Exploring the Post Mortem Interval (PMI) Estimation Model by circRNA circRnf169 in Mouse Liver Tissue
Abstract
1. Introduction
2. Results
2.1. Screening of Reference Genes
2.2. Selection and Validation of the Biomarker in Liver Tissue
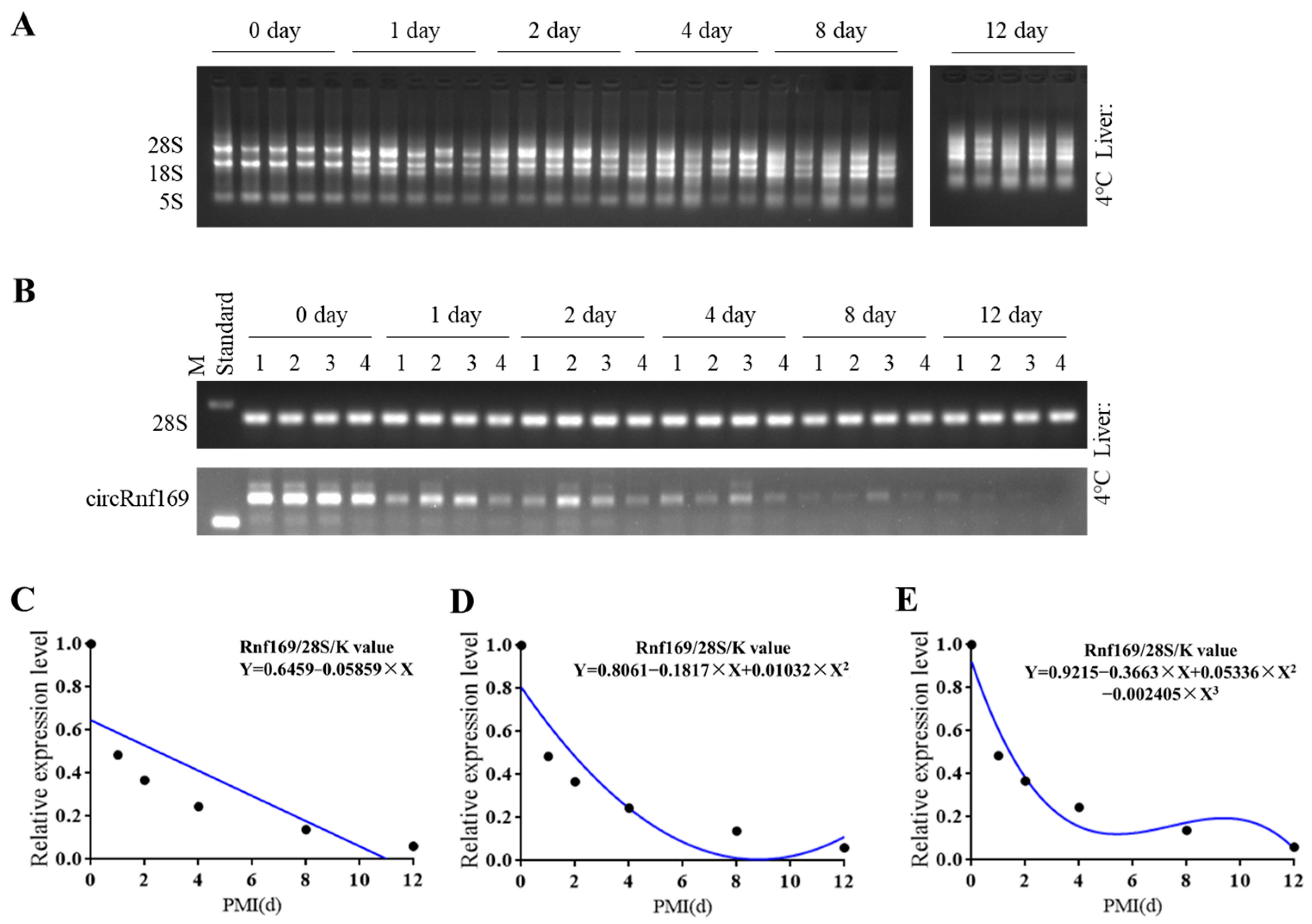
2.3. Construction of the Mouse PMI Estimation Models by Semi-Quantitative PCR
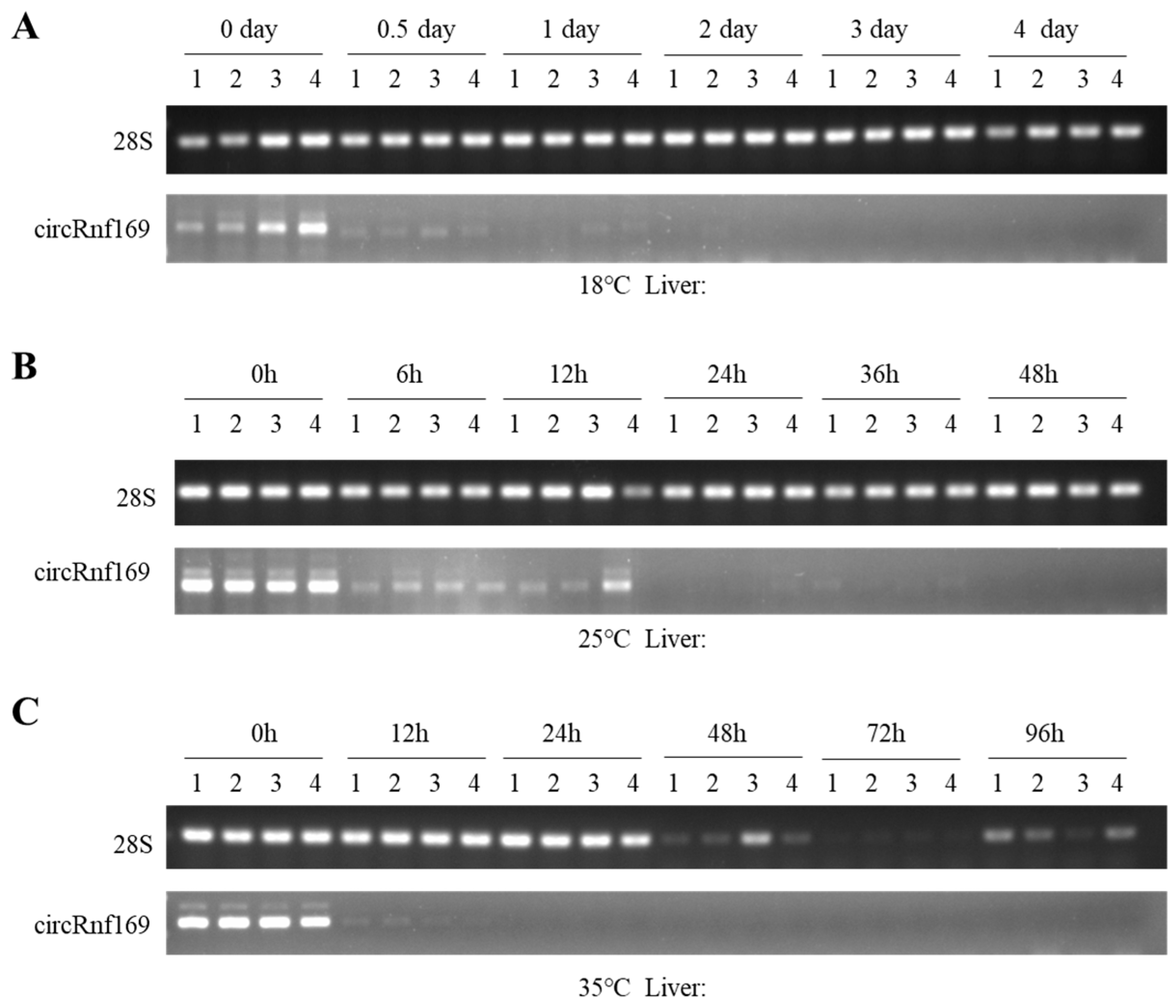
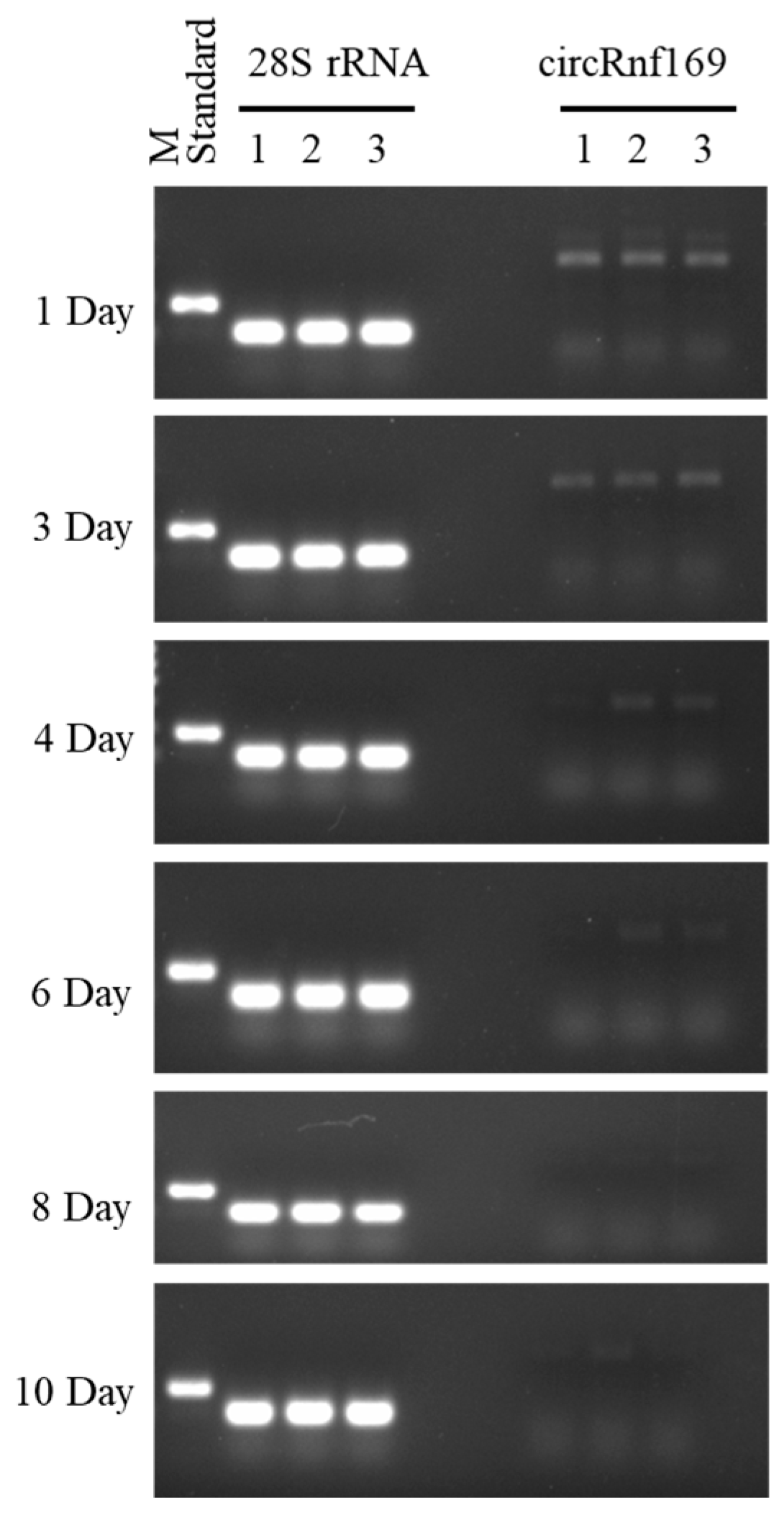
2.4. Validation of the PMI Estimation Model
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Total RNA Extraction
4.3. Reverse Transcription
4.4. Selection of Circular RNAs and Internal Reference Genes
4.5. Primer Design and Semi-Quantitative PCR
4.6. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Z.; Zhang, F.; Wang, L.; Yuan, H.; Guan, D.; Zhao, R. Advances in artificial intelligence-based microbiome for PMI estimation. Front. Microbiol. 2022, 13, 1034051. [Google Scholar] [CrossRef]
- Xiang, Q.Q.; Chen, L.F.; Su, Q.; Du, Y.K.; Liang, P.Y.; Kang, X.D.; Shi, H.; Xu, Q.Y.; Zhao, J.; Liu, C.; et al. Research Progress on Microbial Community Succession in the Postmortem Interval Estimation. Fa Yi Xue Za Zhi 2023, 39, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Woess, C.; Huck, C.W.; Badzoka, J.; Kappacher, C.; Arora, R.; Lindtner, R.A.; Zelger, P.; Schirmer, M.; Rabl, W.; Pallua, J. Raman spectroscopy for postmortem interval estimation of human skeletal remains: A scoping review. J. Biophotonics 2023, 16, e202300189. [Google Scholar] [CrossRef] [PubMed]
