Molecular Signatures of Early-Onset Bipolar Disorder and Schizophrenia: Transcriptomic and Machine-Learning Insights into Calcium and cAMP Signaling, Including Sex-Specific Patterns
Abstract
1. Introduction
2. Results
2.1. Gene Filtering and Expression Distribution
Batch Effect Correction and PCA Analysis
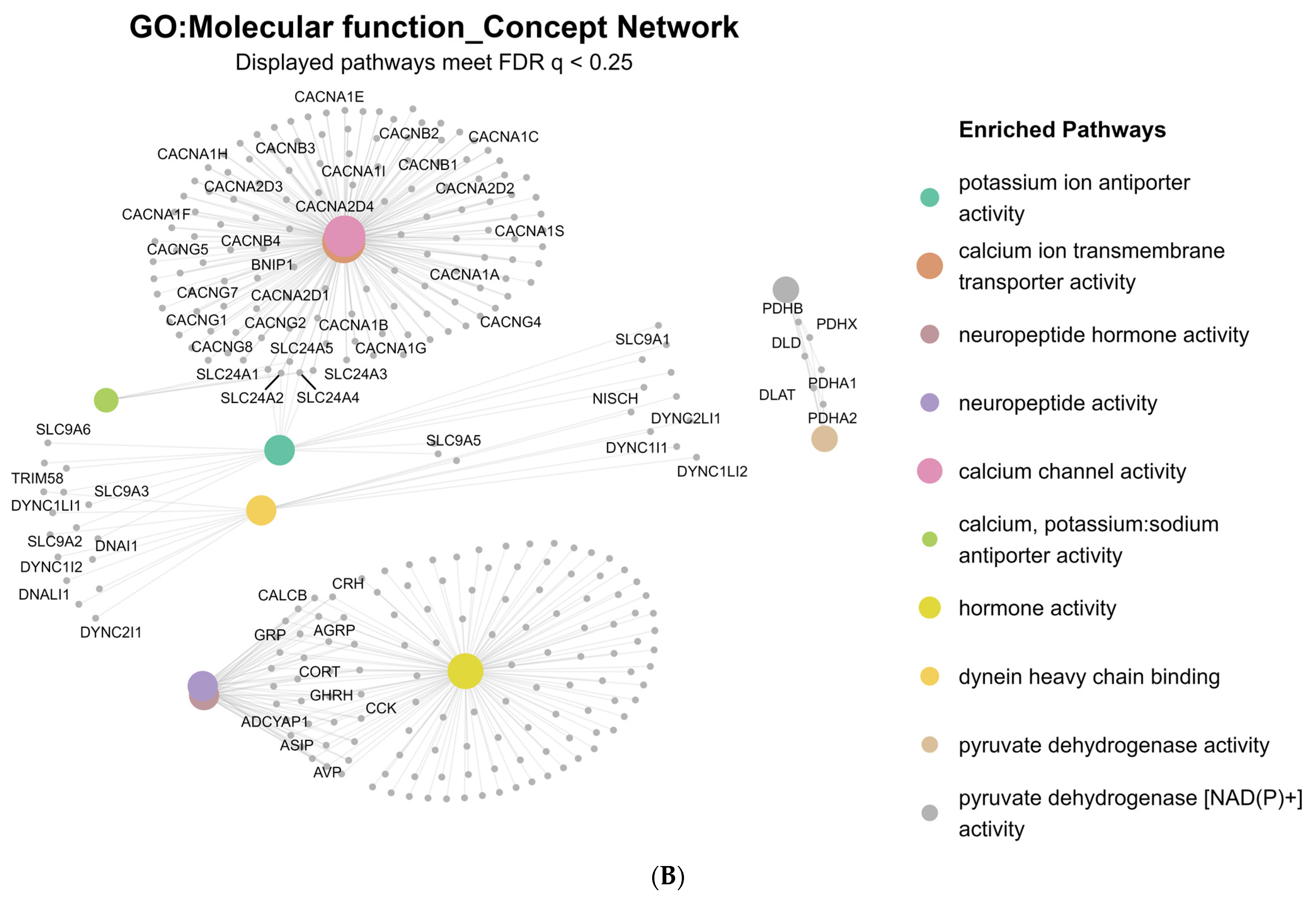
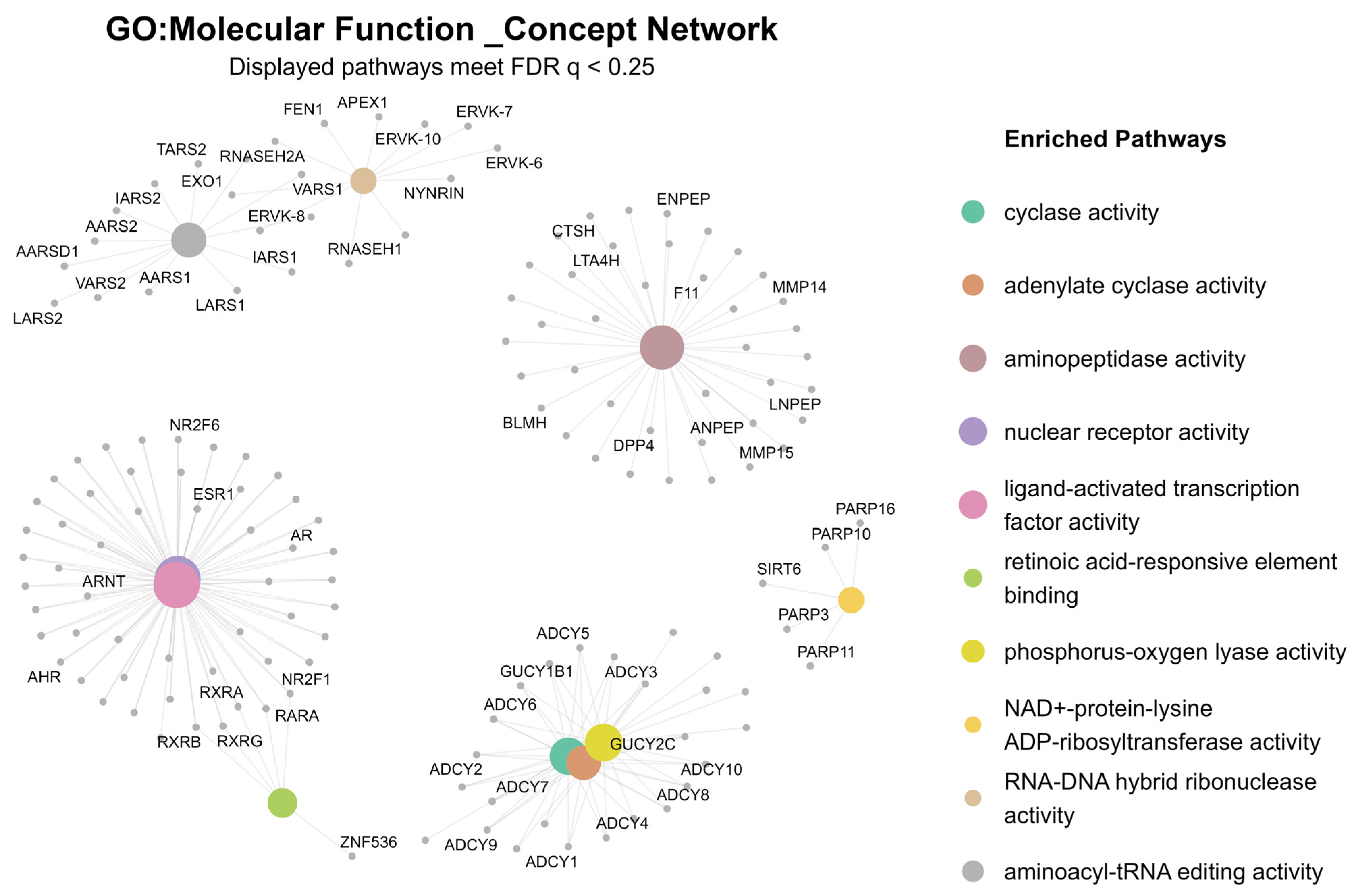
2.2. Differential Gene Expression and Gene Set Enrichment Analysis
2.3. Sex-Stratified Transcriptomic Analysis
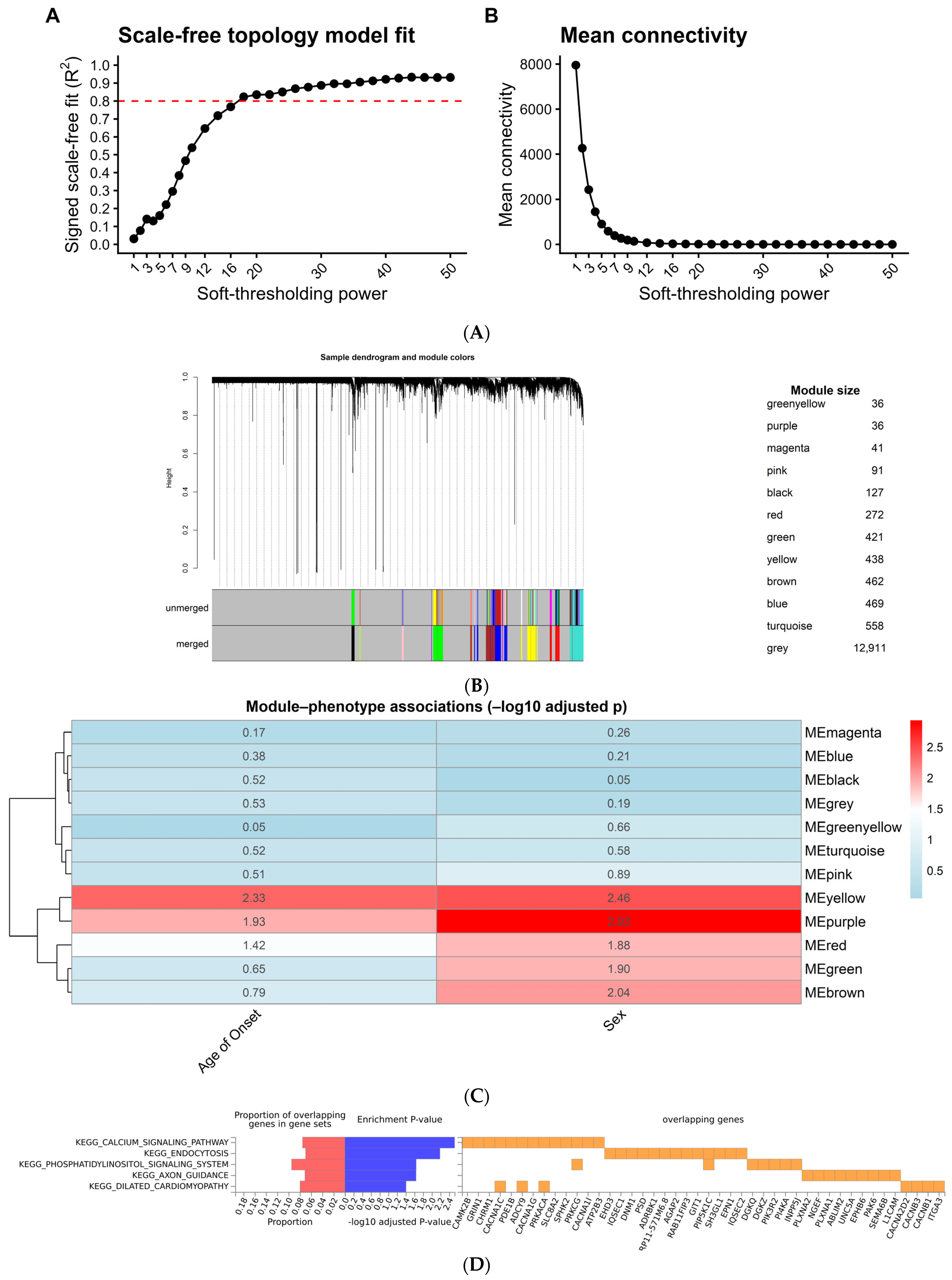
