Genome-Wide Identification and Expression Analyses of Glycoside Hydrolase Family 18 Genes During Nodule Symbiosis in Glycine max
Abstract
1. Introduction
2. Results
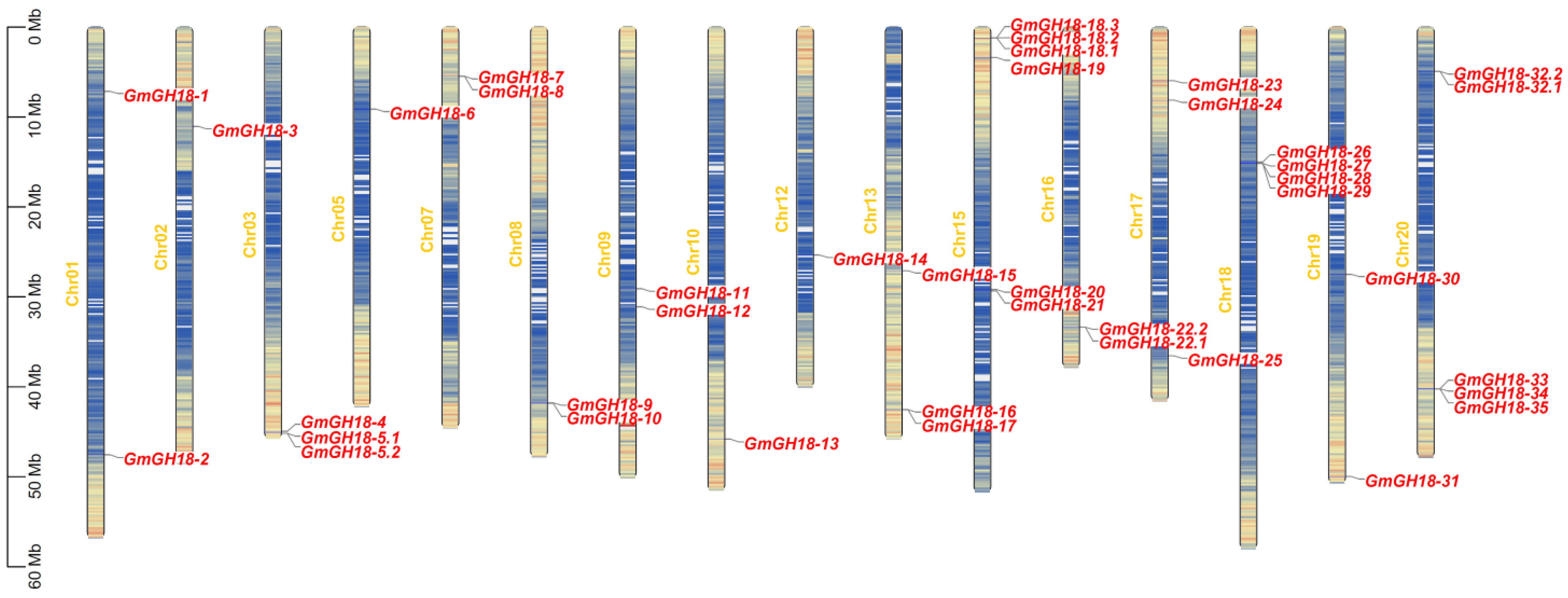
2.1. Chromosomal Distribution of G. max GH18 Genes
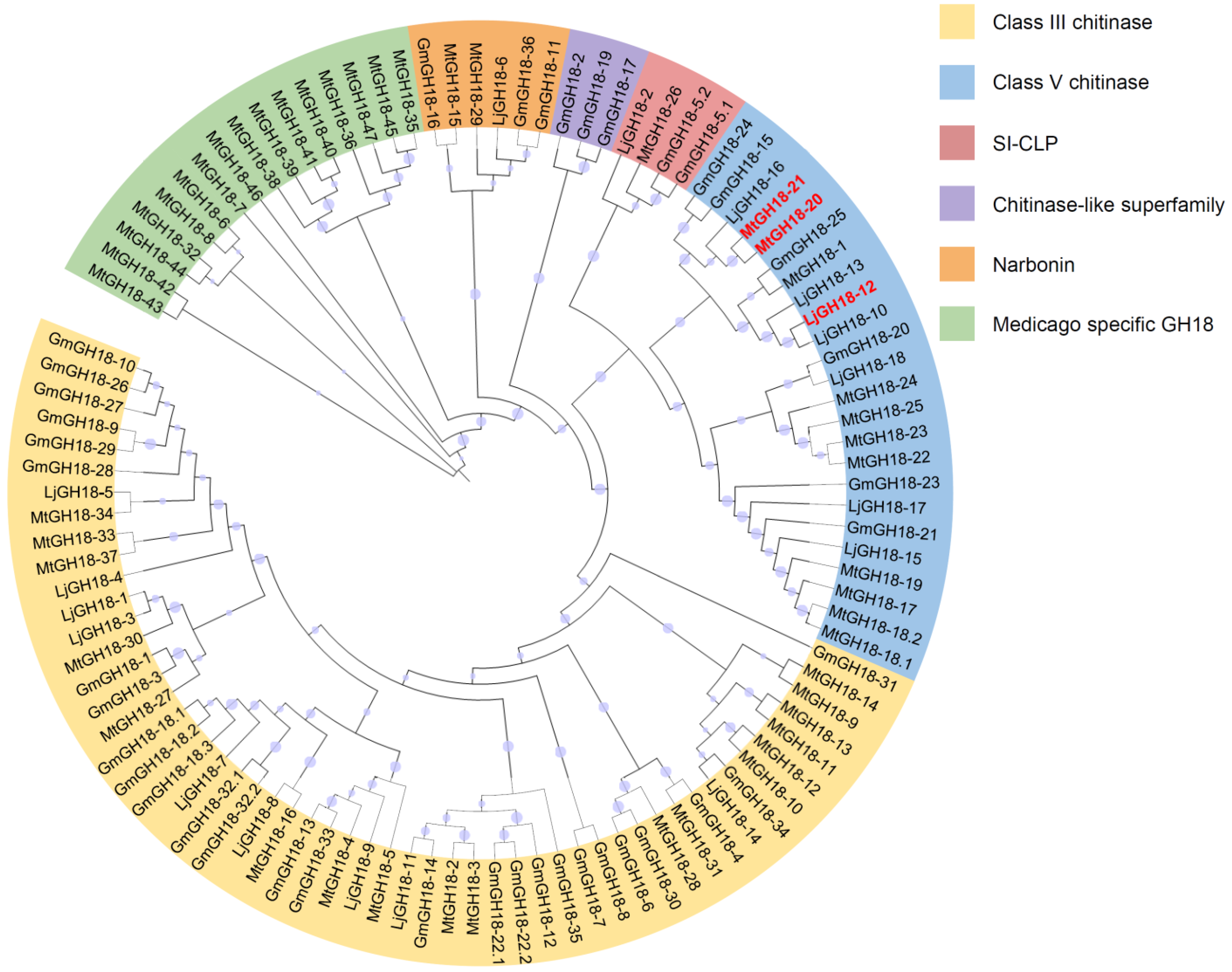
2.2. Analysis of Phylogenetic Tree, Conserved Motifs and Conserved Domains
2.3. Physicochemical Property Analysis of GH18 Proteins in G. max
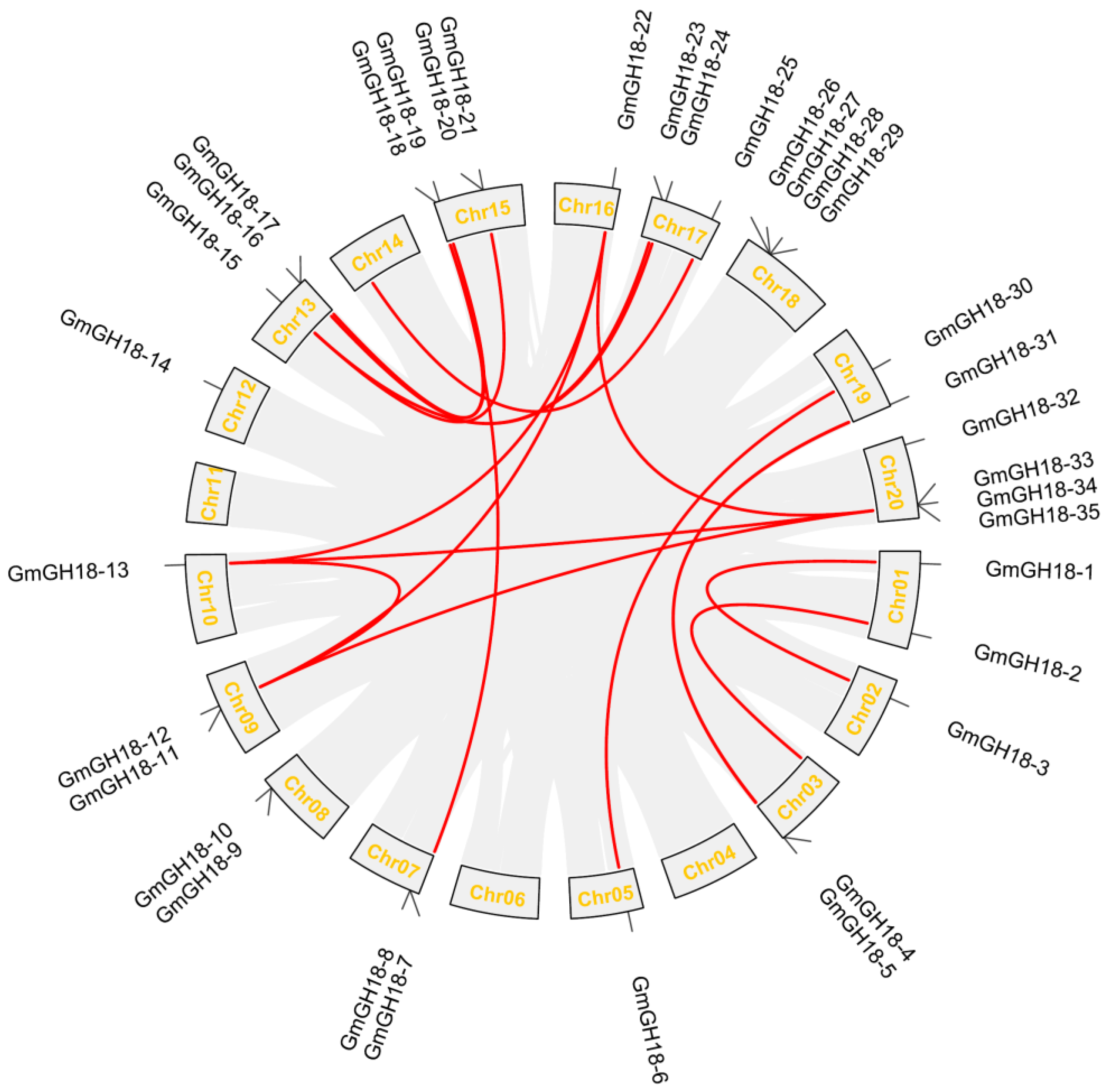
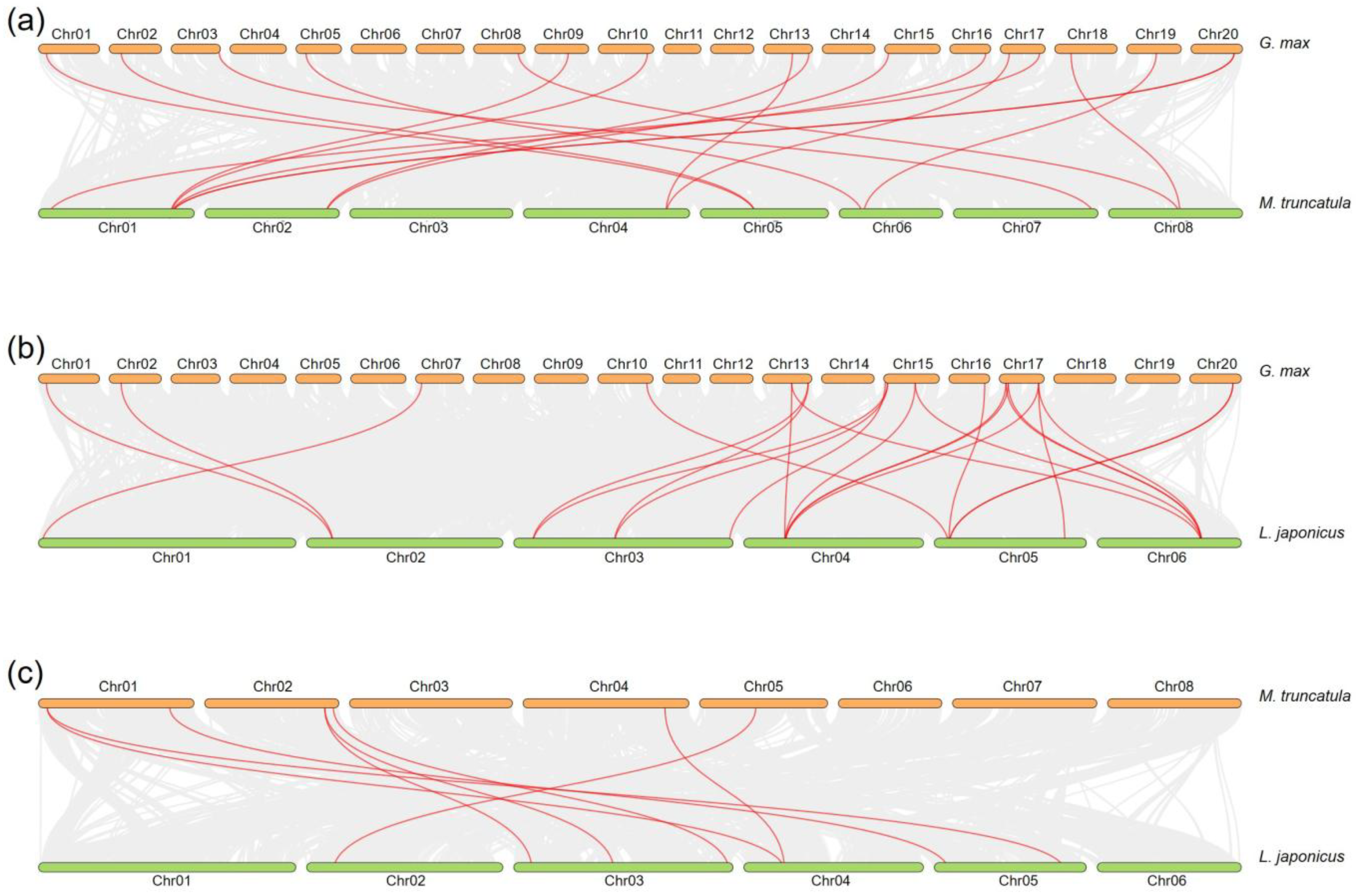
2.4. Collinearity Analysis of GH18 Genes in Soybean, Medicago, and Lotus
2.5. Analysis of Cis-Acting Elements in G. max GH18 Gene Promoter Regions
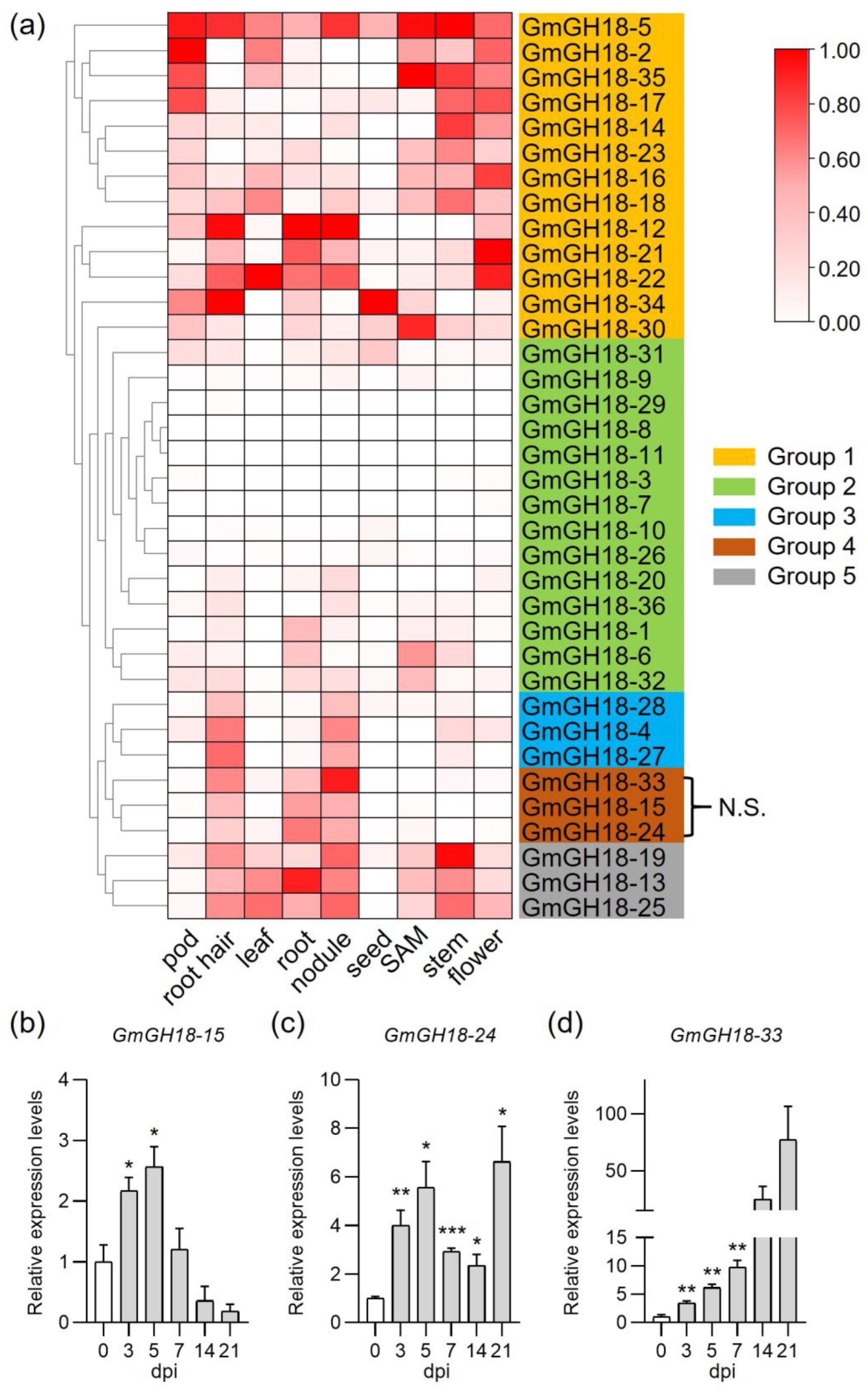
2.6. The Expression of GH18 Genes During Nodule Symbiosis
3. Discussion
4. Materials and Methods
4.1. Sequence Acquisition
4.2. Analysis of Chromosomal Localization
4.3. Construction of Phylogenetic Tree
4.4. Identification of Conserved Motifs and Domains
4.5. Analysis of Physicochemical Properties
4.6. Collinearity Analysis
4.7. Analysis of Cis-Acting Elements and Transcription Factor Binding Sites
4.8. Gene Expression Analysis and Validation by Quantitative Real Time PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, W.; Jiang, X.; Yang, Q. Glycoside hydrolase family 18 chitinases: The known and the unknown. Biotechnol. Adv. 2020, 43, 107553. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.S.; Iqbal, A.; Bibi, A.; Khalil, I.; Ul Islam, Z.; Jan, F.; Khalid, A.; Abdalla, A.N.; Wadood, A. Plant chitinases: Types, structural classification, antifungal potential and transgenic expression in plants for enhanced disease resistance. Plant Cell Tissue Organ Cult. 2024, 156, 75. [Google Scholar] [CrossRef]
- Mahajan, G.; Sharma, V.; Gupta, R. Chitinase: A potent biocatalyst and its diverse applications. Biocatal. Biotransform. 2024, 42, 85–109. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Sharma, A.; Arya, S.K.; Singh, J.; Kapoor, B.; Bhatti, J.S.; Suttee, A.; Singh, G. Prospects of chitinase in sustainable farming and modern biotechnology: An update on recent progress and challenges. Biotechnol. Genet. Eng. Rev. 2024, 40, 310–340. [Google Scholar] [CrossRef]
