A Dual-Gene Colorimetric LAMP Assay for Genus-Level Detection of Salmonella and Specific Identification of the Non-Motile Serovar S. gallinarum Gallinarum
Abstract
1. Introduction
2. Results
2.1. Specificity of the Assay
2.2. Sensitivity of the Assay
2.3. Comparison with Real-Time PCR
2.4. Comparison with Bacteriological Isolation on Cloacal Swabs from Infected Birds
3. Discussion
4. Materials and Methods
4.1. Salmonella gallinarum Challenge and Cloacal Swab Collection
4.2. Bacteriological Isolation of Salmonella gallinarum from Cloacal Swabs
4.3. DNA Collection
4.3.1. Heat-Treatment
4.3.2. DNA Extraction
4.4. Colorimetric Buffer Preparation
4.5. Primer Design
4.6. Colorimetric LAMP Reaction Composition
4.7. The Assay’s Specificity
4.8. The Assay’s Sensitivity
4.9. Confirmation of Amplification in LAMP Reactions
4.10. Real-Time PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| S. gallinarum | Salmonella enterica serovar Gallinarum |
| LAMP | Loop-mediated isothermal amplification |
| invA | A conserved Salmonella invAsion gene in SPI-1 (genus-specific marker) |
| TRX | Taxon-Restricted eXclusive sequence (serovar-specific marker for S. gallinarum) |
| CFUs | Colony-Forming Units (measure of viable bacteria) |
| PCR | Polymerase Chain Reaction |
| qPCR/real-time PCR | Quantitative real-time PCR |
| DPI | Days Post-Infection |
| NC | Negative Control |
| Ld | Ladder (molecular weight size marker in gel electrophoresis) |
| API 20E | Analytical Profile Index 20E system (biochemical ID kit for Enterobacteriaceae) |
| CESASPV | Comité d’Éthique en Sciences et Santé Animales et Santé Publique Vétérinaire (IAV Hassan II) |
| IAV Hassan II | Institut Agronomique et Vétérinaire Hassan II (Morocco) |
| ISO 6579-1:2017 | International Organization for Standardization method for Salmonella detection |
| PBS | Phosphate-Buffered Saline |
| ARRIVE guidelines | Animal Research: Reporting of In Vivo Experiments |
| SPI-1 | Salmonella Pathogenicity Island-1 |
| F3/B3/FIP/BIP/LF/LB | LAMP primer types (Forward/Backward outer, Forward/Backward Inner Primer, Loop Forward/Backward) |
| BstY Polymerase | DNA polymerase enzyme used in isothermal amplification |
| Tris-HCl | Tris(hydroxymethyl)aminomethane hydrochloride buffer |
| MgSO4 | Magnesium sulfate |
| KCl | Potassium chloride |
| KOH | Potassium hydroxide |
| (NH4)2SO4 | Ammonium sulfate |
| FRET-PCR | Fluorescence Resonance Energy Transfer PCR |
References
- Barrow, P.A.; Neto, O.C.F. Pullorum Disease and Fowl Typhoid—New Thoughts on Old Diseases: A Review. Avian Pathol. 2011, 40, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Shivaprasad, H.L. Fowl Typhoid and Pullorum Disease. Rev. Sci. Tech. 2000, 19, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Wigley, P. Salmonella enterica Serovar Gallinarum: Addressing Fundamental Questions in Bacteriology Sixty Years on from the 9R Vaccine. Avian Pathol. 2017, 46, 119–124. [Google Scholar] [CrossRef]
- CABI. Fowl Typhoid; CABI Compendium: Wallingford, UK, 2019; p. 82899. [Google Scholar] [CrossRef]
- Word Organisation for Animal Health. Fowl Typhoid (Infection with Salmonella Gallinarum); WOAH—World Organisation for Animal Health: Paris, France, 2018; Available online: https://www.woah.org/en/disease/fowl-typhoid/ (accessed on 27 September 2025).
