Application of an Integrated Single-Cell and Three-Dimensional Spheroid Culture Platform for Investigating Drug Resistance Heterogeneity and Epithelial–Mesenchymal Transition (EMT) in Lung Cancer Subclones
Abstract
1. Introduction
2. Results
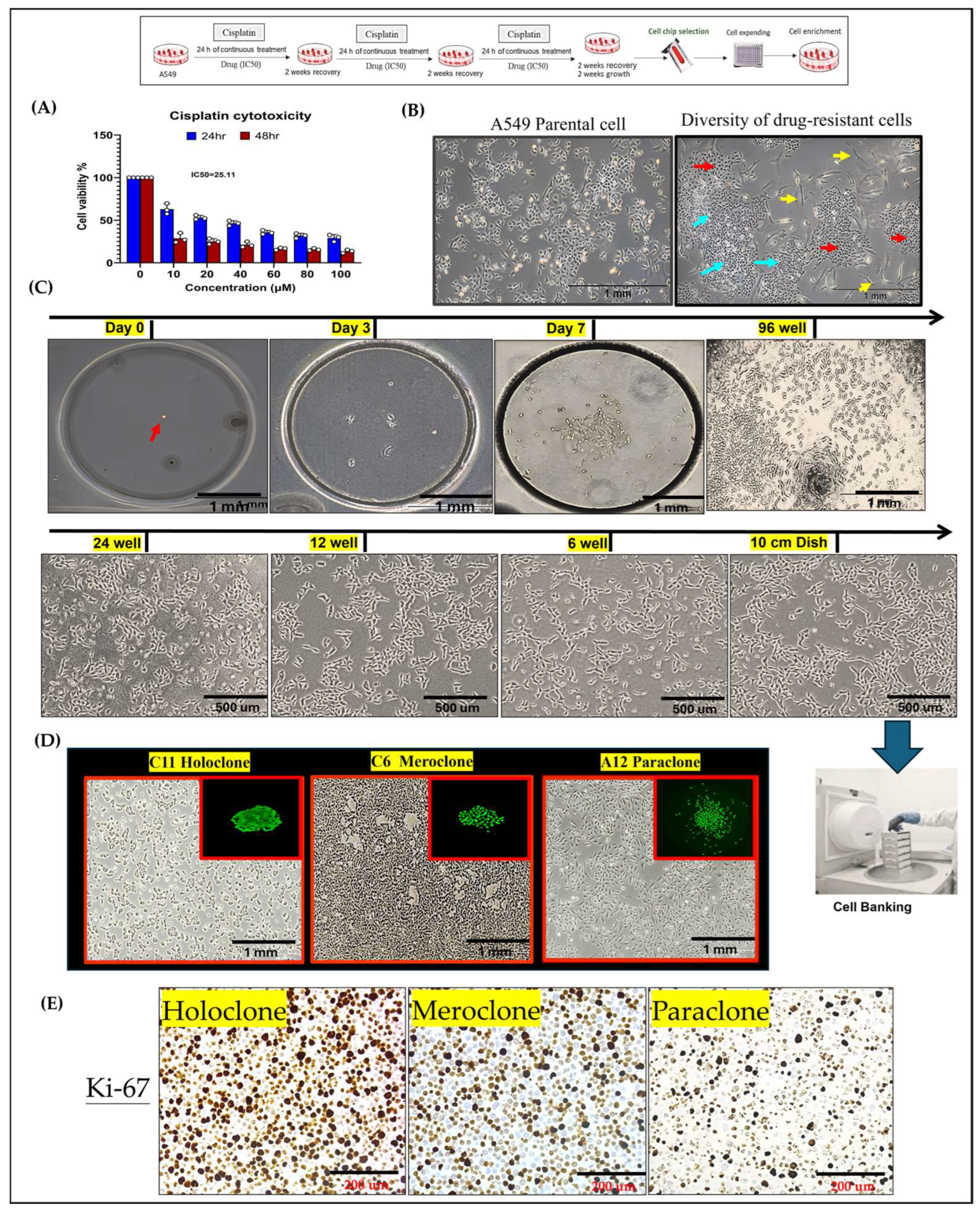
2.1. Integration of Single-Cell Culture Technology for the Establishment and Preservation of Phenotypically Heterogeneous Monoclonal Cell Lines
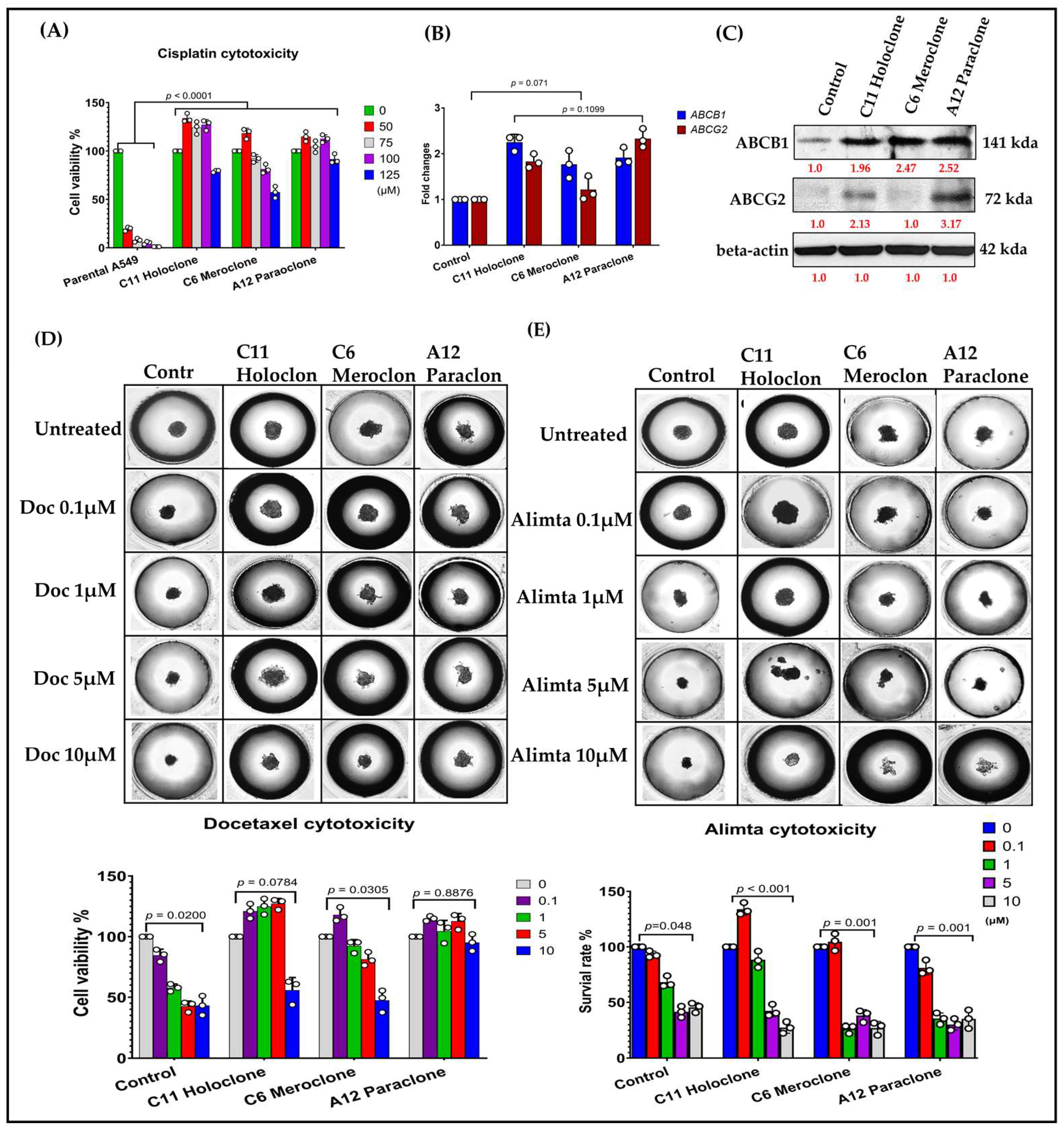
2.2. Phenotypic Heterogeneity of Different Drug-Resistant Subclonal Cells and Their Impact on Chemotherapy Drug Sensitivity
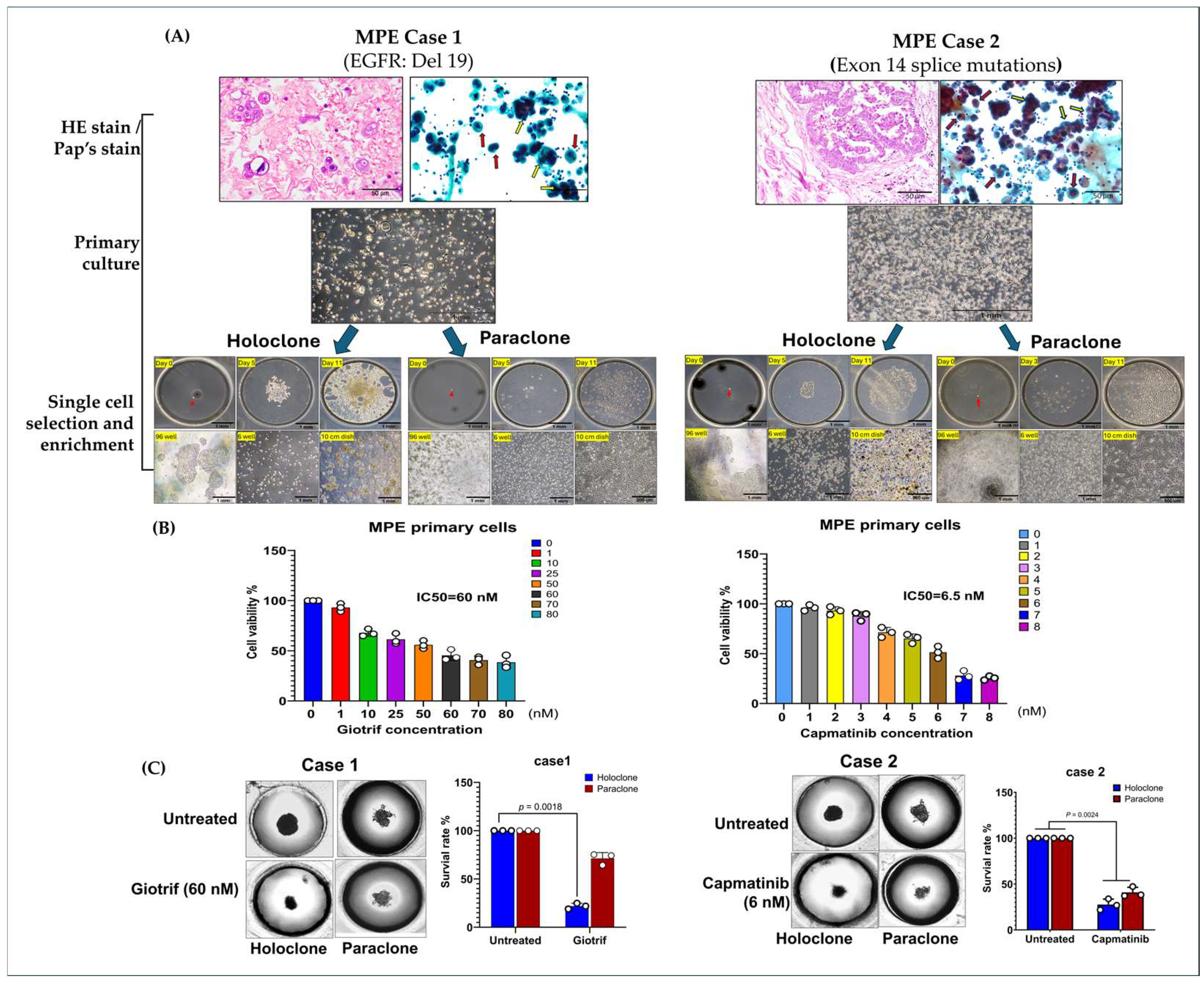
2.3. Evidence of Cellular Phenotypic Heterogeneity and Tumor Spheroid Drug Prediction in Primary MPE Cultures
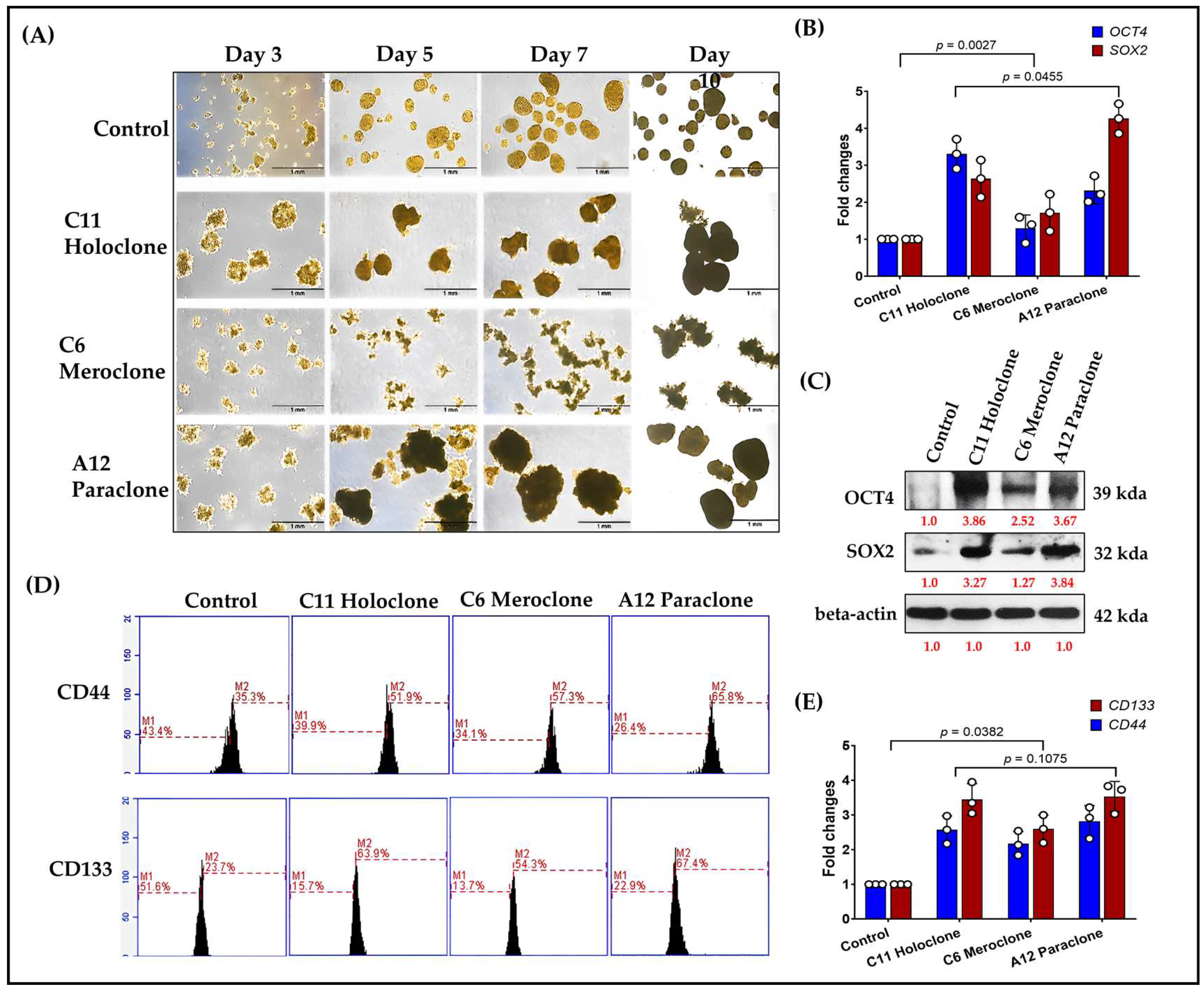
2.4. Multilevel Analysis of Stem Cell Characteristics and Biomarker Expression in Drug-Resistant Subclonal Cells
2.5. Molecular Characterization of EMT Transformation and Enhanced Invasive Capability in Drug-Resistant Subclonal Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Reagent, Drug-Resistant Cancer Cell Lines, and MPE Sample Prepare
4.2. Single-Cell Isolation and Culture Procedure
4.3. 3D Tumorsphere Culture
4.4. Growth Curves of 3D Tumorsphere Detected by CCK8 Assay
4.5. Reverse Transcription Quantitative PCR (RT-qPCR)
4.6. Western Blot Analysis
4.7. Migration and Invasion Assay
4.8. Flow Cytometry
4.9. Immunohistochemistry
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zheng, Z. Understanding the global cancer statistics 2022: Growing cancer burden. Sci. China Life Sci. 2024, 67, 2274–2276. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Xia, C.; Li, H.; Yan, X.; Yang, F.; Cao, M.; Zhang, S.; Teng, Y.; He, S.; Cao, M.; et al. Disparities in 36 cancers across 185 countries: Secondary analysis of global cancer statistics. Front. Med. 2024, 18, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Lim, Z.F.; Ma, P.C. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J. Hematol. Oncol. 2019, 12, 134. [Google Scholar] [CrossRef]