- Xie, Z.-P.; Staehelin, C.; Wiemken, A.; Boller, T. Ethylene responsiveness of soybean cultivars characterized by leaf senescence, chitinase induction and nodulation. J. Plant Physiol. 1996, 149, 690–694. [Google Scholar] [CrossRef]
- Xie, Z.-P.; Staehelin, C.; Wiemken, A.; Broughton, W.J.; Müller, J.; Boller, T. Symbiosis-stimulated chitinase isoenzymes of soybean (Glycine max (L.) Merr.). J. Exp. Bot. 1999, 50, 327–333. [Google Scholar] [CrossRef][Green Version]
- Grover, A. Plant chitinases: Genetic diversity and physiological roles. Crit. Rev. Plant Sci. 2012, 31, 57–73. [Google Scholar] [CrossRef]
- Meins, F.; Fritig, B.; Linthorst, H.J.; Mikkelsen, J.D.; Neuhaus, J.-M.; Ryals, J. Plant chitinase genes. Plant Mol. Biol. Rep. 1994, 12, S22–S28. [Google Scholar] [CrossRef]
- Neuhaus, J.-M.; Fritig, B.; Linthorst, H.; Meins, F.; Mikkelsen, J.; Ryals, J. A revised nomenclature for chitinase genes. Plant Mol. Biol. Rep. 1996, 14, 102–104. [Google Scholar] [CrossRef]
- Melchers, L.S.; Apotheker-de Groot, M.; van der Knaap, J.A.; Ponstein, A.S.; Sela-Buurlage, M.B.; Bol, J.F.; Cornelissen, B.J.; van den Elzen, P.J.; Linthorst, H.J. A new class of tobacco chitinases homologous to bacterial exo-chitinases displays antifungal activity. Plant J. 1994, 5, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Brunner, F.; Stintzi, A.; Fritig, B.; Legrand, M. Substrate specificities of tobacco chitinases. Plant J. 1998, 14, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Ohnuma, T.; Numata, T.; Osawa, T.; Mizuhara, M.; Lampela, O.; Juffer, A.H.; Skriver, K.; Fukamizo, T. A class V chitinase from Arabidopsis thaliana: Gene responses, enzymatic properties, and crystallographic analysis. Planta 2011, 234, 123–137. [Google Scholar] [CrossRef]
- Schlöffel, M.A.; Käsbauer, C.; Gust, A.A. Interplay of plant glycan hydrolases and LysM proteins in plant—Bacteria interactions. Int. J. Med. Microbiol. 2019, 309, 252–257. [Google Scholar] [CrossRef]