- Kingdom of Morocco. Law No. 49–99 on the Sanitary Protection of Poultry Holdings and the Marketing of Poultry and Poultry Products (Dahir No. 1-02-119 of 13 June 2002); Official Bulletin of the Kingdom of Morocco: Rabat, Morocco, 2002. [Google Scholar]
- Malorny, B.; Paccassoni, E.; Fach, P.; Bunge, C.; Martin, A.; Helmuth, R. Diagnostic Real-Time PCR for Detection of Salmonella in Food. Appl. Environ. Microbiol. 2004, 70, 7046–7052. [Google Scholar] [CrossRef] [PubMed]
- Thilakarathna, S.H.; Chui, L. A Pilot Study to Detect Viable Salmonella spp. in Diarrheal Stool Using Viability Real-Time PCR as a Culture-Independent Diagnostic Tool in a Clinical Setting. Int. J. Mol. Sci. 2023, 24, 9979. [Google Scholar] [CrossRef]
- Cho, I.-H.; Ku, S. Current Technical Approaches for the Early Detection of Foodborne Pathogens: Challenges and Opportunities. Int. J. Mol. Sci. 2017, 18, 2078. [Google Scholar] [CrossRef]
- Chaudhary, D.; Poudel, S.; Jia, L.; Sukumaran, A.T.; Zhang, X.; Cheng, W.-H.; Kiess, A.S.; Macklin, K.S.; Zhang, L. Optimization of a Loop-Mediated Isothermal Amplification (Lamp) Assay for the Rapid Detection of Clostridium Perfringens. J. Appl. Poult. Res. 2025, 34, 100513. [Google Scholar] [CrossRef]
- Kamel, M.; Davidson, J.L.; Schober, J.M.; Fraley, G.S.; Verma, M.S. A Paper-Based Loop-Mediated Isothermal Amplification Assay for Highly Pathogenic Avian Influenza. Sci. Rep. 2025, 15, 12110. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-Mediated Isothermal Amplifi-cation of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Mori, Y.; Notomi, T. Loop-Mediated Isothermal Amplification (LAMP): A Rapid, Accurate, and Cost-Effective Diagnostic Method for Infectious Diseases. J. Infect. Chemother. 2009, 15, 62–69. [Google Scholar] [CrossRef]