- Bhat, G.R.; Sethi, I.; Sadida, H.Q.; Rah, B.; Mir, R.; Algehainy, N.; Albalawi, I.A.; Masoodi, T.; Subbaraj, G.K.; Jamal, F.; et al. Cancer cell plasticity: From cellular, molecular, and genetic mechanisms to tumor heterogeneity and drug resistance. Cancer Metastasis Rev. 2024, 43, 197–228. [Google Scholar] [CrossRef]
- Zhang, A.; Miao, K.; Sun, H.; Deng, C.X. Tumor heterogeneity reshapes the tumor microenvironment to influence drug resistance. Int. J. Biol. Sci. 2022, 18, 3019–3033. [Google Scholar] [CrossRef]
- Li, S.; Lv, J.; Li, Z.; Zhang, Q.; Lu, J.; Huo, X.; Guo, M.; Liu, X.; Li, C.; Wang, J.; et al. Overcoming multi-drug resistance in SCLC: A synergistic approach with venetoclax and hydroxychloroquine targeting the lncRNA LYPLAL1-DT/BCL2/BECN1 pathway. Mol. Cancer 2024, 23, 243. [Google Scholar] [CrossRef]
- Huang, Y.H.; Chiu, L.Y.; Tseng, J.S.; Hsu, K.H.; Chen, C.H.; Sheu, G.T.; Yang, T.Y. Attenuation of PI3K-Akt-mTOR Pathway to Reduce Cancer Stemness on Chemoresistant Lung Cancer Cells by Shikonin and Synergy with BEZ235 Inhibitor. Int. J. Mol. Sci. 2024, 25, 616. [Google Scholar] [CrossRef]
- Brosseau, S.; Abreu, P.; Bouchez, C.; Charon, L.; Kieffer, Y.; Gentric, G.; Picant, V.; Veith, I.; Camonis, J.; Descroix, S.; et al. YAP/TEAD involvement in resistance to paclitaxel chemotherapy in lung cancer. Mol. Cell. Biochem. 2025, 480, 231–248. [Google Scholar] [CrossRef]
- Wang, D.; Lippard, S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005, 4, 307–320. [Google Scholar] [CrossRef]
- Galluzzi, L.; Senovilla, L.; Vitale, I.; Michels, J.; Martins, I.; Kepp, O.; Castedo, M.; Kroemer, G. Molecular mechanisms of cisplatin resistance. Oncogene 2012, 31, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.W.; Pouliot, L.M.; Hall, M.D.; Gottesman, M.M. Cisplatin resistance: A cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol. Rev. 2012, 64, 706–721. [Google Scholar] [CrossRef] [PubMed]
- Makovec, T. Cisplatin and beyond: Molecular mechanisms of action and drug resistance development in cancer chemotherapy. Radiol. Oncol. 2019, 53, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Wang, Y.; Yang, Z.; Chen, P.; Xu, J.; Tian, Y.; Fan, T.; Xiao, C.; Bai, G.; Li, L.; et al. Genetic trajectory and clonal evolution of multiple primary lung cancer with lymph node metastasis. Cancer Gene Ther. 2023, 30, 507–520. [Google Scholar] [CrossRef]