2.4. Weighted Gene Co-Expression Network Analysis (WGCNA)
2.5. WGCNA for Bipolar Disorder Samples
2.6. Cell Type Proportion Analysis
2.7. Predicting Early-Onset Schizophrenia and Bipolar Disorder Using Machine Learning for Transcriptomic Feature Selection
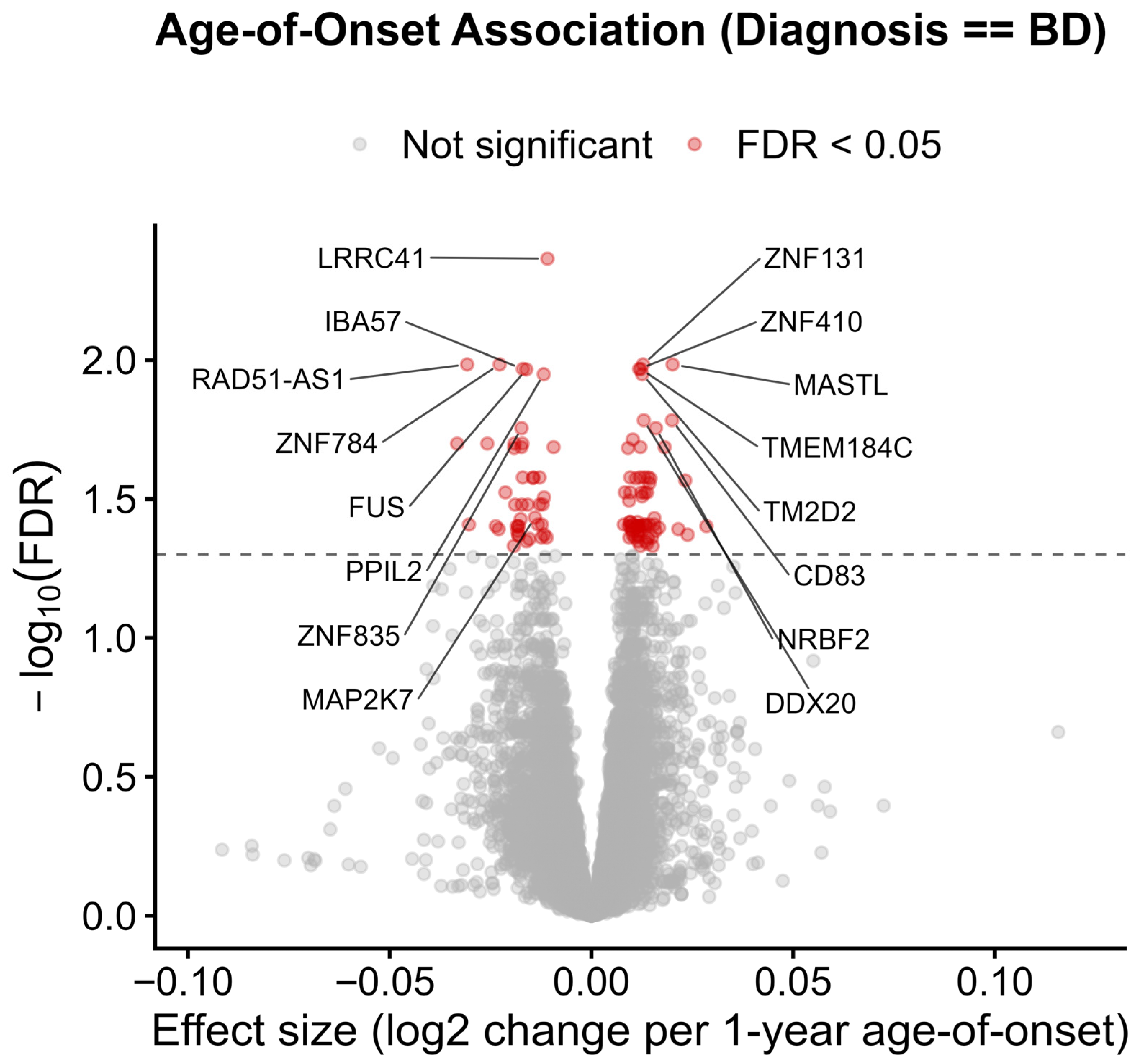
2.8. Interaction Model: Age of Onset × Primary Diagnosis Uncovers Disorder-Specific Transcriptional Signatures
3. Discussion
3.1. Calcium Channel Dysregulation and Psychiatric Onset
3.2. Calcium- and cAMP-Mediated Molecular Mechanisms in Early-Onset Illness
3.3. Sex-Specific Transcriptomic Signatures: cAMP and Adenylate Cyclase Activity
3.4. Co-Expression Network Analysis: Systems-Level Evidence of Calcium Dysregulation
3.5. Predictive Modeling Reveals Key Biomarkers
3.6. Disorder-Specific Transcriptomic Signatures in Bipolar Disorder
3.7. External Replication in Independent GEO Bipolar Disorder Cohorts
- (i)
- FDR < 0.05 in Fisher’s test;
- (ii)
- FDR < 0.05 in signed Stouffer’s test;
- (iii)
- concordant direction of effect in both datasets (Supplementary Figure S7).