- Kreitlow, A.; Becker, A.; Schotte, U.; Malorny, B.; Plötz, M.; Abdulmawjood, A. Establishment and Validation of a Loop-Mediated Isothermal Amplification (LAMP) Assay Targeting the ttrRSBCA Locus for Rapid Detection of Salmonella spp. in Food. Food Control 2021, 126, 107973. [Google Scholar] [CrossRef]
- Shang, Y.; Ye, Q.; Cai, S.; Wu, Q.; Pang, R.; Yang, S.; Xiang, X.; Wang, C.; Zha, F.; Ding, Y.; et al. Loop-Mediated Isothermal Am-plification (LAMP) for Rapid Detection of Salmonella in Foods Based on New Molecular Targets. LWT 2021, 142, 110999. [Google Scholar] [CrossRef]
- Yang, Q.; Domesle, K.J.; Ge, B. Loop-Mediated Isothermal Amplification for Salmonella Detection in Food and Feed: Current Ap-plications and Future Directions. Foodborne Pathog. Dis. 2018, 15, 309–331. [Google Scholar] [CrossRef] [PubMed]
- FDA. Salmonella Loop-Mediated Isothermal Amplification (LAMP) Protocol; FDA: Silver Spring, MD, USA, 2024. [Google Scholar]
- Wei, S.; Su, Z.; Bu, X.; Shi, X.; Pang, B.; Zhang, L.; Li, J.; Zhao, C. On-Site Colorimetric Detection of Salmonella typhimurium. npj Sci. Food 2022, 6, 48. [Google Scholar] [CrossRef]
- Regal, P.; Doval, A.; García-Ramos, I.; Cepeda, A.; Garrido-Maestu, A.; Lamas, A. Loop-Mediated Isothermal Amplification-Based Workflow for the Detection and Serotyping of Salmonella spp. in Environmental Poultry Flock Samples. Foods 2024, 13, 4069. [Google Scholar] [CrossRef]