- Russo, A.; Scilla, K.A.; Mehra, R.; Gittens, A.; McCusker, M.G.; de Miguel-Perez, D.; Gomez, J.E.; Peleg, A.; Del Re, M.; Rolfo, C.D. Tracking Clonal Evolution of EGFR-Mutated Non-Small Cell Lung Cancer Through Liquid Biopsy: Management of C797S Acquired Mutation. Clin. Lung Cancer 2023, 24, 660–665. [Google Scholar] [CrossRef]
- Moore, P.C.; Henderson, K.W.; Classon, M. The epigenome and the many facets of cancer drug tolerance. Adv. Cancer Res. 2023, 158, 1–39. [Google Scholar] [CrossRef]
- Franca, G.S.; Baron, M.; King, B.R.; Bossowski, J.P.; Bjornberg, A.; Pour, M.; Rao, A.; Patel, A.S.; Misirlioglu, S.; Barkley, D.; et al. Cellular adaptation to cancer therapy along a resistance continuum. Nature 2024, 631, 876–883. [Google Scholar] [CrossRef]
- Chmielecki, J.; Gray, J.E.; Cheng, Y.; Ohe, Y.; Imamura, F.; Cho, B.C.; Lin, M.C.; Majem, M.; Shah, R.; Rukazenkov, Y.; et al. Candidate mechanisms of acquired resistance to first-line osimertinib in EGFR-mutated advanced non-small cell lung cancer. Nat. Commun. 2023, 14, 1070. [Google Scholar] [CrossRef]
- Yao, W.; Wang, Z.; Ma, H.; Lin, Y.; Liu, X.; Li, P.; He, X. Epithelial-mesenchymal plasticity (EMP) in wound healing: Exploring EMT mechanisms, regulatory network, and therapeutic opportunities. Heliyon 2024, 10, e34269. [Google Scholar] [CrossRef]
- Sisto, M.; Lisi, S. Epigenetic Regulation of EMP/EMT-Dependent Fibrosis. Int. J. Mol. Sci. 2024, 25, 2775. [Google Scholar] [CrossRef]
- Mishra, A.B.; Nishank, S.S. Therapeutic targeting approach on epithelial-mesenchymal plasticity to combat cancer metastasis. Med. Oncol. 2023, 40, 190. [Google Scholar] [CrossRef] [PubMed]
- Haerinck, J.; Goossens, S.; Berx, G. The epithelial-mesenchymal plasticity landscape: Principles of design and mechanisms of regulation. Nat. Rev. Genet. 2023, 24, 590–609. [Google Scholar] [CrossRef] [PubMed]
- Acloque, H.; Yang, J.; Theveneau, E. Epithelial-to-mesenchymal plasticity from development to disease: An introduction to the special issue. Genesis 2024, 62, e23581. [Google Scholar] [CrossRef] [PubMed]