3.8. Limitations
4. Materials and Methods
4.1. Dataset
4.2. Data Processing and Normalization
- Primary Model (Full Cohort):
- β0: Intercept term;
- β1: Effect of age of onset (continuous variable);
- β2: Effect of primary diagnosis (categorical: BD vs. SCZ);
- β3–β9: Effects of covariates: age at death, sex, race, postmortem interval (PMI), brain pH, and RNA integrity number (RIN).
- Interaction Model (Full Cohort):
- β0: Intercept term;
- β1: Effect of age of onset (continuous variable);
- β2: Effect of primary diagnosis (categorical: BD vs. SCZ);
- β3: Interaction term: how the relationship between age of onset and gene expression differs by diagnosis;
- β4–β9: Effects of covariates: age at death, sex, race, postmortem interval (PMI), brain pH, and RNA integrity number (RIN).
4.2.1. Weighted Gene Co-Expression Network Analysis (WGCNA)
Data Preprocessing and Gene Filtering
Network Construction
Topological Overlap Matrix (TOM) Transformation
Module Detection
Module–Trait Relationship Analysis
4.2.2. Estimation of Bulk RNA-seq-Derived Relative Cell Type Proportions (rCTPs)
- Differential Gene Expression Analysis Results: We compared the significant genes identified from our DGE analysis with the MGP dataset to assess overlap between gene expression patterns and estimated cell type proportions. Fisher’s exact test was used to determine the statistical significance of the overlap.
- WGCNA Modules: We performed correlation analyses between module eigengenes (derived from WGCNA) and the rCTPs to identify modules that showed significant associations with specific cell types.
4.2.3. Functional Enrichment Analysis
Gene Set Enrichment Analysis (GSEA)
4.3. Machine Learning Model for Age of Onset Prediction
4.3.1. Cross-Validation Strategy
4.3.2. Two-Stage Machine Learning Pipeline
Stage 1: Transcriptomic Feature Selection Using LASSO Regression
Stage 2: Classification of Early vs. Late Onset
4.3.3. Evaluation Metrics
5. Conclusions
Clinical Implications and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell 2018, 173, 1705–1715.e16. [Google Scholar] [CrossRef]
- Craddock, N.; Owen, M.J. The Kraepelinian dichotomy—going, going … but still not gone. Br. J. Psychiatry 2010, 196, 92–95. [Google Scholar] [CrossRef]
- Robinson, N.; Bergen, S.E. Environmental Risk Factors for Schizophrenia and Bipolar Disorder and Their Relationship to Genetic Risk: Current Knowledge and Future Directions. Front. Genet. 2021, 12, 686666. [Google Scholar] [CrossRef]
- Dines, M.; Kes, M.; Ailán, D.; Cetkovich-Bakmas, M.; Born, C.; Grunze, H. Bipolar disorders and schizophrenia: Discrete disorders? Front. Psychiatry 2024, 15, 1352250. [Google Scholar] [CrossRef]
- Carlson, G.A.; Pataki, C. Understanding Early Age of Onset: A Review of the Last 5 Years. Curr. Psychiatry Rep. 2016, 18, 114. [Google Scholar] [CrossRef]
- Carter, T.D.C.; Mundo, E.; Parikh, S.V.; Kennedy, J.L. Early age at onset as a risk factor for poor outcome of bipolar disorder. J. Psychiatr. Res. 2003, 37, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Duffy, A.; Carlson, G.; Dubicka, B.; Hillegers, M.H.J. Pre-pubertal bipolar disorder: Origins and current status of the controversy. Int. J. Bipolar Disord. 2020, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Majuri, T.; Haapea, M.; Nordström, T.; Säynäjäkangas, V.; Moilanen, K.; Tolonen, J.; Ala-Mursula, L.; Miettunen, J.; Jääskeläinen, E. Effect of onset age on the long-term outcome of early-onset psychoses and other mental disorders: A register-based Northern Finland Birth Cohort 1986 study. Eur. Child Adolesc. Psychiatry 2023, 33, 1741–1753. [Google Scholar] [CrossRef] [PubMed]
- McCutcheon, R.A.; Keefe, R.S.E.; McGuire, P.K. Cognitive impairment in schizophrenia: Aetiology, pathophysiology, and treatment. Mol. Psychiatry 2023, 28, 1902–1918, Erratum in Mol. Psychiatry 2023, 28, 1919.. [Google Scholar] [CrossRef]
- Kendhari, J.; Shankar, R.; Young-Walker, L. A Review of Childhood-Onset Schizophrenia. Focus J. Life Long Learn. Psychiatry 2016, 14, 328–332. [Google Scholar] [CrossRef]
- Riecher-Rössler, A.; Butler, S.; Kulkarni, J. Sex and gender differences in schizophrenic psychoses—A critical review. Arch. Women’s Ment. Health 2018, 21, 627–648. [Google Scholar] [CrossRef]
- Seney, M.L.; Sibille, E. Sex differences in mood disorders: Perspectives from humans and rodent models. Biol. Sex Differ. 2014, 5, 17. [Google Scholar] [CrossRef]
- Menculini, G.; Steardo, L.; Sciarma, T.; D’Angelo, M.; Lanza, L.; Cinesi, G.; Cirimbilli, F.; Moretti, P.; Verdolini, N.; De Fazio, P.; et al. Sex Differences in Bipolar Disorders: Impact on Psychopathological Features and Treatment Response. Front. Psychiatry 2022, 13, 926594. [Google Scholar] [CrossRef]
- Abel, K.M.; Drake, R.; Goldstein, J.M. Sex differences in schizophrenia. Int. Rev. Psychiatry 2010, 22, 417–428. [Google Scholar] [CrossRef]
