Expanding the Genetic Spectrum of Non-Syndromic Cleft Lip and Palate Through Whole-Exome Sequencing
Abstract
1. Introduction
2. Results
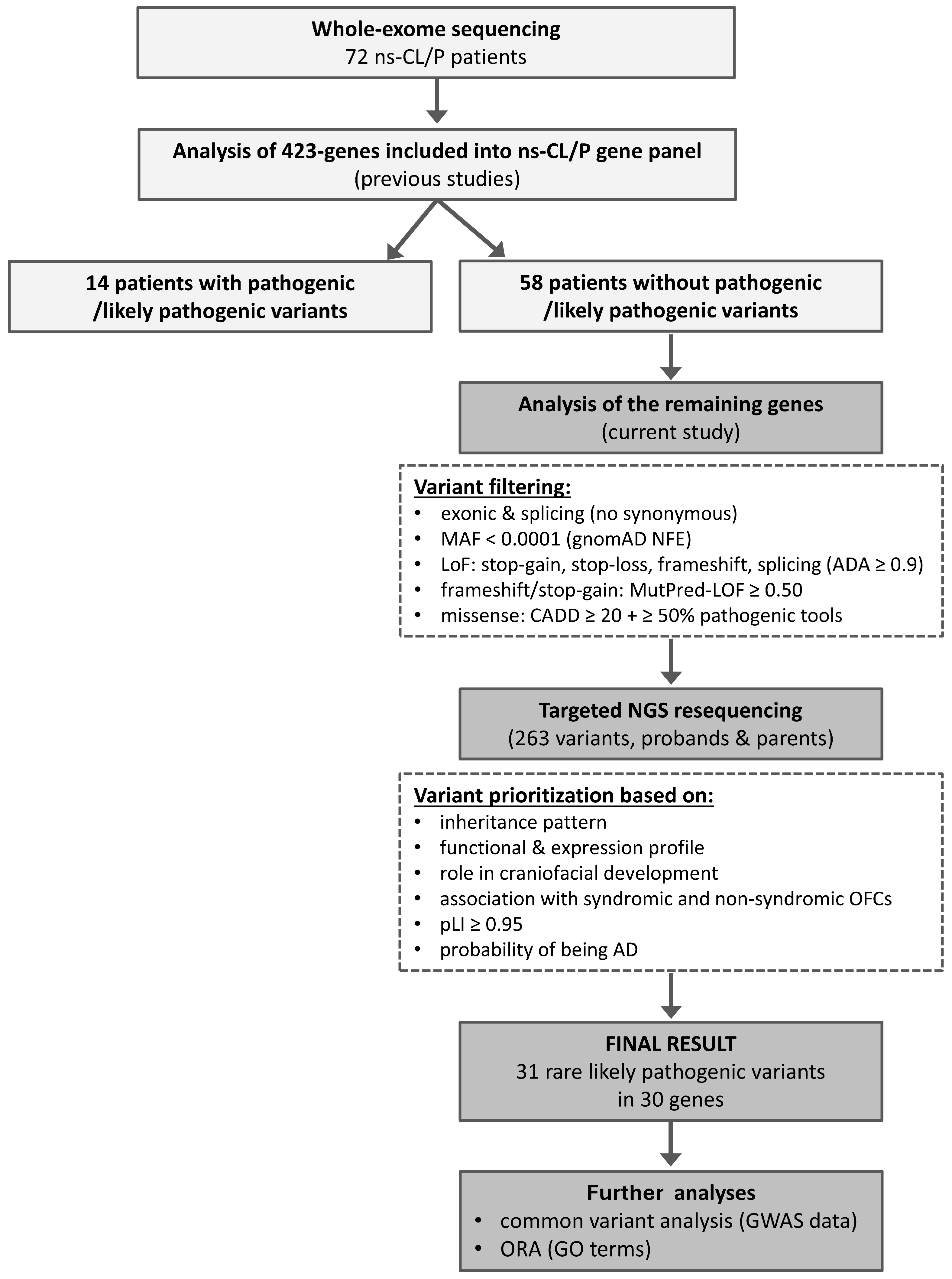
2.1. Whole-Exome Sequencing (WES) Results
2.2. Results of Common Variants Analysis—GWAS Data
2.3. Enrichment of Gene Ontology (GO) Categories
3. Discussion
4. Materials and Methods
4.1. Workflow of the Study
4.2. Study Population
4.3. Whole Exome Sequencing (WES)
4.4. Variant Selection and Prioritizing
4.5. Confirmation and Segregation Analyses
4.6. Common Variants Analysis—GWAS Data
4.7. Over Representation Analysis (ORA)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Autosomal Dominant |
| ADA | Adaptive Boosting |
| CADD | Combined Annotation Dependent Depletion |
| DANN | Deleterious Annotation of Genetic Variants Using Neural Networks |
| DNV | De Novo variant |
| GO | Gene Ontology |
| GWAS | Genome-Wide Association Study |
| LD | Linkage Disequilibrium |
| LoF | Loss of Function |
| MAF | Minor Allele Frequency |
| NFE | European non-Finnish |
| NGS | Next-Generation Sequencing |
| ns-CL | Non-Syndromic Cleft Lip |
| ns-CL/P | Non-Syndromic Cleft Lip with or without Cleft Palate |
| ns-CLP | Non-Syndromic Cleft Lip with Cleft Palate |
| ORA | Overrepresentation Enrichment Analysis |
| P/LP | Pathogenic or Likely Pathogenic |
| pLI | Probability of Being Loss-of-Function Intolerant |
| SNV | Single Nucleotide Variants |
| VUS | Variant of Uncertain Significance |
| WES | Whole-Exome Sequencing |
| WGS | Whole-Genome Sequencing |
References
- Mossey, P.A.; Little, J.; Munger, R.G.; Dixon, M.J.; Shaw, W.C. Cleft lip and palate. Lancet 2009, 374, 1773–1785. [Google Scholar] [CrossRef]
- Nasreddine, G.; El Hajj, J.; Ghassibe-Sabbagh, M. Orofacial clefts embryology, classification, epidemiology, and genetics. Mutat. Res. Rev. Mutat. Res. 2021, 787, 108373. [Google Scholar] [CrossRef]
- Ludwig, K.U.; Ahmed, S.T.; Böhmer, A.C.; Sangani, N.B.; Varghese, S.; Klamt, J.; Schuenke, H.; Gültepe, P.; Hofmann, A.; Rubini, M.; et al. Meta-analysis Reveals Genome-Wide Significance at 15q13 for Nonsyndromic Clefting of Both the Lip and the Palate, and Functional Analyses Implicate GREM1 As a Plausible Causative Gene. PLoS Genet. 2016, 12, e1005914. [Google Scholar] [CrossRef] [PubMed]
- Wehby, G.L.; Cassell, C.H. The impact of orofacial clefts on quality of life and healthcare use and costs. Oral Dis. 2010, 16, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.J.; Marazita, M.L.; Beaty, T.H.; Murray, J.C. Cleft lip and palate: Understanding genetic and environmental influences. Nat. Rev. Genet. 2011, 12, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Babai, A.; Irving, M. Orofacial Clefts: Genetics of Cleft Lip and Palate. Genes 2023, 14, 1603. [Google Scholar] [CrossRef]
- Alvizi, L.; Ke, X.; Brito, L.A.; Seselgyte, R.; Moore, G.E.; Stanier, P.; Passos-Bueno, M.R. Differential methylation is associated with non-syndromic cleft lip and palate and contributes to penetrance effects. Sci. Rep. 2017, 7, 2441. [Google Scholar] [CrossRef]
- Sharp, G.C.; Ho, K.; Davies, A.; Stergiakouli, E.; Humphries, K.; McArdle, W.; Sandy, J.; Davey Smith, G.; Lewis, S.J.; Relton, C.L. Distinct DNA methylation profiles in subtypes of orofacial cleft. Clin. Epigenetics 2017, 9, 63. [Google Scholar] [CrossRef]
- Xu, Z.; Lie, R.T.; Wilcox, A.J.; Saugstad, O.D.; Taylor, J.A. A comparison of DNA methylation in newborn blood samples from infants with and without orofacial clefts. Clin. Epigenetics 2019, 11, 40. [Google Scholar] [CrossRef]
- Zhao, A.D.; Huang, Y.J.; Zhang, H.F.; Tang, W.; Zhang, M.F. Study on DNA methylation profiles in non-syndromic cleft lip/palate based on bioinformatics. Shanghai Kou Qiang Yi Xue 2019, 28, 57–62. (In Chinese) [Google Scholar]
- Im, H.; Song, Y.; Kim, J.K.; Park, D.K.; Kim, D.S.; Kim, H.; Shin, J.O. Molecular Regulation of Palatogenesis and Clefting: An Integrative Analysis of Genetic, Epigenetic Networks, and Environmental Interactions. Int. J. Mol. Sci. 2025, 26, 1382. [Google Scholar] [CrossRef]
- Li, L.; Meng, T.; Jia, Z.; Zhu, G.; Shi, B. Single nucleotide polymorphism associated with nonsyndromic cleft palate influences the processing of miR-140. Am. J. Med. Genet. A 2010, 152A, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Stüssel, L.G.; Hollstein, R.; Laugsch, M.; Hochfeld, L.M.; Welzenbach, J.; Schröder, J.; Thieme, F.; Ishorst, N.; Romero, R.O.; Weinhold, L.; et al. MiRNA-149 as a Candidate for Facial Clefting and Neural Crest Cell Migration. J. Dent. Res. 2022, 101, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Grosen, D.; Bille, C.; Petersen, I.; Skytthe, A.; Hjelmborg, J.; Pedersen, J.K.; Murray, J.C.; Christensen, K. Risk of oral clefts in twins. Epidemiology 2011, 22, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Sivertsen, A.; Wilcox, A.J.; Skjaerven, R.; Vindenes, H.A.; Abyholm, F.; Harville, E.; Lie, R.T. Familial risk of oral clefts by morphological type and severity: Population based cohort study of first degree relatives. BMJ 2008, 336, 432–434. [Google Scholar] [CrossRef]
- Little, J.; Bryan, E. Congenital anomalies in twins. Semin. Perinatol. 1986, 10, 50–64. [Google Scholar]