- Salzer, P.; Bonanomi, A.; Beyer, K.; Vögeli-Lange, R.; Aeschbacher, R.A.; Lange, J.; Wiemken, A.; Kim, D.; Cook, D.R.; Boller, T. Differential expression of eight chitinase genes in Medicago truncatula roots during mycorrhiza formation, nodulation, and pathogen infection. Mol. Plant-Microbe Interact. 2000, 13, 763–777. [Google Scholar] [CrossRef]
- Frettinger, P.; Herrmann, S.; Lapeyrie, F.; Oelmüller, R.; Buscot, F. Differential expression of two class III chitinases in two types of roots of Quercus robur during pre-mycorrhizal interactions with Piloderma croceum. Mycorrhiza 2006, 16, 219–223. [Google Scholar] [CrossRef]
- Li, H.Y.; Yang, G.D.; Shu, H.R.; Yang, Y.T.; Ye, B.X.; Nishida, I.; Zheng, C.C. Colonization by the arbuscular mycorrhizal fungus Glomus versiforme induces a defense response against the root-knot nematode Meloidogyne incognita in the grapevine (Vitis amurensis Rupr.), which includes transcriptional activation of the class III chitinase gene VCH3. Plant Cell Physiol. 2006, 47, 154–163. [Google Scholar]
- Goormachtig, S.; Lievens, S.; Van de Velde, W.; Van Montagu, M.; Holsters, M. Srchi13, a novel early nodulin from Sesbania rostrata, is related to acidic class III chitinases. Plant Cell 1998, 10, 905–915. [Google Scholar] [CrossRef]
- Fortunato, A.; Santos, P.; Graça, I.; Gouveia, M.M.; Martins, S.M.; Pinto Ricardo, C.P.; Pawlowski, K.; Ribeiro, A. Isolation and characterization of cgchi3, a nodule-specific gene from Casuarina glauca encoding a class III chitinase. Physiol. Plant. 2007, 130, 418–426. [Google Scholar] [CrossRef]
- Tian, Y.; Liu, W.; Cai, J.; Zhang, L.Y.; Wong, K.B.; Feddermann, N.; Boller, T.; Xie, Z.P.; Staehelin, C. The nodulation factor hydrolase of Medicago truncatula: Characterization of an enzyme specifically cleaving rhizobial nodulation signals. Plant Physiol. 2013, 163, 1179–1190. [Google Scholar] [CrossRef]
