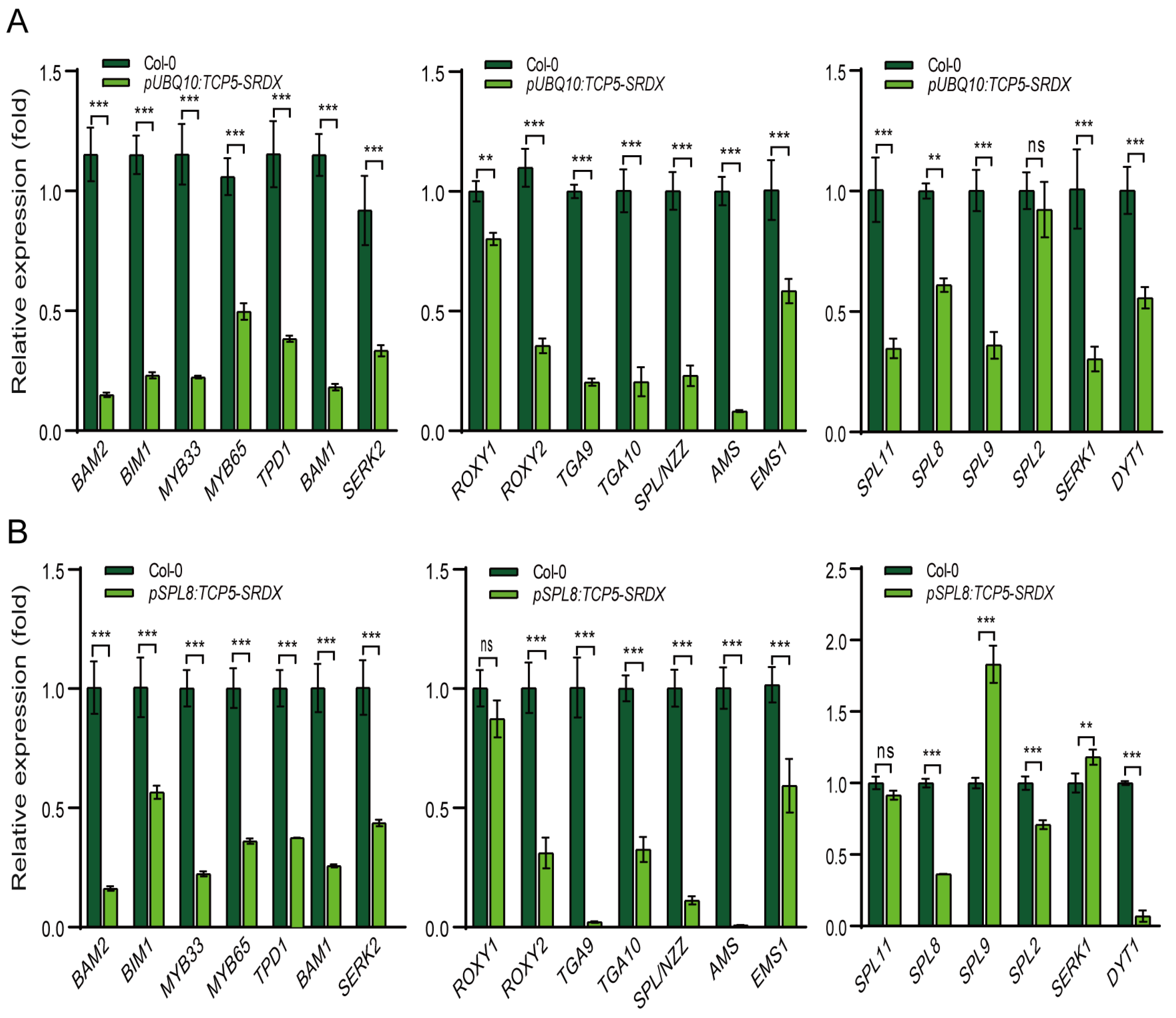
2.4. TCP5 Overexpression Affected the Expressions of Genes Involved in Secondary Cell Wall Formation and Other Key Anther Genes
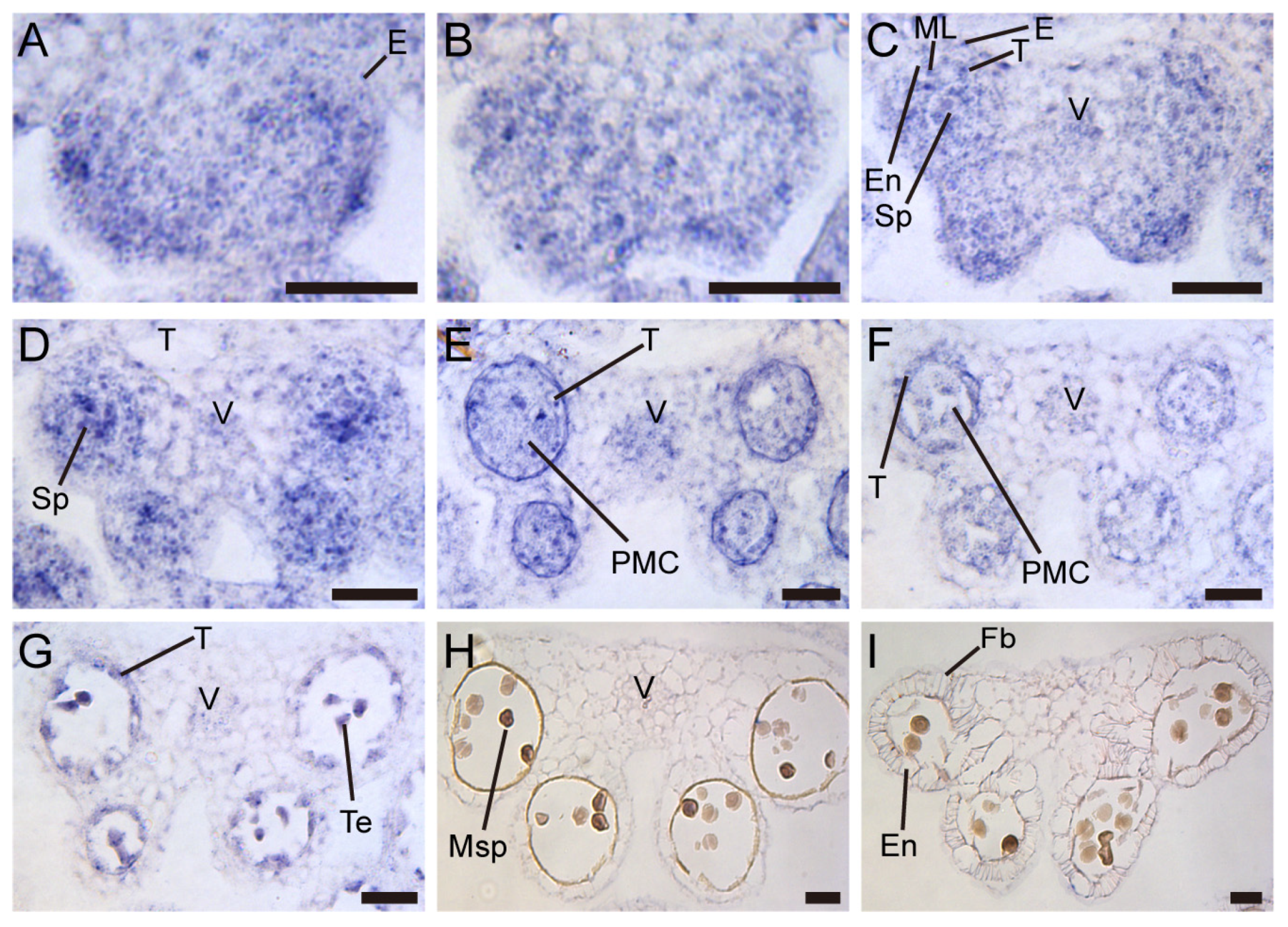
2.5. TCP5 Was Expressed in Early Anthers
2.6. TCP5-SRDX Transgenic Plants Also Had Male Fertility Defects
2.7. TCP5-SRDX Transgenic Plants Exhibited Altered Expression of Early-Anther Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Plasmid Construction and Transgenic Plants
4.3. Histology, Histochemistry, and Microscopy
4.4. In Situ Hybridization
4.5. RNA Extraction and qRT-PCR Analysis
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- González-Grandío, E.; Cubas, P. TCP Transcription Factors: Evolution, Structure, and Biochemical Function. In Plant Transcription Factors; González, D., Ed.; Elsevier Inc.: London, UK, 2015; pp. 139–151. [Google Scholar] [CrossRef]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999, 18, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Manassero, N.G.U.; Viola, I.L.; Welchen, E.; Gonzalez, D.H. TCP transcription factors: Architectures of plant form. Biomol. Concepts 2013, 4, 111–127. [Google Scholar] [CrossRef]
- Viola, I.L.; Gonzalez, D.H. TCP transcription factors in plant reproductive development: Juggling multiple roles. Biomolecules 2023, 13, 750. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Zheng, Y.; Lu, W.; Li, J.; Duan, Y.; Zhang, S.; Wang, Y. Roles of miR319-regulated TCPs in plant development and response to abiotic stress. Crop J. 2021, 9, 17–28. [Google Scholar] [CrossRef]
- Schommer, C.; Debernardi, J.M.; Bresso, E.G.; Rodriguez, R.E.; Palatnik, J.F. Repression of cell proliferation by miR319-regulated TCP4. Mol. Plant 2014, 7, 1533–1544. [Google Scholar] [CrossRef]
- Huang, T.; Irish, V.F. Temporal control of plant organ growth by TCP transcription factors. Curr. Biol. 2015, 25, 1765–1770. [Google Scholar] [CrossRef]
- Zhang, W.; Cochet, F.; Ponnaiah, M.; Lebreton, S.; Matheron, L.; Pionneau, C.; Boudsocq, M.; Resentini, F.; Huguet, S.; Blazquez, M.A.; et al. The MPK8-TCP14 pathway promotes seed germination in Arabidopsis. Plant J. 2019, 100, 677–692. [Google Scholar] [CrossRef]
- Xu, H.; Lantzouni, O.; Bruggink, T.; Benjamins, R.; Lanfermeijer, F.; Denby, K.; Schwechheimer, C.; Bassel, G.W. A molecular signal integration network underpinning Arabidopsis seed germination. Curr. Biol. 2020, 30, 3703–3712. [Google Scholar] [CrossRef]
- Tatematsu, K.; Nakabayashi, K.; Kamiya, Y.; Nambara, E. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 2008, 53, 42–52. [Google Scholar] [CrossRef]