- Balaga, K.B.; Pavon, R.D.N.; Calayag, A.M.B.; Justo, C.A.C.; Adao, D.E.V.; Rivera, W.L. Development of a Closed-Tube, Calcein-Based Loop-Mediated Isothermal Amplification Assay to Detect Salmonella spp. in Raw Meat Samples. J. Microbiol. Methods 2024, 220, 106922. [Google Scholar] [CrossRef]
- Skenndri, S.; Nassik, S.; Lakhmi, R.; Anneggah, B.E.; Lahkak, F.E.; Moumen, A.; Abdellaoui Maane, I. A Colorimetric LAMP Assay for Salmonella spp. Detection: Towards a DNA Extraction-Free Approach for Pathogen Screening. Foods 2025, 14, 521. [Google Scholar] [CrossRef]
- EFSA Panel on Animal Health and Welfare (AHAW); More, S.; Bøtner, A.; Butterworth, A.; Calistri, P.; Depner, K.; Edwards, S.; Garin-Bastuji, B.; Good, M.; Gortázar Schmidt, C.; et al. Assessment of Listing and Categorisation of Animal Diseases within the Framework of the Animal Health Law (Regulation (EU) No 2016/429): Salmonella Infection in Poultry with Serotypes of Animal Health Relevance (S. pullorum, S. gallinarum and S. arizonae). EFSA J. 2017, 15, e04954. [Google Scholar] [CrossRef]
- Pavon, R.D.N.; Rivera, W.L. Loop-Mediated Isothermal Amplification Assay for Visual Detection of Salmonella enterica Serovar Typhimurium in Food Animal Meat Products. Foods 2025, 14, 1731. [Google Scholar] [CrossRef]
- Xiong, D.; Song, L.; Geng, S.; Tao, J.; An, S.; Pan, Z.; Jiao, X. One-Step PCR Detection of Salmonella Pullorum/Gallinarum Using a Novel Target: The Flagellar Biosynthesis Gene flhB. Front. Microbiol. 2016, 7, 1863. [Google Scholar] [CrossRef]