- Rahimov, F.; Marazita, M.L.; Visel, A.; Cooper, M.E.; Hitchler, M.J.; Rubini, M.; Domann, F.E.; Govil, M.; Christensen, K.; Bille, C.; et al. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat. Genet. 2008, 40, 1341–1347. [Google Scholar] [CrossRef]
- Birnbaum, S.; Ludwig, K.U.; Reutter, H.; Herms, S.; Steffens, M.; Rubini, M.; Baluardo, C.; Ferrian, M.; Almeida de Assis, N.; Alblas, M.A.; et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat. Genet. 2009, 41, 473–477. [Google Scholar] [CrossRef]
- Mostowska, A.; Hozyasz, K.K.; Wojcicki, P.; Biedziak, B.; Paradowska, P.; Jagodzinski, P.P. Association between genetic variants of reported candidate genes or regions and risk of cleft lip with or without cleft palate in the polish population. Birth Defects Res. A Clin. Mol. Teratol. 2010, 88, 538–545. [Google Scholar] [CrossRef]
- Awotoye, W.; Mossey, P.A.; Hetmanski, J.B.; Gowans, L.J.J.; Eshete, M.A.; Adeyemo, W.L.; Alade, A.; Zeng, E.; Adamson, O.; Naicker, T.; et al. Whole-genome sequencing reveals de-novo mutations associated with nonsyndromic cleft lip/palate. Sci. Rep. 2022, 12, 11743. [Google Scholar] [CrossRef]
- Dąbrowska, J.; Biedziak, B.; Szponar-Żurowska, A.; Budner, M.; Jagodziński, P.P.; Płoski, R.; Mostowska, A. Identification of novel susceptibility genes for non-syndromic cleft lip with or without cleft palate using NGS-based multigene panel testing. Mol. Genet. Genom. 2022, 297, 1315–1327. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Yue, J.; Xue, L.; Xu, Y.; Ding, Q.; Xiao, W. Using whole exome sequencing to identify susceptibility genes associated with nonsyndromic cleft lip with or without cleft palate. Mol. Genet. Genom. 2023, 298, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.; Newbury, D.F.; Kini, U. Analysis of exome data in a UK cohort of 603 patients with syndromic orofacial clefting identifies causal molecular pathways. Hum. Mol. Genet. 2023, 32, 1932–1942. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Chang, J.W.; Zhong, N.N.; Huang, Z.; Yue, H.; Cao, H.; Wu, Z.; He, M.; Bian, Z. Exome analyses unravel the genetic architecture of Mendelian dominant nonsyndromic orofacial clefts. Genomics 2025, 117, 111039. [Google Scholar] [CrossRef]
- Cox, L.L.; Cox, T.C.; Moreno Uribe, L.M.; Zhu, Y.; Richter, C.T.; Nidey, N.; Standley, J.M.; Deng, M.; Blue, E.; Chong, J.X.; et al. Mutations in the Epithelial Cadherin-p120-Catenin Complex Cause Mendelian Non-Syndromic Cleft Lip with or without Cleft Palate. Am. J. Hum. Genet. 2018, 102, 1143–1157. [Google Scholar] [CrossRef]
- Bishop, M.R.; Diaz Perez, K.K.; Sun, M.; Ho, S.; Chopra, P.; Mukhopadhyay, N.; Hetmanski, J.B.; Taub, M.A.; Moreno-Uribe, L.M.; Valencia-Ramirez, L.C.; et al. Genome-wide Enrichment of De Novo Coding Mutations in Orofacial Cleft Trios. Am. J. Hum. Genet. 2020, 107, 124–136. [Google Scholar] [CrossRef]
- Ishorst, N.; Henschel, L.; Thieme, F.; Drichel, D.; Sivalingam, S.; Mehrem, S.L.; Fechtner, A.C.; Fazaal, J.; Welzenbach, J.; Heimbach, A.; et al. Identification of de novo variants in nonsyndromic cleft lip with/without cleft palate patients with low polygenic risk scores. Mol. Genet. Genom. Med. 2023, 11, e2109. [Google Scholar] [CrossRef]
- Aylward, A.; Cai, Y.; Lee, A.; Blue, E.; Rabinowitz, D.; Haddad, J., Jr.; University of Washington Center for Mendelian Genomics. Using Whole Exome Sequencing to Identify Candidate Genes with Rare Variants In Nonsyndromic Cleft Lip and Palate. Genet. Epidemiol. 2016, 40, 432–441. [Google Scholar] [CrossRef]
- Li, M.; Wang, H. Pathway analysis identified a significant association between cell-cell adherens junctions-related genes and non-syndromic cleft lip/palate in 895 Asian case-parent trios. Arch. Oral Biol. 2022, 136, 105384. [Google Scholar] [CrossRef]
- Itai, T.; Yan, F.; Liu, A.; Dai, Y.; Iwaya, C.; Curtis, S.W.; Leslie, E.J.; Simon, L.M.; Jia, P.; Chen, X.; et al. Investigating gene functions and single-cell expression profiles of de novo variants in orofacial clefts. HGG Adv. 2024, 5, 100313. [Google Scholar] [CrossRef]
- Lou, S.; Ma, L.; Kan, S.; Yu, X.; Wang, Y.; Yang, F.; Zhu, G.; Fan, L.; Li, D.; Wang, H.; et al. Association Study of Genetic Variants in Autophagy Pathway and Risk of Non-syndromic Cleft Lip With or Without Cleft Palate. Front. Cell. Dev. Biol. 2020, 8, 576. [Google Scholar] [CrossRef]
- Luo, W.; Cao, H.; Zuo, Y.; He, M. Functional Analysis of IFT172 Mutations in Families with Non-syndromic Cleft Lip with or without Palate. J. Oral Sci. Res. 2025, 41, 517–520. [Google Scholar] [CrossRef]
- Pengelly, R.J.; Arias, L.; Martínez, J.; Upstill-Goddard, R.; Seaby, E.G.; Gibson, J.; Ennis, S.; Collins, A.; Briceño, I. Deleterious coding variants in multi-case families with non-syndromic cleft lip and/or palate phenotypes. Sci. Rep. 2016, 6, 30457. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.D.; Lin, Y.S.; Shi, B.; Jia, Z.L. Identifying New Susceptibility Genes of Non-Syndromic Orofacial Cleft Based on Syndromes Accompanied With Craniosynostosis. Cleft. Palate Craniofac. J. 2025, 22, 10556656251313842. [Google Scholar] [CrossRef] [PubMed]