- van den Berge, M.; Wiskerke, D.; Gerretsen, R.R.; Tabak, J.; Sijen, T. DNA and RNA profiling of excavated human remains with varying postmortem intervals. Int. J. Legal Med. 2016, 130, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, I.; Grassi, S.; Castiglione, F.; Bartoli, C.; De Saint Pierre, B.; Focardi, M.; Oliva, A.; Pinchi, V. Dental DNA as an Indicator of Post-Mortem Interval (PMI): A Pilot Research. Int. J. Mol. Sci. 2022, 23, 12896. [Google Scholar] [CrossRef]
- Lv, Y.H.; Ma, K.J.; Zhang, H.; He, M.; Zhang, P.; Shen, Y.W.; Jiang, N.; Ma, D.; Chen, L. A time course study demonstrating mRNA, microRNA, 18S rRNA, and U6 snRNA changes to estimate PMI in deceased rat’s spleen. J. Forensic Sci. 2014, 59, 1286–1294. [Google Scholar] [CrossRef]
- Tu, C.; Du, T.; Ye, X.; Shao, C.; Xie, J.; Shen, Y. Using miRNAs and circRNAs to estimate PMI in advanced stage. Leg. Med. 2019, 38, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, P.; Ma, K.J.; Lv, Y.H.; Li, W.C.; Luo, C.L.; Li, L.L.; Shen, Y.W.; He, M.; Jiang, J.Q.; et al. The selection of endogenous genes in human postmortem tissues. Sci. Justice 2013, 53, 115–120. [Google Scholar] [CrossRef]
- Yu, C.Y.; Liu, H.J.; Hung, L.Y.; Kuo, H.C.; Chuang, T.J. Is an observed non-co-linear RNA product spliced in trans, in cis or just in vitro? Nucleic Acids Res. 2014, 42, 9410–9423. [Google Scholar] [CrossRef]
- Chuang, T.J.; Chen, Y.J.; Chen, C.Y.; Mai, T.L.; Wang, Y.D.; Yeh, C.S.; Yang, M.Y.; Hsiao, Y.T.; Chang, T.H.; Kuo, T.C.; et al. Integrative transcriptome sequencing reveals extensive alternative trans-splicing and cis-backsplicing in human cells. Nucleic Acids Res. 2018, 46, 3671–3691. [Google Scholar] [CrossRef] [PubMed]