- He, Z.; Zhao, X.; Kong, F.; Zuo, Z.; Liu, X. TCP2 positively regulates HY5/HYH and photomorphogenesis in Arabidopsis. J. Exp. Bot. 2016, 67, 775–785. [Google Scholar] [CrossRef]
- Zhou, Y.; Xun, Q.; Zhang, D.; Lv, M.; Ou, Y.; Li, J. TCP transcription factors associate with PHYTOCHROME INTERACTING FACTOR 4 and CRYPTOCHROME 1 to regulate thermomorphogenesis in Arabidopsis thaliana. iScience 2019, 15, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Challa, K.R.; Aggarwal, P.; Nath, U. Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 2016, 28, 2117–2130. [Google Scholar] [CrossRef] [PubMed]
- Camoirano, A.; Arce, A.L.; Ariel, F.D.; Alem, A.L.; Gonzalez, D.H.; Viola, I.L. Class I TCP transcription factors regulate trichome branching and cuticle development in Arabidopsis. J. Exp. Bot. 2020, 71, 5438–5453. [Google Scholar] [CrossRef]
- Vadde, B.V.L.; Challa, K.R.; Sunkara, P.; Hegde, A.S.; Nath, U. The TCP4 transcription factor directly activates TRICHOMELESS1 and 2 and suppresses trichome initiation. Plant Physiol. 2019, 181, 1587–1599. [Google Scholar] [CrossRef]
- Baulies, J.L.; Bresso, E.G.; Goldy, C.; Palatnik, J.F.; Schommer, C. Potent inhibition of TCP transcription factors by miR319 ensures proper root growth in Arabidopsis. Plant Mol. Biol. 2022, 108, 93–103. [Google Scholar] [CrossRef]
- Yu, H.; Zhang, L.; Wang, W.; Tian, P.; Wang, W.; Wang, K.; Gao, Z.; Liu, S.; Zhang, Y.; Irish, V.F.; et al. TCP5 controls leaf margin development by regulating KNOX and BEL-like transcription factors in Arabidopsis. J. Exp. Bot. 2021, 72, 1809–1821. [Google Scholar] [CrossRef]
- Koyama, T.; Sato, F.; Ohme-Takagi, M. A role of TCP1 in the longitudinal elongation of leaves in Arabidopsis. Biosci. Biotechnol. Biochem. 2010, 74, 2145–2147. [Google Scholar] [CrossRef]
- Kieffer, M.; Master, V.; Waites, R.; Davies, B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 2011, 68, 147–158. [Google Scholar] [CrossRef]
- Danisman, S.; Van der Wal, F.; Dhondt, S.; Waites, R.; De Folter, S.; Bimbo, A.; Van Dijk, A.D.; Muino, J.M.; Cutri, L.; Dornelas, M.C.; et al. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 2012, 159, 1511–1523. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Martinez, J.A.; Sinha, N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front. Plant Sci. 2013, 4, 406. [Google Scholar] [CrossRef]