- Breakspear, A.; Liu, C.; Roy, S.; Stacey, N.; Rogers, C.; Trick, M.; Morieri, G.; Mysore, K.S.; Wen, J.; Oldroyd, G.E.; et al. The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell 2014, 26, 4680–4701. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Zhang, L.Y.; Liu, W.; Tian, Y.; Xiong, J.S.; Wang, Y.H.; Li, R.J.; Li, H.M.; Wen, J.; Mysore, K.S.; et al. Role of the Nod Factor Hydrolase MtNFH1 in regulating Nod factor levels during rhizobial infection and in mature nodules of Medicago truncatula. Plant Cell 2018, 30, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Li, R.J.; Zhang, C.X.; Fan, S.Y.; Wang, Y.H.; Wen, J.; Mysore, K.S.; Xie, Z.P.; Staehelin, C. The Medicago truncatula hydrolase MtCHIT5b degrades Nod factors of Sinorhizobium meliloti and cooperates with MtNFH1 to regulate the nodule symbiosis. Front. Plant Sci. 2022, 13, 1034230. [Google Scholar] [CrossRef] [PubMed]
- Malolepszy, A.; Kelly, S.; Sørensen, K.K.; James, E.K.; Kalisch, C.; Bozsoki, Z.; Panting, M.; Andersen, S.U.; Sato, S.; Tao, K.; et al. A plant chitinase controls cortical infection thread progression and nitrogen-fixing symbiosis. eLife 2018, 7, e38874. [Google Scholar] [CrossRef]
- Biswas, B.; Gresshoff, P.M. The role of symbiotic nitrogen fixation in sustainable production of biofuels. Int. J. Mol. Sci. 2014, 15, 7380–7397. [Google Scholar] [CrossRef]
- Lv, P.; Zhang, C.; Xie, P.; Yang, X.; El-Sheikh, M.A.; Hefft, D.I.; Ahmad, P.; Zhao, T.; Bhat, J.A. Genome-wide identification and expression analyses of the chitinase gene family in response to white mold and drought stress in soybean (Glycine max). Life 2022, 12, 1340. [Google Scholar] [CrossRef]
- Chen, J.Y.; Sang, H.; Chilvers, M.I.; Wu, C.H.; Chang, H.X. Characterization of soybean chitinase genes induced by rhizobacteria involved in the defense against Fusarium oxysporum. Front. Plant Sci. 2024, 15, 1341181. [Google Scholar] [CrossRef]
- Coulson, A.F. A proposed structure for ‘family 18’ chitinases. A possible function for narbonin. FEBS Lett. 1994, 354, 41–44. [Google Scholar] [CrossRef]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef]