- Caba, L.; Florea, L.; Gug, C.; Dimitriu, D.C.; Gorduza, E.V. Circular RNA-Is the Circle Perfect? Biomolecules 2021, 11, 1755. [Google Scholar] [CrossRef] [PubMed]
- Misir, S.; Wu, N.; Yang, B.B. Specific expression and functions of circular RNAs. Cell Death Differ. 2022, 29, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Feng, J.; Lei, L.; Hu, J.; Xia, L.; Wang, J.; Xiang, Y.; Liu, L.; Zhong, S.; Han, L.; et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief. Bioinform. 2017, 18, 984–992. [Google Scholar] [CrossRef]
- Wenzlow, N.; Mills, D.; Byrd, J.; Warren, M.; Long, M.T. Review of the current and potential use of biological and molecular methods for the estimation of the postmortem interval in animals and humans. J. Vet. Diagn. Investig. 2023, 35, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef]
- Kubes, P.; Jenne, C. Immune Responses in the Liver. Annu. Rev. Immunol. 2018, 36, 247–277. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.L.; Nakib, D.; Perciani, C.T.; MacParland, S.A. The immune niche of the liver. Clin. Sci. 2021, 135, 2445–2466. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Shen, H.; He, J.; Jin, B.; Tian, Y.; Li, W.; Hou, L.; Zhao, W.; Nan, J.; Zhao, J.; et al. Cytoskeleton remodeling mediated by circRNA-YBX1 phase separation suppresses the metastasis of liver cancer. Proc. Natl. Acad. Sci. USA 2023, 120, e2220296120. [Google Scholar] [CrossRef]
- Zhao, C.; Qian, S.; Tai, Y.; Guo, Y.; Tang, C.; Huang, Z.; Gao, J. Proangiogenic role of circRNA-007371 in liver fibrosis. Cell Prolif. 2023, 56, e13432. [Google Scholar] [CrossRef]