- Toyokawa, G.; Bersani, F.; Bironzo, P.; Picca, F.; Tabbo, F.; Haratake, N.; Takenaka, T.; Seto, T.; Yoshizumi, T.; Novello, S.; et al. Tumor plasticity and therapeutic resistance in oncogene-addicted non-small cell lung cancer: From preclinical observations to clinical implications. Crit. Rev. Oncol. Hematol. 2023, 184, 103966. [Google Scholar] [CrossRef]
- Shi, Z.D.; Pang, K.; Wu, Z.X.; Dong, Y.; Hao, L.; Qin, J.X.; Wang, W.; Chen, Z.S.; Han, C.H. Tumor cell plasticity in targeted therapy-induced resistance: Mechanisms and new strategies. Signal Transduct. Target. Ther. 2023, 8, 113. [Google Scholar] [CrossRef]
- Ramisetty, S.; Subbalakshmi, A.R.; Pareek, S.; Mirzapoiazova, T.; Do, D.; Prabhakar, D.; Pisick, E.; Shrestha, S.; Achuthan, S.; Bhattacharya, S.; et al. Leveraging Cancer Phenotypic Plasticity for Novel Treatment Strategies. J. Clin. Med. 2024, 13, 3337. [Google Scholar] [CrossRef]
- McDonald, P.C.; Dedhar, S. Persister cell plasticity in tumour drug resistance. Semin. Cell Dev. Biol. 2024, 156, 1–10. [Google Scholar] [CrossRef]
- He, J.; Qiu, Z.; Fan, J.; Xie, X.; Sheng, Q.; Sui, X. Drug tolerant persister cell plasticity in cancer: A revolutionary strategy for more effective anticancer therapies. Signal Transduct. Target. Ther. 2024, 9, 209. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, X.; Deng, H.; Liu, L.; Xiang, Y.; Feng, J. Overcoming cancer drug-resistance calls for novel strategies targeting abnormal alternative splicing. Pharmacol. Ther. 2024, 261, 108697. [Google Scholar] [CrossRef]
- Xu, X.; Liu, Y.; Gong, Q.; Ma, L.; Wei, W.; Zhao, L.; Luo, Z. PARP1 promotes EGFR-TKI drug-resistance via PI3K/AKT pathway in non-small-cell lung cancer. Cancer Chemother. Pharmacol. 2024, 94, 209–221. [Google Scholar] [CrossRef]