- Carceller, H.; Hidalgo, M.R.; Escartí, M.J.; Nacher, J.; de la Iglesia-Vayá, M.; García-García, F. The impact of sex on gene expression in the brain of schizophrenic patients: A systematic review and meta-analysis of transcriptomic studies. Biol. Sex Differ. 2024, 15, 59. [Google Scholar] [CrossRef]
- Gandal, M.J.; Zhang, P.; Hadjimichael, E.; Walker, R.L.; Chen, C.; Liu, S.; Won, H.; Van Bakel, H.; Varghese, M.; Wang, Y.; et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 2018, 362, eaat8127. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Amiri, M.; Meng, Q.; Unnithan, R.R.; French, C. Calcium Signalling in Neurological Disorders, with Insights from Miniature Fluorescence Microscopy. Cells 2024, 14, 4. [Google Scholar] [CrossRef] [PubMed]
- Shokrgozar, A.; Rahimi, M.; Shoraka, S. Identification of key genes and pathways in schizophrenia: A bioinformatics analysis based on GWAS and GEO. Front. Psychiatry 2025, 16, 1464676. [Google Scholar] [CrossRef] [PubMed]
- Nanou, E.; Catterall, W.A. Calcium Channels, Synaptic Plasticity, and Neuropsychiatric Disease. Neuron 2018, 98, 466–481. [Google Scholar] [CrossRef]
- Brini, M.; Calì, T.; Ottolini, D.; Carafoli, E. Neuronal calcium signaling: Function and dysfunction. Cell. Mol. Life Sci. 2014, 71, 2787–2814. [Google Scholar] [CrossRef]
- Heberle, B.A.; Fox, K.L.; Libermann, L.L.; Xavier, S.R.M.; Dallarosa, G.T.; Santos, R.C.; Fardo, D.W.; Viola, T.W.; Ebbert, M.T.W. Systematic review and meta-analysis of bulk RNAseq studies in human Alzheimer’s disease brain tissue. Alzheimer’s Dement. 2025, 21, e70025. [Google Scholar] [CrossRef]
- Fromer, M.; Roussos, P.; Sieberts, S.K.; Johnson, J.S.; Kavanagh, D.H.; Perumal, T.M.; Ruderfer, D.M.; Oh, E.C.; Topol, A.; Shah, H.R.; et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016, 19, 1442–1453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Valeri, J.; Eladawi, M.A.; Gisabella, B.; Garrett, M.R.; Vallender, E.J.; McCullumsmith, R.; Pantazopoulos, H.; O’Donovan, S.M. Transcriptomic Analysis of the Amygdala in Subjects with Schizophrenia, Bipolar Disorder and Major Depressive Disorder Reveals Differentially Altered Metabolic Pathways. Schizophr. Bull. 2024, sbae193. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.; Dao, D.T.; Terrillion, C.E.; Arad, M.; Smith, R.J.; Soldatov, N.M.; Gould, T.D. CACNA1C (Cav1.2) in the pathophysiology of psychiatric disease. Prog. Neurobiol. 2012, 99, 1–14. [Google Scholar] [CrossRef]
- Hirakawa, H.; Terao, T. The genetic association between bipolar disorder and dementia: A qualitative review. Front. Psychiatry 2024, 15, 1414776. [Google Scholar] [CrossRef]
- Sarangi, S.; Sharma, S.; Nahak, S.K.; Panda, A.K. Association of CACNA1C polymorphisms (rs1006737, rs4765905, rs2007044) with schizophrenia: A meta-analysis and trial sequential analysis. Schizophr. Res. 2024, 274, 247–256. [Google Scholar] [CrossRef]
- Gogos, A.; Ney, L.J.; Seymour, N.; Van Rheenen, T.E.; Felmingham, K.L. Sex differences in schizophrenia, bipolar disorder, and post-traumatic stress disorder: Are gonadal hormones the link? Br. J. Pharmacol. 2019, 176, 4119–4135. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, G.E.; Ma, Y.; Montgomery, K.S.; Bendl, J.; Jaiswal, M.K.; Kozlenkov, A.; Peters, M.A.; Dracheva, S.; Fullard, J.F.; Chess, A.; et al. Sex Differences in the Human Brain Transcriptome of Cases with Schizophrenia. Biol. Psychiatry 2022, 91, 92–101. [Google Scholar] [CrossRef]
- Rubinow, D.R.; Schmidt, P.J. Sex differences and the neurobiology of affective disorders. Neuropsychopharmacology 2018, 44, 111–128. [Google Scholar] [CrossRef]
- Klein, S.L.; Flanagan, K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef]
- Deng, Z.; Cai, W.; Liu, J.; Deng, A.; Yang, Y.; Tu, J.; Yuan, C.; Xiao, H.; Gao, W. Co-expression modules construction by WGCNA and identify potential hub genes and regulation pathways of postpartum depression. Front. Biosci. 2021, 26, 1019–1030. [Google Scholar] [CrossRef]
- Lin, W.; Wang, Y.; Chen, Y.; Wang, Q.; Gu, Z.; Zhu, Y. Role of Calcium Signaling Pathway-Related Gene Regulatory Networks in Ischemic Stroke Based on Multiple WGCNA and Single-Cell Analysis. Oxidative Med. Cell. Longev. 2021, 2021, 8060477. [Google Scholar] [CrossRef]
- Li, D.; Wang, Y.; Wang, J.; Tang, Q. Identification of key proteins in early-onset Alzheimer’s disease based on WGCNA. Front. Aging Neurosci. 2024, 16, 1412222. [Google Scholar] [CrossRef]
- Bolton, S.; Joyce, D.W.; Gordon-Smith, K.; Jones, L.; Jones, I.; Geddes, J.; Saunders, K.E.A. Psychosocial markers of age at onset in bipolar disorder: A machine learning approach. BJPsych Open 2022, 8, e133. [Google Scholar] [CrossRef] [PubMed]
- Xiong, M.; Lin, L.; Jin, Y.; Kang, W.; Wu, S.; Sun, S. Comparison of Machine Learning Models for Brain Age Prediction Using Six Imaging Modalities on Middle-Aged and Older Adults. Sensors 2023, 23, 3622. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, G.E.; Bendl, J.; Voloudakis, G.; Montgomery, K.S.; Sloofman, L.; Wang, Y.-C.; Shah, H.R.; Hauberg, M.E.; Johnson, J.S.; Girdhar, K.; et al. CommonMind Consortium provides transcriptomic and epigenomic data for Schizophrenia and Bipolar Disorder. Sci. Data 2019, 6, 180. [Google Scholar] [CrossRef]