- Guo, X.; Xi, L.; Li, L.; Guo, J.; Jin, W.; Chang, C.; Zhang, J.; Xu, C.; Chen, G. circRNA-14723 promotes hepatocytes proliferation in rat liver regeneration by sponging rno-miR-16-5p. J. Cell Physiol. 2020, 235, 8176–8186. [Google Scholar] [CrossRef]
- Laplace, K.; Baccino, E.; Peyron, P.A. Estimation of the time since death based on body cooling: A comparative study of four temperature-based methods. Int. J. Legal Med. 2021, 135, 2479–2487. [Google Scholar] [CrossRef] [PubMed]
- Wescott, D.J. Recent advances in forensic anthropology: Decomposition research. Forensic Sci. Res. 2018, 3, 327–342. [Google Scholar] [CrossRef] [PubMed]
- Obafunwa, J.O.; Roe, A.; Higley, L. A review of the estimation of postmortem interval using forensic entomology. Med. Sci. Law. 2025, 65, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Sguazzi, G.; Mickleburgh, H.L.; Ghignone, S.; Voyron, S.; Renò, F.; Migliario, M.; Sellitto, F.; Lovisolo, F.; Camurani, G.; Ogbanga, N.; et al. Microbial DNA in human nucleic acid extracts: Recoverability of the microbiome in DNA extracts stored frozen long-term and its potential and ethical implications for forensic investigation. Forensic Sci. Int. Genet. 2022, 59, 102686. [Google Scholar] [CrossRef] [PubMed]
- Lü, Y.H.; Li, Z.H.; Tuo, Y.; Liu, L.; Li, K.; Bian, J.; Ma, J.L.; Chen, L. Correlation between RNA Degradation Patterns of Rat’s Brain and Early PMI at Different Temperatures. Fa Yi Xue Za Zhi 2016, 32, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Qian, J.; Fu, J. Research progress and potential application of microRNA and other non-coding RNAs in forensic medicine. Int. J. Legal Med. 2024, 138, 329–350. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, C.; Wei, Y.; Zhao, Y.; Wang, C.; Lu, C.; Feng, J.; Li, S.; Cong, B. Circular RNA as a Potential Biomarker for Forensic Age Prediction. Front. Genet. 2022, 13, 825443. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, J.; Wang, N.; Liu, J.; Geng, J.; Zhu, J.; Cong, B.; Sun, H.; Wu, R. Integrative lncRNA, circRNA, and mRNA analysis reveals expression profiles of six forensic body fluids/tissue. Int. J. Legal Med. 2024, 138, 731–742. [Google Scholar] [CrossRef] [PubMed]
- An, L.; Dong, C.; Li, J.; Chen, J.; Yuan, J.; Huang, J.; Chan, K.M.; Yu, C.H.; Huen, M.S.Y. RNF169 limits 53BP1 deposition at DSBs to stimulate single-strand annealing repair. Proc. Natl. Acad. Sci. USA 2018, 115, E8286–E8295. [Google Scholar] [CrossRef]
- Wang, J.; Chen, H.; Deng, Q.; Chen, Y.; Wang, Z.; Yan, Z.; Wang, Y.; Tang, H.; Liang, H.; Jiang, Y. High expression of RNF169 is associated with poor prognosis in pancreatic adenocarcinoma by regulating tumour immune infiltration. Front. Genet. 2022, 13, 1022626. [Google Scholar] [CrossRef]
- Su, X.; Lu, L.; Li, Y.; Zhen, C.; Hu, G.; Jiang, K.; Yan, Y.; Xu, Y.; Wang, G.; Shi, M.; et al. Reference gene selection for quantitative real-time PCR (qRT-PCR) expression analysis in Galium aparine L. PLoS ONE 2020, 15, e0226668. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Ma, Y.; Guo, Q. Expression stability of internal reference gene in response to Trichoderma polysporum infection in Avena fatua L. Curr. Genet. 2021, 67, 909–918. [Google Scholar] [CrossRef] [PubMed]