- Landis, J.B.; Soltis, D.E.; Li, Z.; Marx, H.E.; Barker, M.S.; Tank, D.C.; Soltis, P.S. Impact of whole-genome duplication events on diversification rates in angiosperms. Am. J. Bot. 2018, 105, 348–363. [Google Scholar] [CrossRef]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Sterck, L.; Rombauts, S.; Sato, S.; Cheung, F.; Gouzy, J.; Wang, X.; Mudge, J.; Vasdewani, J.; Schiex, T.; et al. Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc. Natl. Acad. Sci. USA 2006, 103, 14959–14964. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, Y.; Nakano, S.; Tamoi, M.; Sakuda, S.; Fukamizo, T. Chitinase gene expression in response to environmental stresses in Arabidopsis thaliana: Chitinase inhibitor allosamidin enhances stress tolerance. Biosci. Biotechnol. Biochem. 2009, 73, 1066–1071. [Google Scholar] [CrossRef]
- Vaghela, B.; Vashi, R.; Rajput, K.; Joshi, R. Plant chitinases and their role in plant defense: A comprehensive review. Enzym. Microb. Technol. 2022, 159, 110055. [Google Scholar] [CrossRef]
- De Jong, A.J.; Cordewener, J.; Lo Schiavo, F.; Terzi, M.; Vandekerckhove, J.; Van Kammen, A.; De Vries, S.C. A carrot somatic embryo mutant is rescued by chitinase. Plant Cell 1992, 4, 425–433. [Google Scholar] [CrossRef]
- Perret, X.; Staehelin, C.; Broughton, W.J. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 2000, 64, 180–201. [Google Scholar] [CrossRef]
- Heidstra, R.; Geurts, R.; Franssen, H.; Spaink, H.P.; Van Kammen, A.; Bisseling, T. Root hair deformation activity of nodulation factors and their fate on Vicia sativa. Plant Physiol. 1994, 105, 787–797. [Google Scholar] [CrossRef]
- Staehelin, C.; Schultze, M.; Kondorosi, E.; Kondorosi, A. Lipo-chitooligosaccharide nodulation signals from Rhizobium meliloti induce their rapid degradation by the host plant alfalfa. Plant Physiol. 1995, 108, 1607–1614. [Google Scholar] [CrossRef][Green Version]
- Ovtsyna, A.O.; Dolgikh, E.A.; Kilanova, A.S.; Tsyganov, V.E.; Borisov, A.Y.; Tikhonovich, I.A.; Staehelin, C. Nod factors induce Nod factor cleaving enzymes in pea roots. Genetic and pharmacological approaches indicate different activation mechanisms. Plant Physiol. 2005, 139, 1051–1064. [Google Scholar] [CrossRef][Green Version]