- Tao, Q.; Guo, D.; Wei, B.; Zhang, F.; Pang, C.; Jiang, H.; Zhang, J.; Wei, T.; Gu, H.; Qu, L.J.; et al. The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 2013, 25, 421–437. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Sato, F.; Ohme-Takagi, M. Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol. 2017, 175, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Bresso, E.G.; Chorostecki, U.; Rodriguez, R.E.; Palatnik, J.F.; Schommer, C. Spatial control of gene expression by miR319-regulated TCP transcription factors in leaf development. Plant Physiol. 2018, 176, 1694–1708. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, L.; Song, S.; Zhao, Y.N.; Gu, H.H.; Zhu, Z.; Wang, J.; Lu, S. ORANGE interplays with TCP7 to regulate endoreduplication and leaf size. Plant J. 2024, 120, 473–490. [Google Scholar] [CrossRef] [PubMed]
- Finlayson, S.A.; Physiology, C. Arabidopsis TEOSINTE BRANCHED1-LIKE 1 regulates axillary bud outgrowth and is homologous to monocot TEOSINTE BRANCHED1. Plant Cell Physiol. 2007, 48, 667–677. [Google Scholar] [CrossRef]
- Gastaldi, V.; Nicolas, M.; Munoz-Gasca, A.; Cubas, P.; Gonzalez, D.H.; Lucero, L. Class I TCP transcription factors TCP14 and TCP15 promote axillary branching in Arabidopsis by counteracting the action of Class II TCP BRANCHED1. New Phytol. 2024, 243, 1810–1822. [Google Scholar] [CrossRef]
- Li, X.; Zhang, G.; Liang, Y.; Hu, L.; Zhu, B.; Qi, D.; Cui, S.; Zhao, H. TCP7 interacts with Nuclear Factor-Ys to promote flowering by directly regulating SOC1 in Arabidopsis. Plant J. 2021, 108, 1493–1506. [Google Scholar] [CrossRef]
- Li, D.; Zhang, H.; Mou, M.; Chen, Y.; Xiang, S.; Chen, L.; Yu, D. Arabidopsis Class II TCP transcription factors integrate with the FT-FD module to control flowering. Plant Physiol. 2019, 181, 97–111. [Google Scholar] [CrossRef]
- Wang, X.; Xu, X.; Mo, X.; Zhong, L.; Zhang, J.; Mo, B.; Kuai, B. Overexpression of TCP8 delays Arabidopsis flowering through a FLOWERING LOCUS C-dependent pathway. BMC Plant Biol. 2019, 19, 534. [Google Scholar] [CrossRef]
- Wu, J.F.; Tsai, H.L.; Joanito, I.; Wu, Y.C.; Chang, C.W.; Li, Y.H.; Wang, Y.; Hong, J.C.; Chu, J.W.; Hsu, C.P.; et al. LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 2016, 7, 13181. [Google Scholar] [CrossRef]
- Nicolas, M.; Cubas, P. TCP factors: New kids on the signaling block. Curr. Opin. Plant Biol. 2016, 33, 33–41. [Google Scholar] [CrossRef] [PubMed]
- van Es, S.W.; Silveira, S.R.; Rocha, D.I.; Bimbo, A.; Martinelli, A.P.; Dornelas, M.C.; Angenent, G.C.; Immink, R.G.H. Novel functions of the Arabidopsis transcription factor TCP5 in petal development and ethylene biosynthesis. Plant J. 2018, 94, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Irish, V.F. An epigenetic timer regulates the transition from cell division to cell expansion during Arabidopsis petal organogenesis. PLoS Genet. 2024, 20, e1011203. [Google Scholar] [CrossRef] [PubMed]