- Akbarian, S.; Liu, C.; A Knowles, J.; Vaccarino, F.M.; Farnham, P.J.; E Crawford, G.; E Jaffe, A.; Pinto, D.; Dracheva, S.; Geschwind, D.H.; et al. The PsychENCODE project. Nat. Neurosci. 2015, 18, 1707–1712. [Google Scholar] [CrossRef]
- Enwright, J.F.; A Lewis, D. Similarities in Cortical Transcriptome Alterations Between Schizophrenia and Bipolar Disorder Are Related to the Presence of Psychosis. Schizophr. Bull. 2021, 47, 1442–1451. [Google Scholar] [CrossRef]
- Ludtmann, M.H.; Boeckeler, K.; Williams, R.S. Molecular pharmacology in a simple model system: Implicating MAP kinase and phosphoinositide signalling in bipolar disorder. Semin. Cell Dev. Biol. 2011, 22, 105–113. [Google Scholar] [CrossRef]
- Waltereit, R.; Weller, M. Signaling from cAMP/PKA to MAPK and Synaptic Plasticity. Mol. Neurobiol. 2003, 27, 99–106. [Google Scholar] [CrossRef]
- Winchester, C.L.; Ohzeki, H.; Vouyiouklis, D.A.; Thompson, R.; Penninger, J.M.; Yamagami, K.; Norrie, J.D.; Hunter, R.; Pratt, J.A.; Morris, B.J. Converging evidence that sequence variations in the novel candidate gene MAP2K7 (MKK7) are functionally associated with schizophrenia. Hum. Mol. Genet. 2012, 21, 4910–4921. [Google Scholar] [CrossRef]
- Mullins, N.; Forstner, A.J.; O’connell, K.S.; Coombes, B.; Coleman, J.R.I.; Qiao, Z.; Als, T.D.; Bigdeli, T.B.; Børte, S.; Bryois, J.; et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 2021, 53, 817–829. [Google Scholar] [CrossRef]
- Trubetskoy, V.; Pardiñas, A.F.; Qi, T.; Panagiotaropoulou, G.; Awasthi, S.; Bigdeli, T.B.; Bryois, J.; Chen, C.-Y.; Dennison, C.A.; Hall, L.S.; et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 2022, 604, 502–508. [Google Scholar] [CrossRef]
- Wellcome Trust Case Control Consortium; Ferreira, M.A.R.; O’Donovan, M.C.; Meng, Y.A.; Jones, I.R.; Ruderfer, D.M.; Jones, L.; Fan, J.; Kirov, G.; Perlis, R.H.; et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet. 2008, 40, 1056–1058. [Google Scholar] [CrossRef]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group; Sklar, P.; Ripke, S.; Scott, L.J.; Andreassen, O.A.; Cichon, S.; Craddock, N.; Edenberg, H.J.; Nurnberger, J.I.; Rietschel, M.; et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 2011, 43, 977–983. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, Z.; Hui, L.; Sun, P. Effects of CACNA1C and ANK3 on cognitive function in patients with bipolar disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry 2024, 133, 111016. [Google Scholar] [CrossRef]
- Klocke, B.; Krone, K.; Tornes, J.; Moore, C.; Ott, H.; Pitychoutis, P.M. Insights into the role of intracellular calcium signaling in the neurobiology of neurodevelopmental disorders. Front. Neurosci. 2023, 17, 1093099. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Wei, W.; Zhao, L.; Li, M.; Li, X.; Liang, S.; Deng, W.; Du, X.D.; Wang, Q.; Guo, W.-J.; et al. Abnormal Brain Structure Morphology in Early-Onset Schizophrenia. Front. Psychiatry 2022, 13, 925204. [Google Scholar] [CrossRef]
- Si, S.; Bi, A.; Yu, Z.; See, C.; Kelly, S.; Ambrogi, S.; Arango, C.; Baeza, I.; Banaj, N.; Berk, M.; et al. Mapping gray and white matter volume abnormalities in early-onset psychosis: An ENIGMA multicenter voxel-based morphometry study. Mol. Psychiatry 2024, 29, 496–504. [Google Scholar] [CrossRef]
- Illes, P.; Rubini, P.; Ulrich, H.; Yin, H.; Tang, Y. Dysregulation of Astrocytic ATP/Adenosine Release in the Hippocampus Cause Cognitive and Affective Disorders: Molecular Mechanisms, Diagnosis, and Therapy. Medcomm 2025, 6, e70177. [Google Scholar] [CrossRef] [PubMed]
- Sethna, F.; Feng, W.; Ding, Q.; Robison, A.J.; Feng, Y.; Wang, H. Enhanced expression of ADCY1 underlies aberrant neuronal signalling and behaviour in a syndromic autism model. Nat. Commun. 2017, 8, 14359. [Google Scholar] [CrossRef]
- Afjeh, S.S.A.; Shams, J.; Hamednia, S.; Boshehri, B.; Olfat, A.; Omrani, M.D. Investigation of the Impact of an ADCY2 Polymorphism as a Predictive Biomarker in Bipolar Disorder, Suicide Tendency and Response to Lithium Carbonate Therapy: The First Report from Iran. Pharmacogenomics 2020, 21, 1011–1020. [Google Scholar] [CrossRef]
- Koutsouleris, N.; Meisenzahl, E.M.; Davatzikos, C.; Bottlender, R.; Frodl, T.; Scheuerecker, J.; Schmitt, G.; Zetzsche, T.; Decker, P.; Reiser, M.; et al. Use of Neuroanatomical Pattern Classification to Identify Subjects in At-Risk Mental States of Psychosis and Predict Disease Transition. Arch. Gen. Psychiatry 2009, 66, 700–712. [Google Scholar] [CrossRef]