- Zhang, K.; Xi, J.; Wang, Y.; Xue, J.; Li, B.; Huang, Z.; Zheng, Z.; Liang, N.; Wei, Z. A Microfluidic Chip-Based Automated System for Whole-Course Monitoring the Drug Responses of Organoids. Anal. Chem. 2024, 96, 10092–10101. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xiao, X.; Zeng, L.; Zeng, H.; Zheng, Y.; Wang, J.; Li, G.; Dai, W.; He, Y.; Wang, S.; et al. Integrating cfDNA liquid biopsy and organoid-based drug screening reveals PI3K signaling as a promising therapeutic target in colorectal cancer. J. Transl. Med. 2024, 22, 132. [Google Scholar] [CrossRef] [PubMed]
- Piro, G.; Agostini, A.; Larghi, A.; Quero, G.; Carbone, C.; Esposito, A.; Rizzatti, G.; Attili, F.; Alfieri, S.; Costamagna, G.; et al. Pancreatic Cancer Patient-Derived Organoid Platforms: A Clinical Tool to Study Cell- and Non-Cell-Autonomous Mechanisms of Treatment Response. Front. Med. 2021, 8, 793144. [Google Scholar] [CrossRef]
- Li, X.; Fu, G.; Zhang, L.; Guan, R.; Tang, P.; Zhang, J.; Rao, X.; Chen, S.; Xu, X.; Zhou, Y.; et al. Assay establishment and validation of a high-throughput organoid-based drug screening platform. Stem Cell Res. Ther. 2022, 13, 219. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kim, K.; Lee, E.; Lee, K.; Ahn, K.H.; Park, H.; Kim, Y.; Shin, S.; Jeon, S.Y.; Hwang, Y.; et al. Prediction of TKI response in EGFR-mutant lung cancer patients-derived organoids using malignant pleural effusion. npj Precis. Oncol. 2024, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, H.; Xia, W.; Cui, L.; Xu, R.; Lu, H.; Xue, Z.; Zhang, B.; Tian, Z.; Cao, Y.; et al. Isolation and identification of cancer stem cells from PC3 human prostate carcinoma cell line. Int. J. Clin. Exp. Pathol. 2017, 10, 8377–8382. [Google Scholar]
- Meacham, C.E.; Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501, 328–337. [Google Scholar] [CrossRef]
- Luond, F.; Tiede, S.; Christofori, G. Breast cancer as an example of tumour heterogeneity and tumour cell plasticity during malignant progression. Br. J. Cancer 2021, 125, 164–175. [Google Scholar] [CrossRef]
- Almendro, V.; Marusyk, A.; Polyak, K. Cellular heterogeneity and molecular evolution in cancer. Annu. Rev. Pathol. 2013, 8, 277–302. [Google Scholar] [CrossRef]
- Thoma, C.R.; Zimmermann, M.; Agarkova, I.; Kelm, J.M.; Krek, W. 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv. Drug Deliv. Rev. 2014, 69–70, 29–41. [Google Scholar] [CrossRef]