- Chen, W.; Wang, D.; Ke, S.; Cao, Y.; Xiang, W.; Guo, X.; Yang, Q. A soybean cyst nematode suppresses microbial plant symbionts using a lipochitooligosaccharide-hydrolysing enzyme. Nat. Microbiol. 2024, 9, 1993–2005. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Krishnakumar, V.; Bidwell, S.; Rosen, B.; Chan, A.; Zhou, S.; Gentzbittel, L.; Childs, K.L.; Yandell, M.; Gundlach, H.; et al. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genom. 2014, 15, 312. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Jiang, F.; Wu, P.; Wang, K.; Cao, Y. A high-quality genome sequence of model legume Lotus japonicus (MG-20) provides insights into the evolution of root nodule symbiosis. Genes 2020, 11, 483. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.N.; Yang, C.W.; Wu, N.Y.; Wang, H.T.; Tseng, K.C.; Chiu, Y.H.; Lee, T.Y.; Chang, W.C. PlantPAN 4.0: Updated database for identifying conserved non-coding sequences and exploring dynamic transcriptional regulation in plant promoters. Nucleic Acids Res. 2024, 52, D1569–D1578. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.; Mun, T.; Stougaard, J.; Ben, C.; Andersen, S.U. Distinct Lotus japonicus transcriptomic responses to a spectrum of bacteria ranging from symbiotic to pathogenic. Front. Plant Sci. 2018, 9, 1218. [Google Scholar] [CrossRef] [PubMed]


| Gene Symbol | Gene ID | Amino Acids Size | Molecular Weight (kDa) | Isoelectric Point | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Signal Peptide |
|---|---|---|---|---|---|---|---|---|
| GmGH18-1 | Glyma.01G055200.1 | 296 | 31.74 | 5.39 | 35.54 | 83.48 | −0.113 | Yes |
| GmGH18-2 | Glyma.01G142400.1 | 283 | 31.26 | 6.00 | 26.98 | 90.67 | 0.000 | No |
| GmGH18-3 | Glyma.02G113600.1 | 296 | 31.69 | 5.18 | 34.54 | 79.86 | −0.106 | Yes |
| GmGH18-4 | Glyma.03G254300.1 | 303 | 32.59 | 8.97 | 38.48 | 76.90 | −0.231 | Yes |
| GmGH18-5.1 | Glyma.03G256800.1 | 416 | 46.84 | 9.18 | 50.77 | 90.05 | −0.323 | No |
| GmGH18-5.2 | Glyma.03G256800.2 | 418 | 47.02 | 9.10 | 50.37 | 89.86 | −0.325 | No |
| GmGH18-6 | Glyma.05G075000.1 | 298 | 32.64 | 9.41 | 38.72 | 82.21 | −0.115 | Yes |