- Antonucci, L.A.; Penzel, N.; Sanfelici, R.; Pigoni, A.; Kambeitz-Ilankovic, L.; Dwyer, D.; Ruef, A.; Dong, M.S.; Öztürk, Ö.F.; Chisholm, K.; et al. Using combined environmental–clinical classification models to predict role functioning outcome in clinical high-risk states for psychosis and recent-onset depression. Br. J. Psychiatry 2022, 220, 229–245. [Google Scholar] [CrossRef]
- Pancho, A.; Aerts, T.; Mitsogiannis, M.D.; Seuntjens, E. Protocadherins at the Crossroad of Signaling Pathways. Front. Mol. Neurosci. 2020, 13, 117. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, J.L.; Kostadinov, D.; Chen, W.V.; Maniatis, T.; Sanes, J.R. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 2012, 488, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Kostadinov, D.; Sanes, J.R. Protocadherin-dependent dendritic self-avoidance regulates neural connectivity and circuit function. eLife 2015, 4, e08964. [Google Scholar] [CrossRef]
- Flaherty, E.; Maniatis, T. The role of clustered protocadherins in neurodevelopment and neuropsychiatric diseases. Curr. Opin. Genet. Dev. 2020, 65, 144–150. [Google Scholar] [CrossRef]
- Jia, Z.; Wu, Q. Clustered Protocadherins Emerge as Novel Susceptibility Loci for Mental Disorders. Front. Neurosci. 2020, 14, 587819. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Kalman, J.L.; Papiol, S.; Forstner, A.J.; Heilbronner, U.; Degenhardt, F.; Strohmaier, J.; Adli, M.; Adorjan, K.; Akula, N.; Alda, M.; et al. Investigating polygenic burden in age at disease onset in bipolar disorder: Findings from an international multicentric study. Bipolar Disord. 2018, 21, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Caliz, A.D.; Vertii, A.; Fisch, V.; Yoon, S.; Yoo, H.-J.; Keaney, J.F.; Kant, S. Mitogen-activated protein kinase kinase 7 in inflammatory, cancer, and neurological diseases. Front. Cell Dev. Biol. 2022, 10, 979673. [Google Scholar] [CrossRef]
- Lacorazza, H.D. Pharmacological inhibition of the MAP2K7 kinase in human disease. Front. Oncol. 2024, 14, 1486756. [Google Scholar] [CrossRef]
- Ryan, M.M.; E Lockstone, H.; Huffaker, S.J.; Wayland, M.T.; Webster, M.J.; Bahn, S. Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Mol. Psychiatry 2006, 11, 965–978. [Google Scholar] [CrossRef]
- CAP1 Gene—GeneCards|CAP1 Protein|CAP1 Antibody. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CAP1&keywords=CAP1 (accessed on 23 November 2025).
- AK4 Gene—GeneCards|KAD4 Protein|KAD4 Antibody. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=AK4&keywords=AK4 (accessed on 23 November 2025).
- CAB39 Calcium Binding Protein 39 [Homo Sapiens (Human)]—Gene—NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/51719 (accessed on 23 November 2025).
- Caillard, O.; Moreno, H.; Schwaller, B.; Llano, I.; Celio, M.R.; Marty, A. Role of the calcium-binding protein parvalbumin in short-term synaptic plasticity. Proc. Natl. Acad. Sci. USA 2000, 97, 13372–13377. [Google Scholar] [CrossRef]
- Torac, E.; Gaman, L.; Atanasiu, V. The regulator of calcineurin (RCAN1) an important factor involved in atherosclerosis and cardiovascular diseases development. J. Med. Life 2014, 7, 481–487. [Google Scholar] [PubMed]
- Roh, S.; Woo, J.A.; Lakshmana, M.K.; Uhlar, C.; Ankala, V.; Boggess, T.; Liu, T.; Hong, Y.; Mook-Jung, I.; Kim, S.J.; et al. Mitochondrial dysfunction and calcium deregulation by the RanBP9-cofilin pathway. FASEB J. 2013, 27, 4776–4789. [Google Scholar] [CrossRef] [PubMed]
- Bungaro, M.; Garbo, E. NTRK1/2/3: Biology, detection and therapy. Precis. Cancer Med. 2024, 6, 3. [Google Scholar] [CrossRef]
- Shelton, S. RIN Numbers: How They’re Calculated, What They Mean and Why They’re Important. Genohub Blog. Available online: https://blog.genohub.com/2017/12/24/rin-numbers-how-theyre-calculated-what-they-mean-and-why-theyre-important/ (accessed on 21 January 2025).
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13, Erratum in Genome Biol. 2016, 17, 181.. [Google Scholar] [CrossRef]
- Zhang, Y.; Parmigiani, G.; Johnson, W.E. ComBat-seq: Batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2020, 2. [Google Scholar] [CrossRef]
- Kim, J.H. Multicollinearity and misleading statistical results. Korean J. Anesthesiol. 2019, 72, 558–569. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J. Stat. Softw. 2012, 46, 1–17. [Google Scholar] [CrossRef]
- Wu, S.; Xue, T.; Li, Y.; Chen, W.; Ren, Y. Comprehensive bioinformatics analysis identifies hub genes associated with immune cell infiltration in early-onset schizophrenia. BMC Psychiatry 2025, 25, 55. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, L.; Wu, Y.; Yuan, Z.; Zhou, J. Weighted gene co-expression network analysis to identify key modules and hub genes associated with atrial fibrillation. Int. J. Mol. Med. 2019, 45, 401–416. [Google Scholar] [CrossRef]
- Wilson, J. Weighted Gene Correlation Network Analysis (WGCNA) Applied to Microbial Communities. The Bowman Lab. Available online: https://www.polarmicrobes.org/weighted-gene-correlation-network-analysis-wgcna-applied-to-microbial-communities/ (accessed on 17 October 2024).