- Nag, A.; King, S.; Jack, T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 22534–22539. [Google Scholar] [CrossRef]
- Gastaldi, V.; Lucero, L.E.; Ferrero, L.V.; Ariel, F.D.; Gonzalez, D.H. Class-I TCP transcription factors activate the SAUR63 gene subfamily in gibberellin-dependent stamen filament elongation. Plant Physiol. 2020, 182, 2096–2110. [Google Scholar] [CrossRef]
- Lucero, L.E.; Uberti-Manassero, N.G.; Arce, A.L.; Colombatti, F.; Alemano, S.G.; Gonzalez, D.H. TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J. 2015, 84, 267–282. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, N.; Lan, J.; Pan, Y.; Jiang, Y.; Wu, Y.; Chen, X.; Feng, X.; Qin, G. Arabidopsis transcription factor TCP4 controls the identity of the apical gynoecium. Plant Cell 2024, 36, 2668–2688. [Google Scholar] [CrossRef]
- Lan, J.; Wang, N.; Wang, Y.; Jiang, Y.; Yu, H.; Cao, X.; Qin, G. Arabidopsis TCP4 transcription factor inhibits high temperature-induced homeotic conversion of ovules. Nat. Commun. 2023, 14, 5673. [Google Scholar] [CrossRef]
- Gastaldi, V.; Alem, A.L.; Mansilla, N.; Ariel, F.D.; Viola, I.L.; Lucero, L.E.; Gonzalez, D.H. BREVIPEDICELLUS/KNAT1 targets TCP15 to modulate filament elongation during Arabidopsis late stamen development. Plant Physiol. 2023, 191, 29–34. [Google Scholar] [CrossRef]
- Fang, Y.; Guo, D.; Wang, Y.; Wang, N.; Fang, X.; Zhang, Y.; Li, X.; Chen, L.; Yu, D.; Zhang, B.; et al. Rice transcriptional repressor OsTIE1 controls anther dehiscence and male sterility by regulating JA biosynthesis. Plant Cell 2024, 36, 1697–1717. [Google Scholar] [CrossRef]
- Wang, H.; Mao, Y.; Yang, J.; He, Y. TCP24 modulates secondary cell wall thickening and anther endothecium development. Front. Plant Sci. 2015, 6, 436. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Pan, Z.; Kong, W.; Mo, B.; Chen, X.; Yu, Y. miR319-TCPs-TGA9/TGA10/ROXY2 regulatory module controls cell fate specification in early anther development in Arabidopsis. Sci. China Life Sci. 2024, 67, 839–842. [Google Scholar] [CrossRef] [PubMed]
- Grefen, C.; Donald, N.; Hashimoto, K.; Kudla, J.; Schumacher, K.; Blatt, M.R. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 2010, 64, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Sanders, P.M.; Bui, A.Q.; Weterings, K.; McIntire, K.N.; Hsu, Y.-C.; Lee, P.Y.; Truong, M.T.; Beals, T.P.; Goldberg, R.B. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1999, 11, 297–322. [Google Scholar] [CrossRef]