- Xiong, D.; Song, L.; Pan, Z.; Jiao, X. Identification and Discrimination of Salmonella enterica Serovar Gallinarum Biovars Pullorum and Gallinarum Based on a One-Step Multiplex PCR Assay. Front. Microbiol. 2018, 9, 1718. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Iduu, N.V.; Zhang, D.; Chenoweth, K.; Wei, L.; Yang, Y.; Dou, X.; Wang, C. Dual-Emission Fluorescence Resonance Energy Transfer (FRET) PCR Discriminates Salmonella Pullorum and Gallinarum. Microorganisms 2024, 12, 1815. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, H.; Kawana, T.; Fukushima, E.; Suzutani, T. Tolerance of Loop-Mediated Isothermal Amplification to a Culture Medium and Biological Substances. J. Biochem. Biophys. Methods 2007, 70, 499–501. [Google Scholar] [CrossRef] [PubMed]
- Edel, B.; Glöckner, S.; Stoll, S.; Lindig, N.; Boden, K.; Wassill, L.; Simon, S.; Löffler, B.; Rödel, J. Development of a Rapid Diagnostic Test Based on Loop-Mediated Isothermal Amplification to Identify the Most Frequent Non-Typhoidal Salmonella Serovars from Culture. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 461–470. [Google Scholar] [CrossRef]
- Nwe, M.K.; Jangpromma, N.; Taemaitree, L. Evaluation of Molecular Inhibitors of Loop-Mediated Isothermal Amplification (LAMP). Sci. Rep. 2024, 14, 5916. [Google Scholar] [CrossRef]
- Vinayaka, A.C.; Quyen, T.L.; Huynh, V.N.; Madsen, M.; Bang, D.D.; Wolff, A. Rapid Detection of Salmonella enterica in Primary Production Samples by Eliminating DNA Amplification Inhibitors Using an Improved Sample Pre-treatment Method. Microb. Biotechnol 2023, 16, 2105–2113. [Google Scholar] [CrossRef]