- McKie, A.B.; Alsaedi, A.; Vogt, J.; Stuurman, K.E.; Weiss, M.M.; Shakeel, H.; Tee, L.; Morgan, N.V.; Nikkels, P.G.; van Haaften, G.; et al. Germline mutations in RYR1 are associated with foetal akinesia deformation sequence/lethal multiple pterygium syndrome. Acta Neuropathol. Commun. 2014, 2, 148. [Google Scholar] [CrossRef]
- Bharucha-Goebel, D.X.; Santi, M.; Medne, L.; Zukosky, K.; Dastgir, J.; Shieh, P.B.; Winder, T.; Tennekoon, G.; Finkel, R.S.; Dowling, J.J.; et al. Severe congenital RYR1-associated myopathy: The expanding clinicopathologic and genetic spectrum. Neurology 2013, 80, 1584–1589, Erratum in Neurology 2013, 80, 2081. [Google Scholar] [CrossRef]
- Oladayo, A.; Gowans, L.J.J.; Awotoye, W.; Alade, A.; Busch, T.; Naicker, T.; Eshete, M.A.; Adeyemo, W.L.; Hetmanski, J.B.; Zeng, E.; et al. Clinically actionable secondary findings in 130 triads from sub-Saharan African families with non-syndromic orofacial clefts. Mol. Genet. Genom. Med. 2023, 11, e2237. [Google Scholar] [CrossRef]
- Quinodoz, M.; Royer-Bertrand, B.; Cisarova, K.; Di Gioia, S.A.; Superti-Furga, A.; Rivolta, C. DOMINO: Using Machine Learning to Predict Genes Associated with Dominant Disorders. Am. J. Hum. Genet. 2017, 101, 623–629. [Google Scholar] [CrossRef]
- Mostowska, A.; Gaczkowska, A.; Żukowski, K.; Ludwig, K.U.; Hozyasz, K.K.; Wójcicki, P.; Mangold, E.; Böhmer, A.C.; Heilmann-Heimbach, S.; Knapp, M.; et al. Common variants in DLG1 locus are associated with non-syndromic cleft lip with or without cleft palate. Clin. Genet. 2018, 93, 784–793. [Google Scholar] [CrossRef]
- Ang, S.L.; Wierda, A.; Wong, D.; Stevens, K.A.; Cascio, S.; Rossant, J.; Zaret, K.S. The formation and maintenance of the definitive endoderm lineage in the mouse: Involvement of HNF3/forkhead proteins. Development 1993, 119, 1301–1315. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, D.C.; Ruiz i Altaba, A.; Chen, W.S.; Hoodless, P.; Prezioso, V.R.; Jessell, T.M.; Darnell, J.E., Jr. The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell 1994, 78, 575–588. [Google Scholar] [CrossRef]
- Mavromatakis, Y.E.; Lin, W.; Metzakopian, E.; Ferri, A.L.; Yan, C.H.; Sasaki, H.; Whisett, J.; Ang, S.L. Foxa1 and Foxa2 positively and negatively regulate Shh signalling to specify ventral midbrain progenitor identity. Mech. Dev. 2011, 128, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Hammond, N.L.; Dixon, M.J. Revisiting the embryogenesis of lip and palate development. Oral Dis. 2022, 28, 1306–1326. [Google Scholar] [CrossRef] [PubMed]
- Won, H.J.; Kim, J.W.; Won, H.S.; Shin, J.O. Gene Regulatory Networks and Signaling Pathways in Palatogenesis and Cleft Palate: A Comprehensive Review. Cells 2023, 12, 1954. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.G.; Wetherbee, J.J.; Rosenfeld, J.A.; Hersh, J.H. 20p11 deletion in a female child with panhypopituitarism, cleft lip and palate, dysmorphic facial features, global developmental delay and seizure disorder. Am. J. Med. Genet. A 2011, 155A, 186–191. [Google Scholar] [CrossRef]
- Giri, D.; Vignola, M.L.; Gualtieri, A.; Scagliotti, V.; McNamara, P.; Peak, M.; Didi, M.; Gaston-Massuet, C.; Senniappan, S. Novel FOXA2 mutation causes Hyperinsulinism, Hypopituitarism with Craniofacial and Endoderm-derived organ abnormalities. Hum. Mol. Genet. 2017, 26, 4315–4326. [Google Scholar] [CrossRef]
- Shen, H.; McElhinny, A.S.; Cao, Y.; Gao, P.; Liu, J.; Bronson, R.; Griffin, J.D.; Wu, L. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006, 20, 675–688. [Google Scholar] [CrossRef]
- Alves-Guerra, M.C.; Ronchini, C.; Capobianco, A.J. Mastermind-like 1 Is a specific coactivator of beta-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res. 2007, 67, 8690–8698. [Google Scholar] [CrossRef]
- Zhao, Y.; Katzman, R.B.; Delmolino, L.M.; Bhat, I.; Zhang, Y.; Gurumurthy, C.B.; Germaniuk-Kurowska, A.; Reddi, H.V.; Solomon, A.; Zeng, M.S.; et al. The notch regulator MAML1 interacts with p53 and functions as a coactivator. J. Biol. Chem. 2007, 282, 11969–11981. [Google Scholar] [CrossRef]
- Oyama, T.; Harigaya, K.; Sasaki, N.; Okamura, Y.; Kokubo, H.; Saga, Y.; Hozumi, K.; Suganami, A.; Tamura, Y.; Nagase, T.; et al. Mastermind-like 1 (MamL1) and mastermind-like 3 (MamL3) are essential for Notch signaling in vivo. Development 2011, 138, 5235–5246. [Google Scholar] [CrossRef]
- Quaranta, R.; Pelullo, M.; Zema, S.; Nardozza, F.; Checquolo, S.; Lauer, D.M.; Bufalieri, F.; Palermo, R.; Felli, M.P.; Vacca, A.; et al. Maml1 acts cooperatively with Gli proteins to regulate sonic hedgehog signaling pathway. Cell Death Dis. 2017, 8, e2942. [Google Scholar] [CrossRef]
- Watanabe, T.; Oyama, T.; Asada, M.; Harada, D.; Ito, Y.; Inagawa, M.; Suzuki, Y.; Sugano, S.; Katsube, K.; Karsenty, G.; et al. MAML1 enhances the transcriptional activity of Runx2 and plays a role in bone development. PLoS Genet. 2013, 9, e1003132. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhou, L.; Li, M.; Zhao, J.; Liu, Y.; Chen, Y.; Qin, X.; Wang, S.; Chen, H.; Piao, Y.; et al. Genome-wide CRISPR/Cas9 knockout screening uncovers ZNF319 as a novel tumor suppressor critical for breast cancer metastasis. Biochem. Biophys. Res. Commun. 2022, 589, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Mani, A. Pathogenicity of De Novo Rare Variants: Challenges and Opportunities. Circ. Cardiovasc. Genet. 2017, 10, e002013. [Google Scholar] [CrossRef] [PubMed]