- Brown, D.M.; Zeef, L.A.; Ellis, J.; Goodacre, R.; Turner, S.R. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 2005, 17, 2281–2295. [Google Scholar] [CrossRef]
- Hao, Z.; Avci, U.; Tan, L.; Zhu, X.; Glushka, J.; Pattathil, S.; Eberhard, S.; Sholes, T.; Rothstein, G.E.; Lukowitz, W.; et al. Loss of Arabidopsis GAUT12/IRX8 causes anther indehiscence and leads to reduced G lignin associated with altered matrix polysaccharide deposition. Front. Plant Sci. 2014, 5, 357. [Google Scholar] [CrossRef]
- Taylor, N.G. Cellulose biosynthesis and deposition in higher plants. New Phytol. 2008, 178, 239–252. [Google Scholar] [CrossRef]
- Pena, M.J.; Zhong, R.; Zhou, G.K.; Richardson, E.A.; O’Neill, M.A.; Darvill, A.G.; York, W.S.; Ye, Z.H. Arabidopsis irregular xylem8 and irregular xylem9: Implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 2007, 19, 549–563. [Google Scholar] [CrossRef]
- Goujon, T.; Sibout, R.; Eudes, A.; MacKay, J.; Jouanin, L. Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana. Plant Physiol. Biochem. 2003, 41, 677–687. [Google Scholar] [CrossRef]
- Yang, C.; Xu, Z.; Song, J.; Conner, K.; Vizcay Barrena, G.; Wilson, Z.A. Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 2007, 19, 534–548. [Google Scholar] [CrossRef]
- Yang, C.; Song, J.; Ferguson, A.C.; Klisch, D.; Simpson, K.; Mo, R.; Taylor, B.; Mitsuda, N.; Wilson, Z.A. Transcription factor MYB26 is key to spatial specificity in anther secondary thickening formation. Plant Physiol. 2017, 175, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Steiner-Lange, S.; Unte, U.S.; Eckstein, L.; Yang, C.; Wilson, Z.A.; Schmelzer, E.; Dekker, K.; Saedler, H. Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J. 2003, 34, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 2005, 17, 2993–3006. [Google Scholar] [CrossRef]
- Jung, K.W.; Oh, S.-I.; Kim, Y.Y.; Yoo, K.S.; Cui, M.H.; Shin, J.S. Arabidopsis Histidine-containing phosphotransfer factor 4 (AHP4) negatively regulates secondary wall thickening of the anther endothecium during flowering. Mol. Cells 2008, 25, 294–300. [Google Scholar] [CrossRef]
- Yang, S.L.; Xie, L.F.; Mao, H.Z.; Puah, C.S.; Yang, W.C.; Jiang, L.; Sundaresan, V.; Ye, D. Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell 2003, 15, 2792–2804. [Google Scholar] [CrossRef]
- Hord, C.L.; Chen, C.; Deyoung, B.J.; Clark, S.E.; Ma, H. The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 2006, 18, 1667–1680. [Google Scholar] [CrossRef]