- Fujimoto, M.; Kitamura, H. Application of the Colorimetric Loop-Mediated Isothermal Amplification (LAMP) Technique for Geno-typing Cre-Driver Mice. J. Vet. Med. Sci. 2022, 84, 507–510. [Google Scholar] [CrossRef]
- Prado, N.O.; Marin, A.M.; Lalli, L.A.; Sanchuki, H.B.S.; Wosniaki, D.K.; Nardin, J.M.; Morales, H.M.P.; Blanes, L.; Zanette, D.L.; Aoki, M.N. Development and Evaluation of a Lyophilization Protocol for Colorimetric RT-LAMP Diagnostic Assay for COVID-19. Sci. Rep. 2024, 14, 10612. [Google Scholar] [CrossRef]
- Raddatz, B.W.; Kim, E.Y.S.; Imamura, L.M.; Steil, G.J.; Santiago, E.B.; Soares, S.P.T.; Ribeiro, V.H.A.; de Almeida, B.M.M.; Rogal, S.R.; Figueredo, M.V.M. Development of an Optimized Colorimetric RT-LAMP for SARS-CoV-2 Assay with Enhanced Procedure Controls for Remote Diagnostics. Sci. Rep. 2022, 12, 21424. [Google Scholar] [CrossRef]
- Yang, N.; Zhang, H.; Han, X.; Liu, Z.; Lu, Y. Advancements and Applications of Loop-Mediated Isothermal Amplification Tech-nology: A Comprehensive Overview. Front. Microbiol. 2024, 15, 1406632. [Google Scholar] [CrossRef]
- Wang, D.-G.; Brewster, J.D.; Paul, M.; Tomasula, P.M. Two Methods for Increased Specificity and Sensitivity in Loop-Mediated Isothermal Amplification. Molecules 2015, 20, 6048–6059. [Google Scholar] [CrossRef]
- Mei, X.; Zhai, X.; Lei, C.; Ye, X.; Kang, Z.; Wu, X.; Xiang, R.; Wang, Y.; Wang, H. Development and Application of a Visual Loop-Mediated Isothermal Amplification Combined with Lateral Flow Dipstick (LAMP-LFD) Method for Rapid Detection of Salmonella Strains in Food Samples. Food Control 2019, 104, 9–19. [Google Scholar] [CrossRef]