- Robinson, K.R.; Curtis, S.W.; Paschall, J.E.; Beaty, T.H.; Butali, A.; Buxó, C.J.; Cutler, D.J.; Epstein, M.P.; Hecht, J.T.; Uribe, L.M.; et al. Distinguishing syndromic and nonsyndromic cleft palate through analysis of protein-altering de novo variants in 816 trios. medRxiv 2025. preprint. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Topczewski, J.; Dale, R.M.; Sisson, B.E. Planar cell polarity signaling in craniofacial development. Organogenesis 2011, 7, 255–259. [Google Scholar] [CrossRef]
- Peng, Z.; Gong, Y.; Liang, X. Role of FAT1 in health and disease. Oncol. Lett. 2021, 21, 398. [Google Scholar] [CrossRef]
- Cai, S.; Si, N.; Wang, Y.; Yin, N. Transcriptomic analysis of the upper lip and primary palate development in mice. Front. Genet. 2023, 13, 1039850. [Google Scholar] [CrossRef]
- Gorivodsky, M.; Mukhopadhyay, M.; Wilsch-Braeuninger, M.; Phillips, M.; Teufel, A.; Kim, C.; Malik, N.; Huttner, W.; Westphal, H. Intraflagellar transport protein 172 is essential for primary cilia formation and plays a vital role in patterning the mammalian brain. Dev. Biol. 2009, 325, 24–32. [Google Scholar] [CrossRef]
- Brugmann, S.A.; Cordero, D.R.; Helms, J.A. Craniofacial ciliopathies: A new classification for craniofacial disorders. Am. J. Med. Genet. A 2010, 152A, 2995–3006. [Google Scholar] [CrossRef]
- Alzarka, B.; Charnaya, O.; Gunay-Aygun, M. Diseases of the primary cilia: A clinical characteristics review. Pediatr. Nephrol. 2025, 40, 611–627. [Google Scholar] [CrossRef]
- Meister, G. Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 2013, 14, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Qian, Q.; Li, J.; Gong, P.; Jiao, X.; Mao, X.; Xiao, B.; Long, L.; Yang, Z. De novo variants in AGO1 recapitulate a heterogeneous neurodevelopmental disorder phenotype. Clin. Genet. 2022, 101, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Takagi, M.; Ono, S.; Kumaki, T.; Nishimura, N.; Murakami, H.; Enomoto, Y.; Naruto, T.; Ueda, H.; Kurosawa, K. Complex congenital cardiovascular anomaly in a patient with AGO1-associated disorder. Am. J. Med. Genet. A 2023, 191, 882–892. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, B.T.; He, X. Frizzled and LRP5/6 receptors for Wnt/β-catenin signaling. Cold Spring Harb. Perspect. Biol. 2012, 4, a007880. [Google Scholar] [CrossRef]
- Hubert, K.A.; Wellik, D.M. Hox genes in development and beyond. Development 2023, 150, dev192476, Correction in Development 2024, 151, dev202770. [Google Scholar] [CrossRef]
- Li, A.; Qin, G.; Suzuki, A.; Gajera, M.; Iwata, J.; Jia, P.; Zhao, Z. Network-based identification of critical regulators as putative drivers of human cleft lip. BMC Med. Genom. 2019, 12, 16. [Google Scholar] [CrossRef]
- Blanton, S.H.; Henry, R.R.; Yuan, Q.; Mulliken, J.B.; Stal, S.; Finnell, R.H.; Hecht, J.T. Folate pathway and nonsyndromic cleft lip and palate. Birth Defects Res. A Clin. Mol. Teratol. 2011, 91, 50–60. [Google Scholar] [CrossRef]
- Zhang, H.; Xie, J.; So, K.K.H.; Tong, K.K.; Sae-Pang, J.J.; Wang, L.; Tsang, S.L.; Chan, W.Y.; Wong, E.Y.M.; Sham, M.H. Hoxb3 Regulates Jag1 Expression in Pharyngeal Epithelium and Affects Interaction with Neural Crest Cells. Front. Physiol. 2021, 11, 612230. [Google Scholar] [CrossRef]
- Cheng, X.; Du, F.; Long, X.; Huang, J. Genetic Inheritance Models of Non-Syndromic Cleft Lip with or without Palate: From Monogenic to Polygenic. Genes 2023, 14, 1859. [Google Scholar] [CrossRef] [PubMed]
- Inchingolo, A.M.; Fatone, M.C.; Malcangi, G.; Avantario, P.; Piras, F.; Patano, A.; Di Pede, C.; Netti, A.; Ciocia, A.M.; De Ruvo, E.; et al. Modifiable Risk Factors of Non-Syndromic Orofacial Clefts: A Systematic Review. Children 2022, 9, 1846. [Google Scholar] [CrossRef] [PubMed]
- Sabbagh, H.J.; Hassan, M.H.; Innes, N.P.; Elkodary, H.M.; Little, J.; Mossey, P.A. Passive smoking in the etiology of non-syndromic orofacial clefts: A systematic review and meta-analysis. PLoS ONE 2015, 10, e0116963. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Li, J.; Li, Y.; Zou, S. Maternal alcohol consumption and oral clefts: A meta-analysis. Br. J. Oral Maxillofac. Surg. 2019, 57, 839–846. [Google Scholar] [CrossRef]
- De-Regil, L.M.; Pena-Rosas, J.P.; Fernandez-Gaxiola, A.C.; Rayco-Solon, P. Effects and safety of periconceptional oral folate supplementation for preventing birth defects. Cochrane Database Syst. Rev. 2015, 12, CD007950. [Google Scholar] [CrossRef]
- Block, S.R.; Watkins, S.M.; Salemi, J.L.; Rutkowski, R.; Tanner, J.P.; Correia, J.A.; Kirby, R.S. Maternal pre-pregnancy body mass index and risk of selected birth defects: Evidence of a dose-response relationship. Paediatr. Peri. Epidemiol. 2013, 27, 521–531. [Google Scholar] [CrossRef]
- Acs, L.; Banyai, D.; Nemes, B.; Nagy, K.; Acs, N.; Banhidy, F.; Rózsa, N. Maternal-related factors in the origin of isolated cleft palate-A population-based case-control study. Orthod. Craniofac. Res. 2020, 23, 174–180. [Google Scholar] [CrossRef]
- Carmichael, S.L.; Shaw, G.M. Maternal corticosteroid use and risk of selected congenital anomalies. Am. J. Med. Genet. 1999, 86, 242–244. [Google Scholar] [CrossRef]