- Sorensen, A.M.; Krober, S.; Unte, U.S.; Huijser, P.; Dekker, K.; Saedler, H. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 2003, 33, 413–423. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, Y.; Timofejeva, L.; Chen, C.; Grossniklaus, U.; Ma, H. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 2006, 133, 3085–3095. [Google Scholar] [CrossRef]
- Xing, S.; Zachgo, S. ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J. 2008, 53, 790–801. [Google Scholar] [CrossRef]
- Xing, S.; Salinas, M.; Hohmann, S.; Berndtgen, R.; Huijser, P. MiR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 2010, 22, 3935–3950. [Google Scholar] [CrossRef]
- Murmu, J.; Bush, M.J.; DeLong, C.; Li, S.; Xu, M.; Khan, M.; Malcolmson, C.; Fobert, P.R.; Zachgo, S.; Hepworth, S.R. Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 2010, 154, 1492–1504. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Quodt, V.; Chandler, J.; Hohmann, S.; Berndtgen, R.; Huijser, P. SPL8 acts together with the brassinosteroid-signaling component BIM1 in controlling Arabidopsis thaliana male fertility. Plants 2013, 2, 416–428. [Google Scholar] [CrossRef]
- Unte, U.S.; Sorensen, A.M.; Pesaresi, P.; Gandikota, M.; Leister, D.; Saedler, H.; Huijser, P. SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 2003, 15, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, C.; Russinova, E.; Hecht, V.; Baaijens, E.; De Vries, S. The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 2005, 17, 3337–3349. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.Z.; Wang, G.F.; Speal, B.; Ma, H. The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 2002, 16, 2021–2031. [Google Scholar] [CrossRef]
- Yang, W.-C.; Ye, D.; Xu, J.; Sundaresan, V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 1999, 13, 2108–2117. [Google Scholar] [CrossRef]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef]
- Hiratsu, K.; Matsui, K.; Koyama, T.; Ohme-Takagi, M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003, 34, 733–739. [Google Scholar] [CrossRef]
- Astrand, J.; Knight, C.; Robson, J.; Talle, B.; Wilson, Z.A. Evolution and diversity of the angiosperm anther: Trends in function and development. Plant Reprod. 2021, 34, 307–319. [Google Scholar] [CrossRef]
- Wei, B.; Zhang, J.; Pang, C.; Yu, H.; Guo, D.; Jiang, H.; Ding, M.; Chen, Z.; Tao, Q.; Gu, H.; et al. The molecular mechanism of sporocyteless/nozzle in controlling Arabidopsis ovule development. Cell Res. 2015, 25, 121–134. [Google Scholar] [CrossRef]