- Kumar, M.; Nassour-Caswell, L.C.; Alrefai, H.; Anderson, J.C.; Schanel, T.L.; Hicks, P.H.; Cardan, R.; Willey, C.D. A High-Throughput Neurosphere-Based Colony Formation Assay to Test Drug and Radiation Sensitivity of Different Patient-Derived Glioblastoma Lines. Cells 2024, 13, 1995. [Google Scholar] [CrossRef] [PubMed]
- Engelken, J.A.; Butelmann, T.; Tribukait-Riemenschneider, F.; Shastri, V.P. Towards a 3D-Printed Millifluidic Device for Investigating Cellular Processes. Micromachines 2024, 15, 1348. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, S.; Oliveira, B.B.; Valente, R.; Ferreira, D.; Luz, A.; Baptista, P.V.; Fernandes, A.R. Breaking the mold: 3D cell cultures reshaping the future of cancer research. Front. Cell Dev. Biol. 2024, 12, 1507388. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, S.M.; Dunagin, M.C.; Torborg, S.R.; Torre, E.A.; Emert, B.; Krepler, C.; Beqiri, M.; Sproesser, K.; Brafford, P.A.; Xiao, M.; et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 2017, 546, 431–435. [Google Scholar] [CrossRef]
- Flavahan, W.A.; Gaskell, E.; Bernstein, B.E. Epigenetic plasticity and the hallmarks of cancer. Science 2017, 357, eaal2380. [Google Scholar] [CrossRef]
- Wang, L.; Yu, Y.; Fang, Y.; Li, Y.; Yu, W.; Wang, Z.; Lv, J.; Wang, R.; Liang, S. Malignant pleural effusion facilitates the establishment and maintenance of tumor organoid biobank with multiple patient-derived lung tumor cell sources. Exp. Hematol. Oncol. 2024, 13, 115. [Google Scholar] [CrossRef]
- Chang, Y.M.; Huang, W.Y.; Yang, S.H.; Jan, C.I.; Nieh, S.; Lin, Y.S.; Chen, S.F.; Lin, Y.C. Interleukin-8/CXCR1 Signaling Contributes to the Progression of Pulmonary Adenocarcinoma Resulting in Malignant Pleural Effusion. Cells 2024, 13, 968. [Google Scholar] [CrossRef]
- Roscilli, G.; De Vitis, C.; Ferrara, F.F.; Noto, A.; Cherubini, E.; Ricci, A.; Mariotta, S.; Giarnieri, E.; Giovagnoli, M.R.; Torrisi, M.R.; et al. Human lung adenocarcinoma cell cultures derived from malignant pleural effusions as model system to predict patients chemosensitivity. J. Transl. Med. 2016, 14, 61. [Google Scholar] [CrossRef]