- Huybrechts, K.F.; Hernandez-Diaz, S.; Straub, L.; Gray, K.J.; Zhu, Y.; Patorno, E.; Desai, R.J.; Mogun, H.; Bateman, B.T. Association of Maternal First-Trimester Ondansetron Use with Cardiac Malformations and Oral Clefts in Offspring. JAMA 2018, 320, 2429–2437. [Google Scholar] [CrossRef]
- Martinelli, M.; Palmieri, A.; Carinci, F.; Scapoli, L. Non-syndromic Cleft Palate: An Overview on Human Genetic and Environmental Risk Factors. Front. Cell Dev. Biol. 2020, 8, 592271. [Google Scholar] [CrossRef]
- Idelfonso-García, O.G.; Alarcón-Sánchez, B.R.; Vásquez-Garzón, V.R.; Baltiérrez-Hoyos, R.; Villa-Treviño, S.; Muriel, P.; Serrano, H.; Pérez-Carreón, J.I.; Arellanes-Robledo, J. Is Nucleoredoxin a Master Regulator of Cellular Redox Homeostasis? Its Implication in Different Pathologies. Antioxidants 2022, 11, 670. [Google Scholar] [CrossRef]
- Scapoli, L.; Carinci, F.; Palmieri, A.; Cura, F.; Baj, A.; Beltramini, G.; Docimo, R.; Martinelli, M. Copy number variation analysis of twin pairs discordant for cleft lip with or without cleft palate. Int. J. Immunopathol. Pharmacol. 2019, 33, 2058738419855873. [Google Scholar] [CrossRef]
- Zieger, H.K.; Weinhold, L.; Schmidt, A.; Holtgrewe, M.; Juranek, S.A.; Siewert, A.; Scheer, A.B.; Thieme, F.; Mangold, E.; Ishorst, N.; et al. Prioritization of non-coding elements involved in non-syndromic cleft lip with/without cleft palate through genome-wide analysis of de novo mutations. HGG Adv. 2022, 4, 100166. [Google Scholar] [CrossRef]
- Leslie, E.J.; Marazita, M.L. Genetics of cleft lip and cleft palate. Am. J. Med. Genet. C Semin. Med. Genet. 2013, 163C, 246–258. [Google Scholar] [CrossRef]
- Gaczkowska, A.; Żukowski, K.; Biedziak, B.; Hozyasz, K.K.; Wójcicki, P.; Zadurska, M.; Budner, M.; Lasota, A.; Szponar-Żurowska, A.; Jagodziński, P.P.; et al. Association of CDKAL1 nucleotide variants with the risk of non-syndromic cleft lip with or without cleft palate. J. Hum. Genet. 2018, 63, 397–406. [Google Scholar] [CrossRef]
- Dąbrowska, J.; Biedziak, B.; Bogdanowicz, A.; Mostowska, A. Identification of Novel Risk Variants of Non-Syndromic Cleft Palate by Targeted Gene Panel Sequencing. J. Clin. Med. 2023, 12, 2051. [Google Scholar] [CrossRef]
- Sawicka-Gutaj, N.; Gruszczyński, D.; Guzik, P.; Mostowska, A.; Walkowiak, J. Ethics of Human Studies in the Light of the Declaration of Helsinki—A Mini-Review. J. Med. Sci. 2022, 91, e700. [Google Scholar] [CrossRef]
| Patient | Gender | Type of Cleft a | Family History b | Gene | Variant Effect | gnomAD | Genotype d | Previous Gene Association with OFC e | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA | Protein | rs Number | Frequency c | Patient | Father | Mother | Syndromic | Non-Syndromic | KO Mice | |||||
| DE NOVO VARIANTS | ||||||||||||||
| CLP_1 | M | CLP/R | NO | FOXA2 | c.779G>C | p.Arg260Pro | novel | NR | HET | wt | wt | |||
| CLP_2 | M | CLP/R | NO | MAML1 | c.193C>T | p.Gln65Ter | novel | NR | HET | wt | wt | |||
| CLP_3 | M | CLP/RL | NO | ZNF319 | c.190C>T | p.Gln64Ter | novel | NR | HET | wt | wt | |||
| VARIANTS INHERITED FROM AFFECTED PARENT | ||||||||||||||
| CLP_4 | M | CLP/RL | YES—father | GDF7 | c.1081C>T | p.Leu361Phe | novel | NR | HET | HET | wt | |||
| CLP_5 | M | CLP/R | YES—mother | FAT1 | c.2681dupA | p.Ile895AspfsTer2 | novel | NR | HET | wt | HET | |||
| CLP_6 | M | CLP/RL | YES—father | VWA8 | c.2003G>A | p.Arg668Gln | rs138075452 | 4.72 × 10−3 | HET | HET | wt | X [28] | ||
| VARIANTS INHERITED FROM HEALTHY PARENT | ||||||||||||||
| CLP_7 | F | CLP/RL | YES—ex. paternal fam. | CTNNA1 | c.2429C>T | p.Ser810Phe | novel | NR | HET | HET | wt | X [29,30] | ||
| CLP_8 | M | CLP/L | YES—ex. maternal fam. | EFNB2 | c.590G>T | p.Ser197Ile | rs250151379 | NR | HET | wt | HET | X [30] | X | |
| CLP_9 | F | CLP/L | YES—ex. maternal fam. | HIF1A | c.806G>A | p.Arg269Gln | rs2044522437 | 6.78 × 10−6 | HET | wt | HET | X [31] | ||
| CLP_10 | M | CLP/RL | YES—ex. family | LAMA5 | c.3520_3521delAG | p.Arg1174AlafsTer19 | novel | NR | HET | HET | wt | X [22] | ||
| CLP_11 | M | CLP/L | YES—sister f | LRP5 | c.3638-1G>A | novel | NR | HET | wt | HET | ||||
| CLP_12 | F | CLP/L | YES—brother f | |||||||||||
| CLP_13 | F | CL/R | NO | AGO1 | c.1823C>T | p.Ser608Phe | rs1353927503 | 8.48 × 10−7 | HET | wt | HET | |||
| CLP_14 | M | CLP/L | NO | ARID1A | c.6428G>A | p.Arg2143His | rs2124148967 | 1.70 × 10−6 | HET | wt | HET | |||
| CLP_15 | M | CL/R | NO | ATN1 | c.2239C>A | p.Pro747Thr | rs782670138 | 7.85 × 10−6 | HET | HET | wt | X | ||
| CLP_16 | M | CLP/L | NO | ATP1A1 | c.1934C>T | p.Ala645Val | rss2101057973 | NR | HET | HET | wt | |||
| CLP_17 | M | CLP/RL | NO | EXT1 | c.493C>G | p.Gln165Glu | rs2130043213 | NR | HET | wt | HET | X | ||
| CLP_18 | M | CLP/L | NO | HAND1 | c.328A>C | p.Ile110Leu | rs905545828 | 1.70 × 10−6 | HET | HET | wt | X | ||
| CLP_19 | F | CLP/RL | NO | HOXB3 | c.374T>G | p.Leu125Arg | rs2068774883 | NR | HET | wt | HET | |||
| CLP_20 g | F | CLP/R | NO | IFT172 | c.4096G>T | p.Asp1366Tyr | rs776240963 | 5.09 × 10−6 | HET | wt | HET | X | X [32] | X |
| CLP_21 | M | CL/R | NO | KRT17 | c.695A>G | p.Gln232Arg | rs1484519975 | 1.7 × 10−6 | HET | wt | HET | X | ||
| CLP_22 | M | CLP/RL | NO | MEN1 | c.1565G>T | p.Arg522Leu | novel | NR | HET | wt | HET | X | ||