| GmGH18-7 | Glyma.07G061600.1 | 289 | 31.30 | 6.31 | 23.17 | 81.38 | −0.068 | No |
| GmGH18-8 | Glyma.07G061800.1 | 173 | 19.46 | 8.42 | 32.33 | 77.28 | −0.296 | No |
| GmGH18-9 | Glyma.08G299700.1 | 300 | 32.00 | 8.08 | 38.61 | 86.57 | 0.024 | Yes |
| GmGH18-10 | Glyma.08G300300.1 | 245 | 25.86 | 4.87 | 35.41 | 87.31 | 0.106 | Yes |
| GmGH18-11 | Glyma.09G121000.1 | 309 | 34.04 | 5.18 | 39.07 | 79.19 | −0.130 | Yes |
| GmGH18-12 | Glyma.09G126200.1 | 292 | 30.88 | 4.07 | 33.02 | 87.57 | −0.016 | Yes |
| GmGH18-13 | Glyma.10G227700.1 | 304 | 32.43 | 7.58 | 39.30 | 85.72 | 0.049 | Yes |
| GmGH18-14 | Glyma.12G156600.1 | 298 | 31.51 | 5.51 | 30.94 | 83.26 | −0.050 | Yes |
| GmGH18-15 | Glyma.13G155800.1 | 379 | 41.07 | 4.78 | 16.18 | 87.81 | 0.141 | Yes |
| GmGH18-16 | Glyma.13G330800.1 | 308 | 34.15 | 5.31 | 34.00 | 83.25 | −0.061 | Yes |
| GmGH18-17 | Glyma.13G330900.1 | 264 | 30.11 | 5.10 | 42.11 | 79.32 | −0.352 | No |
| GmGH18-18.1 | Glyma.15G015100.1 | 624 | 68.80 | 8.29 | 33.35 | 89.58 | −0.072 | Yes |
| GmGH18-18.2 | Glyma.15G015100.2 | 820 | 91.01 | 6.31 | 35.59 | 90.16 | −0.141 | Yes |
| GmGH18-18.3 | Glyma.15G015100.3 | 624 | 68.80 | 8.29 | 33.35 | 89.58 | −0.072 | Yes |
| GmGH18-19 | Glyma.15G043300.1 | 298 | 34.24 | 5.67 | 30.69 | 80.17 | −0.337 | No |
| GmGH18-20 | Glyma.15G206400.1 | 762 | 86.08 | 6.40 | 39.94 | 92.02 | −0.167 | Yes |
| GmGH18-21 | Glyma.15G206800.1 | 365 | 40.09 | 8.92 | 34.77 | 73.29 | −0.304 | Yes |
| GmGH18-22.1 | Glyma.16G173000.1 | 297 | 31.77 | 5.01 | 34.00 | 87.74 | −0.043 | Yes |
| GmGH18-22.2 | Glyma.16G173000.2 | 295 | 31.62 | 5.01 | 34.16 | 88.34 | −0.039 | Yes |
| GmGH18-23 | Glyma.17G076100.1 | 374 | 41.25 | 8.79 | 33.08 | 70.99 | −0.102 | Yes |
| GmGH18-24 | Glyma.17G103500.1 | 377 | 41.06 | 9.11 | 18.08 | 88.81 | 0.158 | Yes |
| GmGH18-25 | Glyma.17G217000.1 | 384 | 43.29 | 6.14 | 32.33 | 79.79 | −0.228 | Yes |
| GmGH18-26 | Glyma.18G120200.1 | 295 | 31.23 | 5.87 | 35.97 | 85.08 | 0.095 | Yes |
| GmGH18-27 | Glyma.18G120700.1 | 295 | 31.27 | 7.50 | 32.96 | 85.08 | 0.045 | Yes |