- Goudarzi, H.; Hida, Y.; Takano, H.; Teramae, H.; Iizasa, H.; Hamada, J. Hypoxia affects in vitro growth of newly established cell lines from patients with malignant pleural mesothelioma. Biomed. Res. 2013, 34, 13–21. [Google Scholar] [CrossRef][Green Version]
- Hocking, A.J.; Mortimer, L.A.; Farrall, A.L.; Russell, P.A.; Klebe, S. Establishing mesothelioma patient-derived organoid models from malignant pleural effusions. Lung Cancer 2024, 191, 107542. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, J.; Wang, B.; Zhao, M.; Wu, Z.; Liu, X. Establishment of an in Vitro Three-Dimensional Vascularized Micro-Tumor Model and Screening of Chemotherapeutic Drugs. Technol. Cancer Res. Treat. 2024, 23, 15330338241286755. [Google Scholar] [CrossRef]
- Ryoo, H.; Kimmel, H.; Rondo, E.; Underhill, G.H. Advances in high throughput cell culture technologies for therapeutic screening and biological discovery applications. Bioeng. Transl. Med. 2024, 9, e10627. [Google Scholar] [CrossRef] [PubMed]
- Petiti, J.; Caria, S.; Revel, L.; Pegoraro, M.; Divieto, C. Standardized Protocol for Resazurin-Based Viability Assays on A549 Cell Line for Improving Cytotoxicity Data Reliability. Cells 2024, 13, 1959. [Google Scholar] [CrossRef] [PubMed]
- Olijnik, A.A.; Rodriguez-Romera, A.; Wong, Z.C.; Shen, Y.; Reyat, J.S.; Jooss, N.J.; Rayes, J.; Psaila, B.; Khan, A.O. Generating human bone marrow organoids for disease modeling and drug discovery. Nat. Protoc. 2024, 19, 2117–2146. [Google Scholar] [CrossRef]
- Hung, H.C.; Mao, T.L.; Fan, M.H.; Huang, G.Z.; Minhalina, A.P.; Chen, C.L.; Liu, C.L. Enhancement of Tumorigenicity, Spheroid Niche, and Drug Resistance of Pancreatic Cancer Cells in Three-Dimensional Culture System. J. Cancer 2024, 15, 2292–2305. [Google Scholar] [CrossRef]
- Gopallawa, I.; Gupta, C.; Jawa, R.; Cyril, A.; Jawa, V.; Chirmule, N.; Gujar, V. Applications of Organoids in Advancing Drug Discovery and Development. J. Pharm. Sci. 2024, 113, 2659–2667. [Google Scholar] [CrossRef]
- Swayden, M.; Iovanna, J.; Soubeyran, P. Pancreatic cancer chemo-resistance is driven by tumor phenotype rather than tumor genotype. Heliyon 2018, 4, e01055. [Google Scholar] [CrossRef]