| CLP_23 | F | CL/R | NO | MYH3 | c.52C>T | p.Arg18Trp | rs750940457 | 5.93 × 10−6 | HET | wt | HET | X | X [33,34] | |
| CLP_24 | M | CLP/RL | NO | NCOR2 | c.650C>T | p.Pro217Leu | rs199588853 | 2.72 × 10−5 | HET | wt | HET | X | ||
| CLP_25 | F | CLP/L | NO | NXN | c.503A>C | p.Lys168Thr | novel | NR | HET | wt | HET | X | X | |
| CLP_26 | F | CLP/R | NO | ROBO1 | c.2026C>T | p.Arg676Trp | novel | NR | HET | HET | wt | X | ||
| CLP_27 | F | CLP/RL | NO | RPGRIP1L | c.413A>G | p.Gln138Arg | rs2544706158 | NR | HET | wt | HET | X | X | |
| CLP_28 | M | CLP | NO | RYR1 | c.9136C>G h | p.Leu3046Val | rs2145659189 | 0.00 | HET | HET | wt | X [35,36] | X [33,37] | X |
| RYR1 | c.9198C>G h | p.Asn3066Lys | rs201863144 | 0.00 | HET | HET | wt | |||||||
| CLP_29 | F | CLP/RL | NO | SLC32A1 | c.466G>T | p.Ala156Ser | rs2515237904 | NR | HET | HET | wt | X | ||
| CLP_30 | F | CLP/R | NO | TENM4 | c.7916G>A | p.Arg2639Gln | rs775067963 | 2.20 × 10−5 | HET | HET | wt | X | ||
| CLP_31 | F | CLP/L | NO | TP53BP2 | c.1757C>A | p.Ala586Asp | rs2464444021 | NR | HET | wt | HET | X | ||
| GENE | VARIANT | ACMG Classification c | In silico Pathogenicity Prediction d | ADA | CADD | DANN | MutPred-LOF h | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual Predictions | Meta Scores | |||||||||||||||||||||||||||||||
| Name | pLi a | Class | AD b | DNA Change | Protein Change | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | Score e | Score f | Score g | ||
| AGO1 | 1.00 | Very likely dominant | 0.999 | c.1823C>T | p.Ser608Phe | VUS | D | D | D | D | D | H | D | D | PS | D | D | D | D | D | P | D | D | T | D | 0.9 | D | D | 29.20 | 1.00 | ||
| ARID1A | 1.00 | Very likely dominant | 1.000 | c.6428G>A | p.Arg2143His | VUS | D | D | D | D | D | M | D | D | UC | D | T | D | D | D | P | D | T | T | D | 0.91 | D | D | 32.00 | 1.00 | ||
| ATN1 | 1.00 | Very likely dominant | 0.943 | c.2239C>A | p.Pro747Thr | VUS | D | D | D | D | D | M | D | D | BP | T | D | D | D | D | P | D | D | D | D | 0.8 | D | D | 26.30 | 1.00 | ||
| ATP1A1 | 1.00 | Very likely dominant | 0.999 | c.1934C>T | p.Ala645Val | VUS | D | D | D | D | D | L | D | D | PS | D | D | D | D | D | P | D | D | D | D | 0.95 | D | D | 27.00 | 1.00 | ||
| CTNNA1 | 0.97 | Very likely dominant | 1.000 | c.2429C>T | p.Ser810Phe | VUS | D | D | D | D | N | M | D | D | PP | D | D | D | D | D | P | D | T | T | D | 0.83 | D | D | 33.00 | 1.00 | ||
| EFNB2 | 1.00 | Very likely dominant | 0.998 | c.590G>T | p.Ser197Ile | VUS | B | B | L | D | PP | T | T | D | D | T | B | D | T | D | D | D | D | 25.40 | 0.99 | |||||||
| EXT1 | 1.00 | Very likely dominant | 0.991 | c.493C>G | p.Gln165Glu | VUS | T | T | P | B | D | L | N | D | UC | T | D | D | D | D | B | D | D | D | D | 0.81 | D | D | 24.70 | 0.58 | ||
| FAT1 | 0.00 | Very likely recessive | 0.178 | c.2681dupA | p.Ile895AspfsTer2 | VUS | 0.54 | |||||||||||||||||||||||||
| FOXA2 | 0.97 | Likely dominant | 0.792 | c.779G>C | p.Arg260Pro | VUS | D | D | D | PS | D | D | D | D | LP | D | D | D | D | 0.99 | D | D | 35.00 | 1.00 | ||||||||
| GDF7 | 0.00 | Very likely dominant | 0.991 | c.1081C>T | p.Leu361Phe | VUS | D | D | D | D | N | M | D | D | UC | D | D | D | D | D | P | N | D | D | D | 0.86 | D | T | 27.00 | 1.00 | ||
| HAND1 | 0.01 | Very likely dominant | 0.839 | c.328A>C | p.Ile110Leu | VUS | T | T | D | D | N | N | N | D | UC | D | D | D | D | D | P | D | D | D | D | 0.9 | D | D | 25.90 | 0.99 | ||
| HIF1A | 1.00 | Very likely dominant | 1.000 | c.806G>A | p.Arg269Gln | VUS | D | D | D | D | D | M | D | D | PM | D | D | D | D | D | P | D | T | T | D | 0.85 | D | D | 28.40 | 1.00 | ||
| HOXB3 | 0.00 | Very likely dominant | 0.988 | c.374T>G | p.Leu125Arg | VUS | T | T | D | D | M | D | D | UC | T | D | D | D | A | D | D | D | 0.87 | D | D | 26.40 | 0.99 | |||||
| IFT172 | 0.00 | Very likely recessive | 0.152 | c.4096G>T | p.Asp1366Tyr | VUS | D | D | D | D | D | M | D | D | PP | T | D | D | D | D | A | D | D | D | D | 0.93 | D | D | 26.30 | 1.00 | ||
| KRT17 | 0.00 | Very likely recessive | 0.152 | c.695A>G | p.Gln232Arg | VUS | D | D | B | B | D | M | D | D | UC | T | D | D | T | D | B | D | D | D | D | 0.85 | D | T | 25.10 | 1.00 | ||
| LAMA5 | 0.00 | Likely recessive | 0.201 | c.3520_3521delAG | p.Arg1174AlafsTer19 | VUS | 0.55 | |||||||||||||||||||||||||
| LRP5 | 0.94 | Very likely dominant | 0.946 | c.3638-1G>A | VUS | 0.999 | 0.99 | |||||||||||||||||||||||||
| MAML1 | 1.00 | Very likely dominant | 0.998 | c.193C>T | p.Gln65Ter | LP | A | D | N | D | D | 37.00 | 1.00 | 0.50 | ||||||||||||||||||
| MEN1 | 1.00 | Very likely dominant | 1.000 | c.1565G>T | p.Arg522Leu | VUS | D | UC | D | D | T | D | D | D | 0.93 | D | D | 23.40 | 1.00 | |||||||||||||
| MYH3 | 0.00 | Likely recessive | 0.400 | c.52C>T | p.Arg18Trp | VUS | D | D | D | H | D | PS | D | D | D | D | D | P | D | D | D | D | D | D | 32.00 | 1.00 | ||||||
| NCOR2 | 1.00 | Very likely dominant | 0.999 | c.650C>T | p.Pro217Leu | VUS | D | D | N | L | D | D | BP | T | D | D | D | D | A | D | T | T | D | 0.74 | T | T | 25.80 | 0.98 | ||||
| NXN | 0.37 | Dominant or recessive | 0.510 | c.503A>C | p.Lys168Thr | VUS | T | T | D | D | L | D | D | PM | D | T | D | D | D | A | D | T | D | D | 0.79 | D | D | 24.00 | 1.00 | |||