- Liu, Y.; Gu, H.-Y.; Zhu, J.; Niu, Y.-M.; Zhang, C.; Guo, G.-L. Identification of Hub Genes and Key Pathways Associated with Bipolar Disorder Based on Weighted Gene Co-expression Network Analysis. Front. Physiol. 2019, 10, 1081. [Google Scholar] [CrossRef]
- Chicco, D.; Agapito, G. Nine quick tips for pathway enrichment analysis. PLoS Comput. Biol. 2022, 18, e1010348. [Google Scholar] [CrossRef]
- Mancarci, B.O.; Toker, L.; Tripathy, S.J.; Li, B.; Rocco, B.; Sibille, E.; Pavlidis, P. Cross-Laboratory Analysis of Brain Cell Type Transcriptomes with Applications to Interpretation of Bulk Tissue Data. eneuro 2017, 4. [Google Scholar] [CrossRef]
- Bioconductor. fgsea. Available online: https://bioconductor.org/packages/fgsea/ (accessed on 17 October 2024).
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Fritz, M.; Holzmann, K.; Castany, S.; Haas, H.; Montag, C.; Engblom, D.; Streb, J.; Dudeck, M. Sex-specific gene expression and weighted co-expression network analysis suggest distinct sex-specific molecular signatures in acutely suicidal MDD-patients without somatic comorbidities. Front. Genet. 2025, 16, 1653768. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef] [PubMed]
- Afjeh, S.; Dey, S.; Gonçalves, V.; Dos Santos, F.; Akbarian, N.; Tripathy, S.; Kiss, D.; Kennedy, J.L. Calcium Pathway Dysregulation in Sex-Specific Differences of Age of Onset in Bipolar Disorder and Schizophrenia: A Transcriptomic and Machine Learning Approach. Biol. Psychiatry 2025, 97, S135. [Google Scholar] [CrossRef]


| Characteristic | Male (n = 191) | Female (n = 178) | Overall (n = 369) |
|---|---|---|---|
| Diagnosis, n (%) | |||
| 148 (77.5%) | 115 (64.6%) | 263 (71.3%) |
| 43 (22.5%) | 63 (35.4%) | 106 (28.7%) |
| Age of Onset (years) | |||
| Mean ± SD | 22.2 ± 8.5 | 22.0 ± 7.9 | 22.1 ± 8.2 |
| Median (min-max) | - | - | 21.0 (7.0–48.0) |
| Age of Death (years) | |||
| Mean ± SD | 46.2 ± 15.3 | 45.5 ± 14.7 | 45.9 ± 14.9 |
| Median (min-max) | - | - | 46.0 (17.0–83.4) |
| RIN | |||
| Mean ± SD | 7.5 ± 0.8 | 7.8 ± 0.7 | 7.7 ± 0.8 |
| Median (min-max) | - | - | 7.7 (5.1–9.4) |
| PMI (hours) | |||
| Mean ± SD | 38.6 ± 26.7 | 36.4 ± 24.1 | 37.4 ± 25.6 |
| Median (min–max) | - | - | 32.0 (5.5–168.0) |
| pH | |||
| Mean ± SD | 6.4 ± 0.4 | 6.4 ± 0.4 | 6.4 ± 0.4 |
| Median (min-max) | - | - | 6.4 (5.7–7.0) |
| White race, n (%) | 142 (74.3%) | 137 (76.9%) | 279 (75.6%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afjeh, S.S.; Dey, S.; Kiss, D.; Sanches, M.; Dos Santos, F.; Pouget, J.G.; Akbarian, N.; Tripathy, S.; Gonçalves, V.F.; Kennedy, J.L. Molecular Signatures of Early-Onset Bipolar Disorder and Schizophrenia: Transcriptomic and Machine-Learning Insights into Calcium and cAMP Signaling, Including Sex-Specific Patterns. Int. J. Mol. Sci. 2025, 26, 12109. https://doi.org/10.3390/ijms262412109
Afjeh SS, Dey S, Kiss D, Sanches M, Dos Santos F, Pouget JG, Akbarian N, Tripathy S, Gonçalves VF, Kennedy JL. Molecular Signatures of Early-Onset Bipolar Disorder and Schizophrenia: Transcriptomic and Machine-Learning Insights into Calcium and cAMP Signaling, Including Sex-Specific Patterns. International Journal of Molecular Sciences. 2025; 26(24):12109. https://doi.org/10.3390/ijms262412109
Chicago/Turabian StyleAfjeh, Sara Sadat, Sohom Dey, Daniel Kiss, Marcos Sanches, Fernanda Dos Santos, Jennie G. Pouget, Niki Akbarian, Shreejoy Tripathy, Vanessa F. Gonçalves, and James L. Kennedy. 2025. "Molecular Signatures of Early-Onset Bipolar Disorder and Schizophrenia: Transcriptomic and Machine-Learning Insights into Calcium and cAMP Signaling, Including Sex-Specific Patterns" International Journal of Molecular Sciences 26, no. 24: 12109. https://doi.org/10.3390/ijms262412109
APA StyleAfjeh, S. S., Dey, S., Kiss, D., Sanches, M., Dos Santos, F., Pouget, J. G., Akbarian, N., Tripathy, S., Gonçalves, V. F., & Kennedy, J. L. (2025). Molecular Signatures of Early-Onset Bipolar Disorder and Schizophrenia: Transcriptomic and Machine-Learning Insights into Calcium and cAMP Signaling, Including Sex-Specific Patterns. International Journal of Molecular Sciences, 26(24), 12109. https://doi.org/10.3390/ijms262412109