- Plava, J.; Cihova, M.; Burikova, M.; Matuskova, M.; Kucerova, L.; Miklikova, S. Recent advances in understanding tumor stroma-mediated chemoresistance in breast cancer. Mol. Cancer 2019, 18, 67. [Google Scholar] [CrossRef]
- Dong, Z.; Wang, Y.; Xu, G.; Liu, B.; Wang, Y.; Reboud, J.; Jajesniak, P.; Yan, S.; Ma, P.; Liu, F.; et al. Genetic and phenotypic profiling of single living circulating tumor cells from patients with microfluidics. Proc. Natl. Acad. Sci. USA 2024, 121, e2315168121. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.-H.; Yu, J.-H.; Lin, Y.-C.; Chang, Y.-M.; Liu, N.-T.; Chen, S.-F. Application of an Integrated Single-Cell and Three-Dimensional Spheroid Culture Platform for Investigating Drug Resistance Heterogeneity and Epithelial–Mesenchymal Transition (EMT) in Lung Cancer Subclones. Int. J. Mol. Sci. 2025, 26, 1766. https://doi.org/10.3390/ijms26041766
Chen S-H, Yu J-H, Lin Y-C, Chang Y-M, Liu N-T, Chen S-F. Application of an Integrated Single-Cell and Three-Dimensional Spheroid Culture Platform for Investigating Drug Resistance Heterogeneity and Epithelial–Mesenchymal Transition (EMT) in Lung Cancer Subclones. International Journal of Molecular Sciences. 2025; 26(4):1766. https://doi.org/10.3390/ijms26041766
Chicago/Turabian StyleChen, Shin-Hu, Jian-Hong Yu, Yu-Chun Lin, Yi-Ming Chang, Nien-Tzu Liu, and Su-Feng Chen. 2025. "Application of an Integrated Single-Cell and Three-Dimensional Spheroid Culture Platform for Investigating Drug Resistance Heterogeneity and Epithelial–Mesenchymal Transition (EMT) in Lung Cancer Subclones" International Journal of Molecular Sciences 26, no. 4: 1766. https://doi.org/10.3390/ijms26041766
APA StyleChen, S.-H., Yu, J.-H., Lin, Y.-C., Chang, Y.-M., Liu, N.-T., & Chen, S.-F. (2025). Application of an Integrated Single-Cell and Three-Dimensional Spheroid Culture Platform for Investigating Drug Resistance Heterogeneity and Epithelial–Mesenchymal Transition (EMT) in Lung Cancer Subclones. International Journal of Molecular Sciences, 26(4), 1766. https://doi.org/10.3390/ijms26041766