| ROBO1 | 0.00 | Dominant or recessive | 0.539 | c.2026C>T | p.Arg676Trp | VUS | D | D | D | D | D | M | D | D | PM | T | T | D | D | D | P | D | T | T | D | 0.81 | D | D | 27.60 | 1.00 | ||
| RPGRIP1L | 0.00 | Very likely recessive | 0.071 | c.413A>G | p.Gln138Arg | VUS | D | D | M | D | BM | T | D | D | T | B | D | D | D | D | 0.67 | D | D | 23.60 | 1.00 | |||||||
| RYR1 | 0.00 | Very likely dominant | 0.987 | c.9136C>G | p.Leu3046Val | VUS | D | P | B | N | M | N | D | PP | T | D | D | D | D | A | D | D | T | D | 0.74 | T | T | 22.20 | 0.99 | |||
| RYR1 | 0.00 | Very likely dominant | 0.987 | c.9198C>G | p.Asn3066Lys | VUS | T | B | B | D | L | D | D | UC | T | D | D | D | D | P | D | D | D | D | 0.72 | T | T | 20.10 | 1.00 | |||
| SLC32A1 | 1.00 | Likely dominant | 0.737 | c.466G>T | p.Ala156Ser | VUS | D | D | D | D | D | M | D | T | PP | D | T | D | D | T | A | D | T | T | D | 0.53 | T | T | 29.70 | 1.00 | ||
| TENM4 | 1.00 | Very likely dominant | 0.878 | c.7916G>A | p.Arg2639Gln | VUS | D | D | D | D | N | M | N | D | UC | T | T | D | D | T | B | D | D | D | D | 0.79 | T | T | 28.60 | 1.00 | ||
| TP53BP2 | 0.00 | Dominant or recessive | 0.438 | c.1757C>A | p.Ala586Asp | VUS | D | D | N | D | D | UC | T | D | D | T | LP | D | T | T | D | 0.52 | D | D | 24.00 | 1.00 | ||||||
| VWA8 | 0.00 | Very likely recessive | 0.092 | c.2003G>A | p.Arg668Gln | VUS (leaning LP) | D | D | D | D | A | M | D | D | UC | T | T | T | D | D | A | D | T | T | T | 0.73 | T | D | 27.90 | 1.00 | ||
| ZNF319 | 0.51 | Likely dominant | 0.744 | c.190C>T | p.Gln64Ter | LP | D | N | D | D | 38.00 | 0.99 | 0.45 | |||||||||||||||||||
| Number of SNVs Plotted | The Most Significant SNV Within a Locus | ||||||
|---|---|---|---|---|---|---|---|
| Gene a | rs Number | Location b | Position Relative to Gene c | MAF d | ptrend Value e | Allelic OR f | |
| AGO1 | 11 | rs6682769 | chr1: 35860595 | 22.6 kb upstream, intergenic | 0.06 | 3.36 × 10−2 | 1.57 |
| ARID1A | 20 | rs4466675 | chr1: 26693521 | 2.5 kb upstream, intergenic | 0.48 | 2.05 × 10−2 | 0.79 |
| ATN1 | 43 | rs10744724 | chr12: 6956118 | 13.8 kb downstream, within PTPN6 | 0.06 | 6.70 × 10−3 | 0.50 |
| ATP1A1 | 29 | rs766429 | chr1: 116495418 | 90.6 kb downstream, intergenic | 0.21 | 1.76 × 10−1 | 0.84 |
| CTNNA1 | 33 | rs12108892 | chr5: 138963941 | 28.9 kb downstream, within SIL1 | 0.39 | 8.13 × 10−2 | 0.83 |
| EFNB2 | 91 | rs7490929 | chr13: 106431374 | 58.4 kb downstream, intergenic | 0.07 | 3.75 × 10−2 | 1.51 |
| EXT1 | 128 | rs7837891 | chr8: 117807339 | exonic (synonymous variant) | 0.44 | 9.56 × 10−4 | 0.70 |
| FAT1 | 93 | rs28647489 | chr4: 186609868 | exonic (missense variant) | 0.14 | 3.18 × 10−2 | 0.71 |
| FOXA2 | 35 | rs2404167 | chr20: 22528795 | 52.2 kb downstream, intergenic | 0.10 | 3.77 × 10−2 | 1.62 |
| GDF7 | 64 | rs340596 | chr2: 20689994 | 10.8 kb downstream, within C2orf43 | 0.43 | 7.43 × 10−3 | 0.76 |
| HAND1 | 86 | rs283438 | chr5: 154494000 | 15.8 kb upstream, intergenic | 0.17 | 3.06 × 10−2 | 1.34 |
| HIF1A | 35 | rs17099248 | chr14: 61832287 | 84.0 kb downstream, intergenic | 0.12 | 2.57 × 10−2 | 0.68 |
| HOXB3 | 52 | rs890435 | chr17: 48642037 | 51.8 kb upstream, intergenic | 0.40 | 1.03 × 10−2 | 1.31 |
| IFT172 | 33 | rs3811644 | chr2: 27579938 | 90.2 kb upstream, within SPATA31H1 | 0.21 | 2.49 × 10−1 | 0.86 |
| KRT17 | 31 | rs7503702 | chr17: 41609234 | 10.2 kb upstream, intergenic | 0.40 | 1.04 × 10−2 | 1.30 |
| LAMA5 | 72 | rs4925238 | chr20: 62431508 | 64.2 kb upstream, intergenic | 0.06 | 1.79 × 10−2 | 1.61 |
| LRP5 | 51 | rs314779 | chr11: 68330358 | intronic | 0.23 | 2.76 × 10−2 | 0.76 |
| MAML1 | 41 | rs28564876 | chr5: 179651067 | 81.8 kb upstream, intergenic | 0.41 | 9.34 × 10−4 | 1.43 |
| MEN1 | 29 | rs10897529 | chr11: 64829444 | 18.9 kb upstream, within CDC42BPG | 0.35 | 2.31 × 10−1 | 1.14 |
| MYH3 | 30 | rs6503319 | chr17: 10670842 | 13.5 kb upstream, intergenic | 0.25 | 3.90 × 10−2 | 1.27 |
| NCOR2 | 154 | rs7132377 | chr12: 124667448 | 99.8 kb upstream, intergenic | 0.09 | 4.63 × 10−3 | 0.56 |
| NXN | 127 | rs8081951 | chr17: 933301 | intronic | 0.18 | 2.52 × 10−4 | 1.62 |
| ROBO1 | 176 | rs1865862 | chr3: 78754700 | intronic | 0.28 | 1.25 × 10−2 | 0.74 |
| RPGRIP1L | 58 | rs1421085 | chr16: 53767042 | 63.2 kb upstream, within FTO | 0.48 | 2.49 × 10−3 | 1.36 |
| RYR1 | 59 | rs4802351 | chr19: 38358156 | 72.5 kb upstream, within CATSPERG | 0.14 | 2.50 × 10−2 | 0.70 |
| SLC32A1 | 41 | rs2902891 | chr20: 38681144 | 43.3 kb upstream, intergenic | 0.30 | 7.65 × 10−2 | 1.22 |
| TENM4 | 312 | rs12362098 | chr11: 78857065 | intronic | 0.16 | 2.07 × 10−3 | 1.50 |
| TP53BP2 | 51 | rs7535882 | chr1: 223857777 | 11.8 kb upstream, intergenic | 0.07 | 1.16 × 10−3 | 1.80 |
| VWA8 | 118 | rs12585194 | chr13: 41977432 | 16.3 kb upstream, within VWA8−AS1 | 0.13 | 1.43 × 10−2 | 1.46 |
| ZNF319 | 48 | rs17241022 | chr16: 57920621 | 74.1 kb downstream, within CNGB1 | 0.10 | 4.40 × 10−2 | 0.68 |
| Biological Process | GO Term | p Value | padj Value a | Genes |
|---|---|---|---|---|
| tissue morphogenesis | GO:0048729 | 1.34 × 10−10 | 1.21 × 10−6 | EFNB2; EXT1; FAT1; GDF7; HAND1; HIF1A; IFT172; KRT17; LAMA5; LRP5; ROBO1; RPGRIP1L |