- Chen, G.H.; Sun, J.Y.; Liu, M.; Liu, J.; Yang, W.C. SPOROCYTELESS is a novel embryophyte-specific transcription repressor that interacts with TPL and TCP proteins in Arabidopsis. J. Genet. Genom. 2014, 41, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Fauser, F.; Schiml, S.; Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014, 79, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Asseck, L.Y.; Mehlhorn, D.G.; Monroy, J.R.; Ricardi, M.M.; Breuninger, H.; Wallmeroth, N.; Berendzen, K.W.; Nowrousian, M.; Xing, S.; Schwappach, B.; et al. Endoplasmic reticulum membrane receptors of the GET pathway are conserved throughout eukaryotes. Proc. Natl. Acad. Sci. USA 2021, 118, e2017636118. [Google Scholar] [CrossRef]
- Shimada, T.L.; Shimada, T.; Hara-Nishimura, I. A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 2010, 61, 519–528. [Google Scholar] [CrossRef]
- Becker, D.; Kemper, E.; Schell, J.; Masterson, R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 1992, 20, 1195–1197. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Alexander, M.P. Differential staining of aborted and nonaborted pollen. Stain. Technol. 1969, 44, 117–122. [Google Scholar] [CrossRef]
- Pride, L.; Agehara, S. Useful image-based techniques for manual and automatic counting using imageJ for horticultural research. EDIS 2021, 2021, HS1405. [Google Scholar] [CrossRef]
- Xing, S.; Rosso, M.G.; Zachgo, S. ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 2005, 132, 1555–1565. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, T.; Tian, P.; Wang, X.; Li, M.; Xing, S. Overexpression of TCP5 or Its Dominant Repressor Form, TCP5-SRDX, Causes Male Infertility in Arabidopsis. Int. J. Mol. Sci. 2025, 26, 1813. https://doi.org/10.3390/ijms26051813
Li T, Tian P, Wang X, Li M, Xing S. Overexpression of TCP5 or Its Dominant Repressor Form, TCP5-SRDX, Causes Male Infertility in Arabidopsis. International Journal of Molecular Sciences. 2025; 26(5):1813. https://doi.org/10.3390/ijms26051813
Chicago/Turabian StyleLi, Tingting, Ping Tian, Xinxin Wang, Mengyao Li, and Shuping Xing. 2025. "Overexpression of TCP5 or Its Dominant Repressor Form, TCP5-SRDX, Causes Male Infertility in Arabidopsis" International Journal of Molecular Sciences 26, no. 5: 1813. https://doi.org/10.3390/ijms26051813
APA StyleLi, T., Tian, P., Wang, X., Li, M., & Xing, S. (2025). Overexpression of TCP5 or Its Dominant Repressor Form, TCP5-SRDX, Causes Male Infertility in Arabidopsis. International Journal of Molecular Sciences, 26(5), 1813. https://doi.org/10.3390/ijms26051813