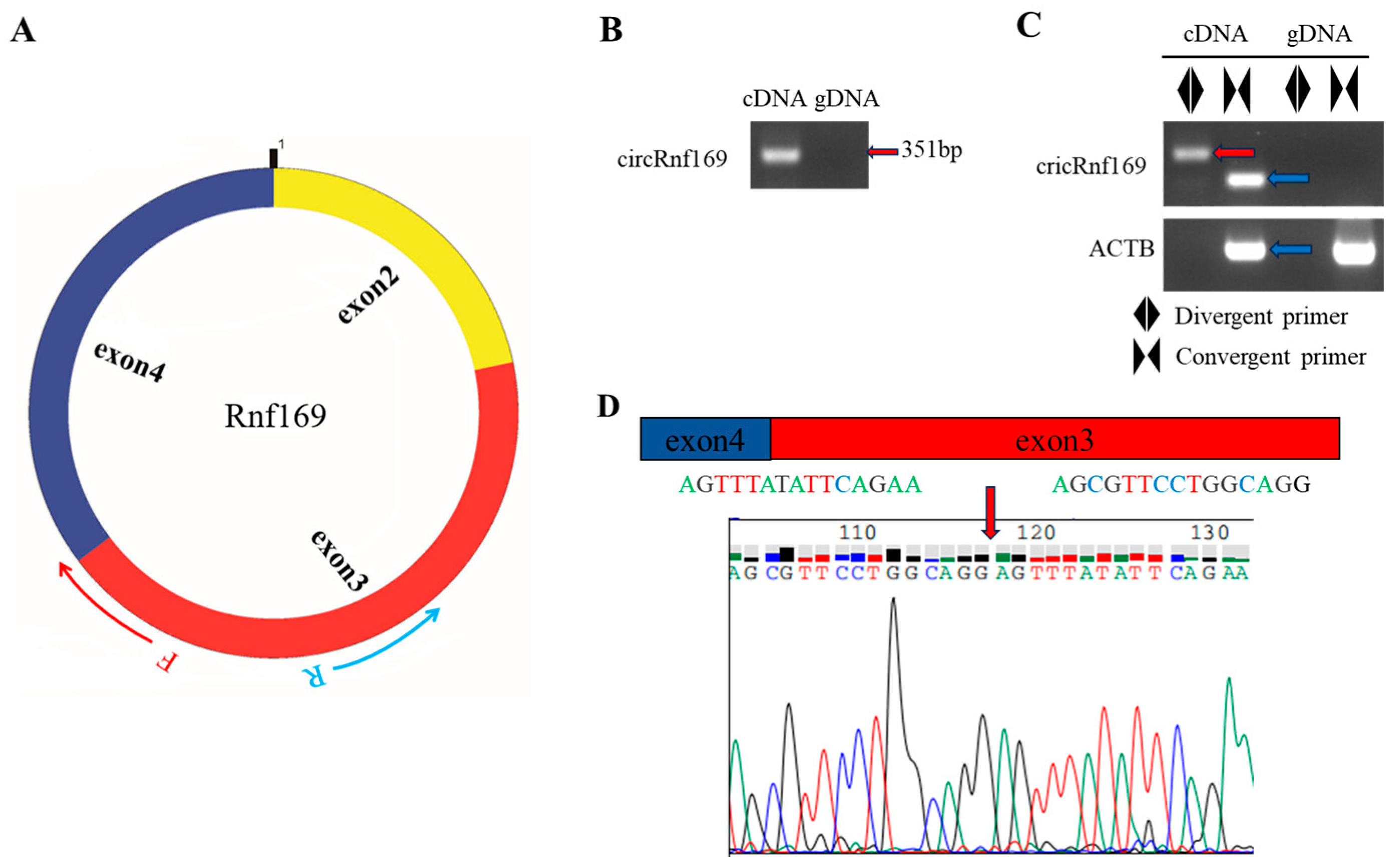
| Marker | Size (bp) | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|---|
| circRnf169-divergent | 351 | CTGAAAACAAGTCTGGAGCAG | TCAGCTTTCCTCTTCCCCATT |
| cricRnf169-convergent | 200 | ATCAAGTTAAGCAAGCCGGG | TGCTCCAGACTTGTTTTCAGC |
| ACTB-divergent | 190 | CTGGCCTGTACACTGACTTGA | GTCATCCATGGCGAACTGGT |
| ACTB-convergent | 392 | TGTTACCAACTGGGACGACA | TCTCAGCTGTGGTGGTGAAG |
| 28S rRNA | 105 | TCATCAGACCCCAGAAAAGG | GTTGATTCGGCAGGTGAGTT |
| Temperature | Reference Gene | Mathematical Models | R2 | p-Value | Equation |
|---|---|---|---|---|---|
| 4 °C | 28S rRNA | Y = 0.6459 − 0.05859 × X | 0.6423 | 0.055 | Linear |
| Y = 0.8061 − 0.1817 × X + 0.01032 × X2 | 0.8395 | 0.064 | Quadratic | ||
| Y = 0.9215 − 0.3663 × X + 0.05336 × X2 − 0.002405 × X3 | 0.9468 | 0.079 | Cubic |
| Temperature | Reference Gene | Real PMI | Estimation PMI | Estimation Error | Error Rate (%) | Equation |
|---|---|---|---|---|---|---|
| 4 °C | 28S rRNA | 1 d | 3.0 d | 2.0 d | 200.0 | Linear |
| 1 d | 2.1 d | 1.1 d | 110.0 | Quadratic | ||
| 1 d | 1.6 d | 0.6 d | 60.0 | Cubic | ||
| 3 d | 7.2 d | 4.2 d | 140.0 | Linear | ||
| 3 d | 4.2 d | 1.2 d | 40.0 | Quadratic | ||
| 3 d | 3.1 d | 0.1 d | 3.3 | Cubic | ||
| 4 d | 10.0 d | 6.0 d | 150.0 | Linear | ||
| 4 d | 6.6 d | 2.6 d | 65.0 | Quadratic | ||
| 4 d | 3.1 d | 0.9 d | 22.5 | Cubic | ||
| 6 d | 10.5 d | 4.5 d | 75.0 | Linear | ||