| embryonic morphogenesis | GO:0048598 | 1.45 × 10−10 | 1.31 × 10−6 | EXT1; FOXA2; GDF7; HAND1; HIF1A; HOXB3; IFT172; LAMA5; LRP5; MYH3; RPGRIP1L; TENM4 |
| animal organ morphogenesis | GO:0009887 | 2.48 × 10−10 | 2.24 × 10−6 | CTNNA1; EFNB2; EXT1; FAT1; GDF7; HAND1; HIF1A; HOXB3; IFT172; LAMA5; LRP5; ROBO1; RPGRIP1L; RYR1 |
| morphogenesis of an epithelium | GO:0002009 | 3.04 × 10−10 | 2.75 × 10−6 | EFNB2; EXT1; FAT1; GDF7; HAND1; HIF1A; IFT172; KRT17; LAMA5; LRP5; RPGRIP1L |
| circulatory system development | GO:0072359 | 9.43 × 10−10 | 8.52 × 10−6 | AGO1; EFNB2; EXT1; HAND1; HIF1A; HOXB3; IFT172; LRP5; MAML1; NXN; ROBO1; RPGRIP1L; RYR1; TENM4 |
| regionalization | GO:0003002 | 1.29 × 10−9 | 1.17 × 10−5 | EXT1; FOXA2; HAND1; HIF1A; HOXB3; IFT172; LAMA5; LRP5; ROBO1; RPGRIP1L |
| tissue development | GO:0009888 | 1.98 × 10−9 | 1.79 × 10−5 | EFNB2; EXT1; FAT1; FOXA2; GDF7; HAND1; HIF1A; HOXB3; IFT172; KRT17; LAMA5; LRP5; MAML1; ROBO1; RPGRIP1L; RYR1; TENM4 |
| pattern specification process | GO:0007389 | 3.33 × 10−9 | 3.00 × 10−5 | EXT1; FOXA2; HAND1; HIF1A; HOXB3; IFT172; LAMA5; LRP5; ROBO1; RPGRIP1L |
| embryo development | GO:0009790 | 1.14 × 10−8 | 1.03 × 10−4 | EXT1; FOXA2; GDF7; HAND1; HIF1A; HOXB3; IFT172; LAMA5; LRP5; MYH3; NXN; RPGRIP1L; TENM4 |
| heart development | GO:0007507 | 2.95 × 10−8 | 2.67 × 10−4 | EFNB2; EXT1; HAND1; HIF1A; IFT172; MAML1; ROBO1; RPGRIP1L; RYR1; TENM4 |
| gland development | GO:0048732 | 4.08 × 10−8 | 3.69 × 10−4 | EXT1; GDF7; HIF1A; HOXB3; LAMA5; LRP5; NCOR2; ROBO1; RPGRIP1L |
| central nervous system development | GO:0007417 | 4.52 × 10−8 | 4.08 × 10−4 | ATN1; EXT1; GDF7; HIF1A; HOXB3; IFT172; LRP5; NCOR2; ROBO1; RPGRIP1L; SLC32A1; TENM4 |
| epithelial tube morphogenesis | GO:0060562 | 6.40 × 10−8 | 5.78 × 10−4 | EFNB2; EXT1; GDF7; HAND1; HIF1A; IFT172; LAMA5; LRP5 |
| tube development | GO:0035295 | 8.68 × 10−8 | 7.84 × 10−4 | AGO1; EFNB2; EXT1; GDF7; HAND1; HIF1A; HOXB3; IFT172; LAMA5; LRP5; ROBO1; RPGRIP1L |
| tube morphogenesis | GO:0035239 | 9.33 × 10−8 | 8.43 × 10−4 | AGO1; EFNB2; EXT1; GDF7; HAND1; HIF1A; HOXB3; IFT172; LAMA5; LRP5; ROBO1 |
| anatomical structure formation involved in morphogenesis | GO:0048646 | 2.19 × 10−7 | 1.97 × 10−3 | AGO1; EFNB2; EXT1; FOXA2; GDF7; HAND1; HIF1A; HOXB3; IFT172; MYH3; ROBO1; TENM4 |
| epithelium development | GO:0060429 | 3.06 × 10−7 | 2.77 × 10−3 | EFNB2; EXT1; FAT1; GDF7; HAND1; HIF1A; IFT172; KRT17; LAMA5; LRP5; ROBO1; RPGRIP1L |
| head development | GO:0060322 | 3.38 × 10−7 | 3.05 × 10−3 | EXT1; GDF7; HIF1A; HOXB3; IFT172; MYH3; NCOR2; ROBO1; RPGRIP1L; SLC32A1 |
| brain development | GO:0007420 | 2.10 × 10−6 | 1.90 × 10−2 | EXT1; GDF7; HIF1A; HOXB3; IFT172; NCOR2; ROBO1; RPGRIP1L; SLC32A1 |
| skeletal system development | GO:0001501 | 2.20 × 10−6 | 1.99 × 10−2 | EXT1; HAND1; HIF1A; HOXB3; IFT172; LAMA5; LRP5; RYR1 |
| camera-type eye morphogenesis | GO:0048593 | 2.73 × 10−6 | 2.47 × 10−2 | FAT1; HIF1A; IFT172; LRP5; RPGRIP1L |
| regulation of cell differentiation | GO:0045595 | 3.88 × 10−6 | 3.50 × 10−2 | ARID1A; CTNNA1; EFNB2; FOXA2; GDF7; HIF1A; HOXB3; LRP5; MAML1; MEN1; ROBO1; TENM4 |
| heart morphogenesis | GO:0003007 | 4.78 × 10−6 | 4.32 × 10−2 | EXT1; HAND1; HIF1A; IFT172; ROBO1; RYR1 |
| morphogenesis of embryonic epithelium | GO:0016331 | 5.31 × 10−6 | 4.80 × 10−2 | GDF7; HAND1; HIF1A; IFT172; LAMA5 |
| All Patients (n = 58) a | ||
|---|---|---|
| N | % | |
| GENDER DISTRIBUTION | ||
| Males | 36 | 62.07 |
| Females | 22 | 37.93 |
| CLEFT TYPE | ||
| Cleft lip with or without cleft palate | 48 | 82.76 |
| Unilateral—right side | 10 | 17.24 |
| Unilateral—left side | 21 | 36.21 |
| Bilateral | 15 | 25.86 |
| Side unknown | 2 | 3.45 |
| Cleft lip only | 10 | 17.24 |
| Unilateral—right side | 3 | 5.17 |
| Unilateral—left side | 2 | 3.45 |
| Bilateral | 1 | 1.72 |
| Side unknown | 4 | 6.90 |
| ASSOCIATED ANOMALIES | 16 | 27.59 |
| Kidney defects b | 2 | 3.45 |
| POSITIVE FAMILY HISTORY | 19 | 32.76 |
| Immediate family (parents, siblings) | 6 | 10.34 |
| Extended family | 13 | 22.41 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biedziak, B.; Dąbrowska, J.; Bogdanowicz, A.; Karbowska, K.; Mostowska, A. Expanding the Genetic Spectrum of Non-Syndromic Cleft Lip and Palate Through Whole-Exome Sequencing. Int. J. Mol. Sci. 2025, 26, 12111. https://doi.org/10.3390/ijms262412111
Biedziak B, Dąbrowska J, Bogdanowicz A, Karbowska K, Mostowska A. Expanding the Genetic Spectrum of Non-Syndromic Cleft Lip and Palate Through Whole-Exome Sequencing. International Journal of Molecular Sciences. 2025; 26(24):12111. https://doi.org/10.3390/ijms262412111
Chicago/Turabian StyleBiedziak, Barbara, Justyna Dąbrowska, Agnieszka Bogdanowicz, Karolina Karbowska, and Adrianna Mostowska. 2025. "Expanding the Genetic Spectrum of Non-Syndromic Cleft Lip and Palate Through Whole-Exome Sequencing" International Journal of Molecular Sciences 26, no. 24: 12111. https://doi.org/10.3390/ijms262412111
APA StyleBiedziak, B., Dąbrowska, J., Bogdanowicz, A., Karbowska, K., & Mostowska, A. (2025). Expanding the Genetic Spectrum of Non-Syndromic Cleft Lip and Palate Through Whole-Exome Sequencing. International Journal of Molecular Sciences, 26(24), 12111. https://doi.org/10.3390/ijms262412111