| GmGH18-28 | Glyma.18G120800.1 | 211 | 22.89 | 4.36 | 29.29 | 89.19 | 0.149 | Yes |
| GmGH18-29 | Glyma.18G120900.1 | 261 | 27.86 | 8.14 | 33.54 | 85.25 | 0.026 | Yes |
| GmGH18-30 | Glyma.19G076200.1 | 316 | 34.75 | 9.42 | 37.34 | 77.53 | −0.238 | Yes |
| GmGH18-31 | Glyma.19G255000.1 | 175 | 20.03 | 8.65 | 45.49 | 90.34 | −0.114 | No |
| GmGH18-32.1 | Glyma.20G035400.1 | 800 | 88.94 | 7.93 | 42.27 | 87.72 | −0.170 | Yes |
| GmGH18-32.2 | Glyma.20G035400.2 | 798 | 88.77 | 7.93 | 42.35 | 87.46 | −0.178 | Yes |
| GmGH18-33 | Glyma.20G164600.1 | 301 | 32.39 | 9.34 | 41.78 | 84.29 | −0.017 | Yes |
| GmGH18-34 | Glyma.20G164700.1 | 333 | 37.03 | 4.87 | 32.39 | 79.97 | −0.163 | Yes |
| GmGH18-35 | Glyma.20G164900.1 | 299 | 32.11 | 4.27 | 38.89 | 83.58 | −0.092 | Yes |
| GmGH18-36 | Glyma.U033800.1 | 302 | 33.54 | 5.44 | 33.17 | 78.15 | −0.118 | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Gou, C.; Zhang, K.; He, M.; Li, L.; Kong, F.; Sun, Z.; Liu, H. Genome-Wide Identification and Expression Analyses of Glycoside Hydrolase Family 18 Genes During Nodule Symbiosis in Glycine max. Int. J. Mol. Sci. 2025, 26, 1649. https://doi.org/10.3390/ijms26041649
Li R, Gou C, Zhang K, He M, Li L, Kong F, Sun Z, Liu H. Genome-Wide Identification and Expression Analyses of Glycoside Hydrolase Family 18 Genes During Nodule Symbiosis in Glycine max. International Journal of Molecular Sciences. 2025; 26(4):1649. https://doi.org/10.3390/ijms26041649
Chicago/Turabian StyleLi, Rujie, Chuanjie Gou, Ke Zhang, Milan He, Lanxin Li, Fanjiang Kong, Zhihui Sun, and Huan Liu. 2025. "Genome-Wide Identification and Expression Analyses of Glycoside Hydrolase Family 18 Genes During Nodule Symbiosis in Glycine max" International Journal of Molecular Sciences 26, no. 4: 1649. https://doi.org/10.3390/ijms26041649
APA StyleLi, R., Gou, C., Zhang, K., He, M., Li, L., Kong, F., Sun, Z., & Liu, H. (2025). Genome-Wide Identification and Expression Analyses of Glycoside Hydrolase Family 18 Genes During Nodule Symbiosis in Glycine max. International Journal of Molecular Sciences, 26(4), 1649. https://doi.org/10.3390/ijms26041649