| 6 d | 7.3 d | 1.3 d | 21.7 | Quadratic | ||
| 6 d | 7.3 d | 1.3 d | 21.7 | Cubic | ||
| 8 d | 10 d | 2.0 d | 25.0 | Linear | ||
| 8 d | 6.5 d | 1.5 d | 18.8 | Quadratic | ||
| 8 d | 7.1 d | 0.9 d | 11.3 | Cubic | ||
| 10 d | 10.2 d | 0.2 d | 2.0 | Linear | ||
| 10 d | 10.8 d | 0.8 d | 8.0 | Quadratic | ||
| 10 d | 12.1 d | 2.1 d | 21.0 | Cubic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, J.; Song, B.; Qian, J.; Cheng, J.; Chiampanichayakul, S.; Anuchapreeda, S.; Fu, J. Exploring the Post Mortem Interval (PMI) Estimation Model by circRNA circRnf169 in Mouse Liver Tissue. Int. J. Mol. Sci. 2025, 26, 1046. https://doi.org/10.3390/ijms26031046
Fu J, Song B, Qian J, Cheng J, Chiampanichayakul S, Anuchapreeda S, Fu J. Exploring the Post Mortem Interval (PMI) Estimation Model by circRNA circRnf169 in Mouse Liver Tissue. International Journal of Molecular Sciences. 2025; 26(3):1046. https://doi.org/10.3390/ijms26031046
Chicago/Turabian StyleFu, Jiewen, Binghui Song, Jie Qian, Jingliang Cheng, Sawitree Chiampanichayakul, Songyot Anuchapreeda, and Junjiang Fu. 2025. "Exploring the Post Mortem Interval (PMI) Estimation Model by circRNA circRnf169 in Mouse Liver Tissue" International Journal of Molecular Sciences 26, no. 3: 1046. https://doi.org/10.3390/ijms26031046
APA StyleFu, J., Song, B., Qian, J., Cheng, J., Chiampanichayakul, S., Anuchapreeda, S., & Fu, J. (2025). Exploring the Post Mortem Interval (PMI) Estimation Model by circRNA circRnf169 in Mouse Liver Tissue. International Journal of Molecular Sciences, 26(3), 1046. https://doi.org/10.3390/ijms26031046