- ISO 6579-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp. International Organization for Standardization: Geneva, Switzerland, 2017.
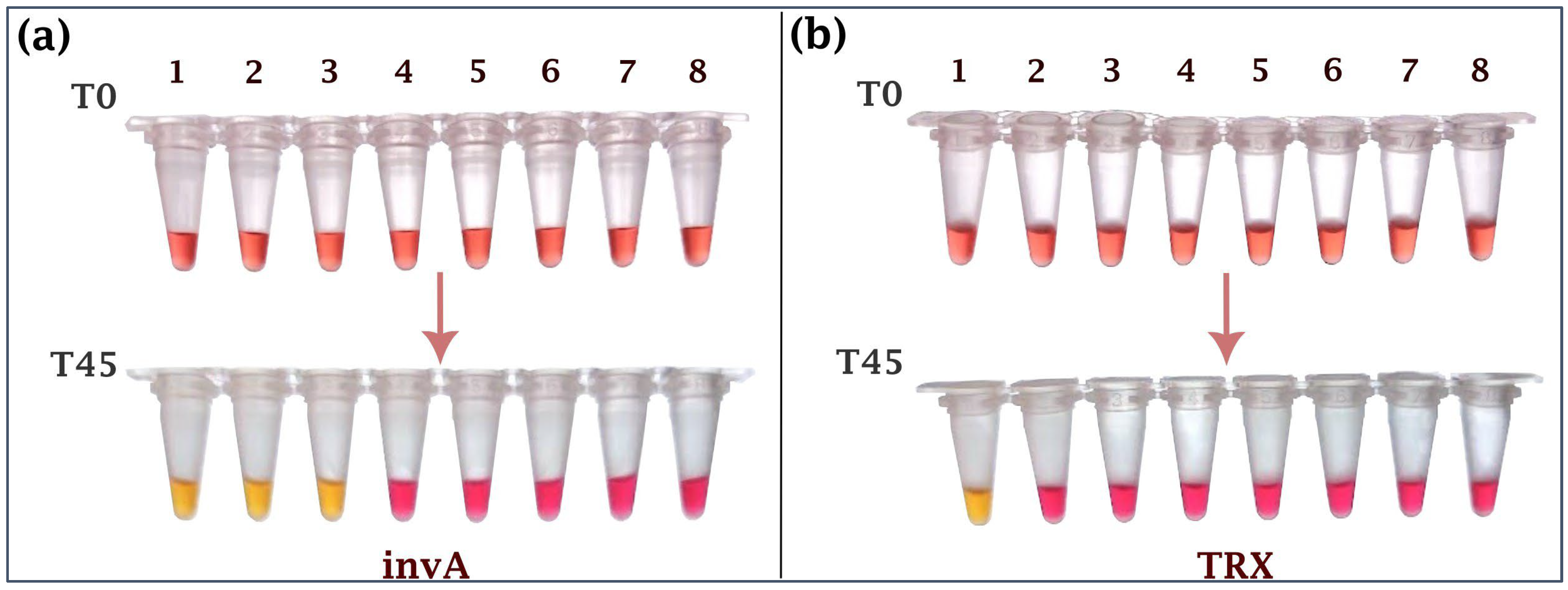
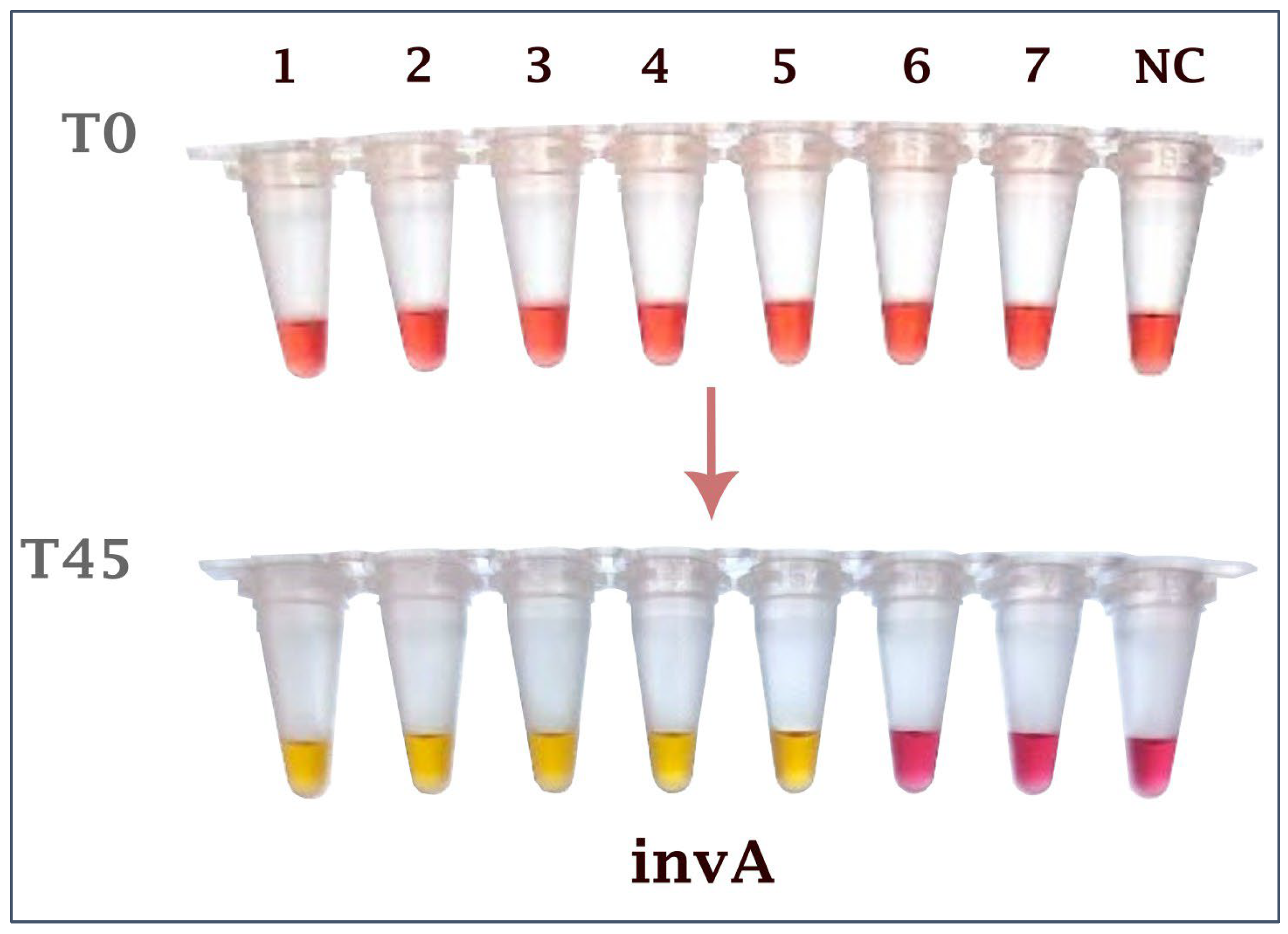

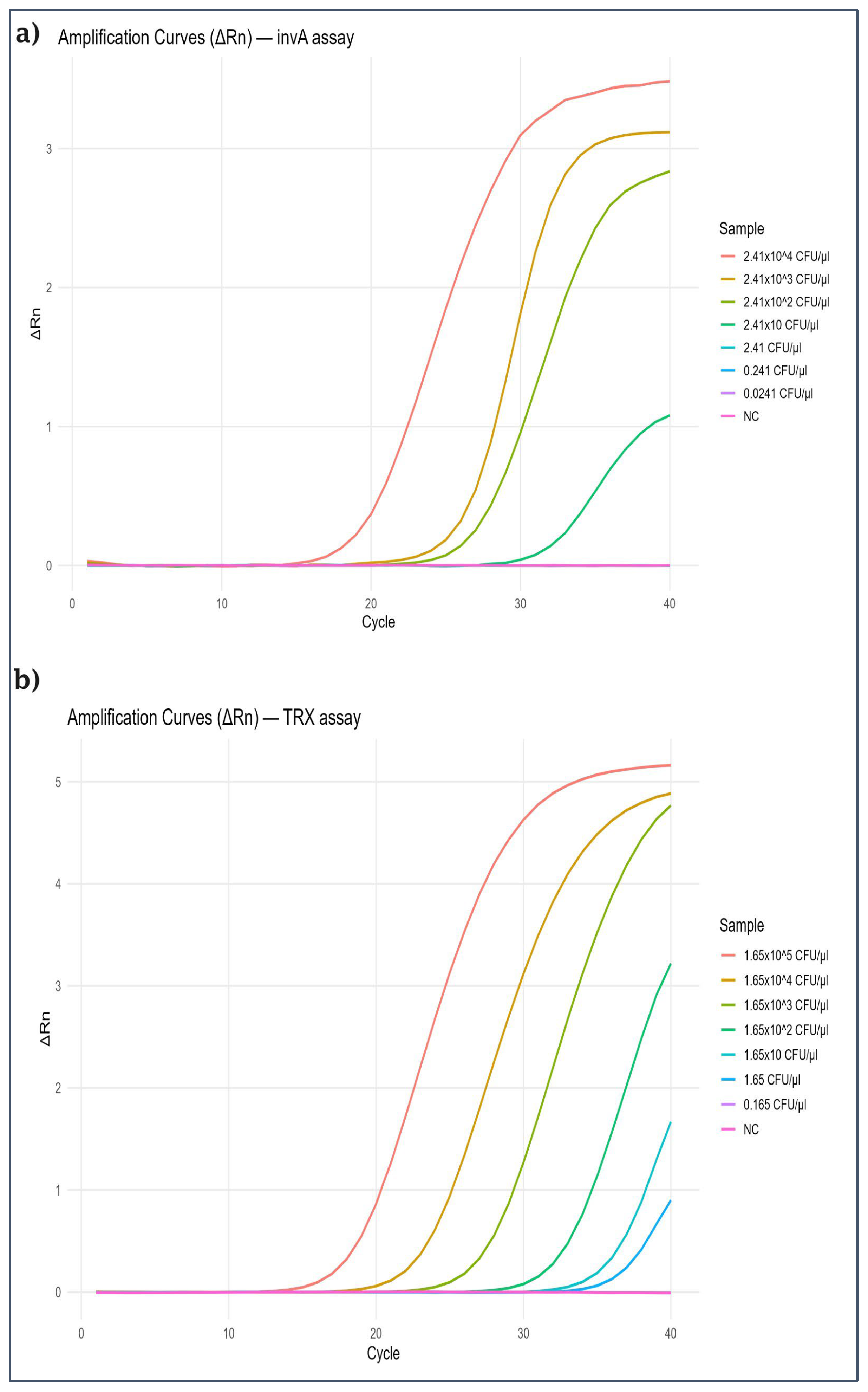

| Bird | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | C | L | C | L | C | L | C | L | C | L | C | L | C | L | C | L | C | L | C | L | C | L | C | L | |
| DPI | 1 | + | + | − | + | + | − | + | − | − | − | − | + | ||||||||||||
| 3 | + | + | − | + | + | + | + | − | + | − | − | − | |||||||||||||
| 5 | + | − | − | − | + | − | + | − | − | + | − | + | |||||||||||||
| Component | Initial Aliquot Concentration [×2] (mM) | Final Concentration in the Reaction [×1] (mM) |
|---|---|---|
| (NH4)2SO4 | 20 | 10 |
| MgSO4 | 20 | 10 |
| KCl | 60 | 30 |
| Phenol red | 0.4 | 0.2 |
| Tris-HCl | 0.8 | 0.4 |
| Gene | Primer | Sequence | Length |
|---|---|---|---|
| invA (NC_011294.1) | F3 | 5′-ACGCGTTCTGAACCTTTGG-3′ | 19 |
| B3 | 5′-CGTTTCCTGCGGTACTGTT-3′ | 19 | |
| FIP | 5′-GCCACGTTCGGGCAATTCGTTATAAACTGGACCACGGTGACA-3′ | 42 | |
| BIP | 5′-AATTTCACCGGCATCGGCTTCACGCTCTTTCGTCTGGCATTA-3′ | 42 | |
| LF | 5′-CGGTGGGTTTTGTTGTCTTCTCTA-3′ | 24 | |
| LB | 5′-TCAAGATAAGACGGCTGGTACTGAT-3′ | 25 | |
| TRX (SPUL_RS24225, NC_016831.1) | F3 | 5′-GGATTGGACCTCAAGTGTA-3′ | 19 |
| B3 | 5′-GTCCCGGCTTTATGAACG-3′ | 18 | |
| FIP | 5′-GTGGGTACTTTGCCGGATGGGGTCTACCATCAGAACTGC-3′ | 39 | |
| BIP | 5′-CGTCCCGTAACATAATTATTGTCGATGATGAGGCTAACAAGGATT-3′ | 45 | |
| LF | 5′-GCACAGTGATTGTGCGTGATG-3′ | 21 | |
| LB | 5′-CCTTAACATCGCTAGGGGATAAGTT-3′ | 25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skenndri, S.; Lahkak, F.E.; El Kamli, T.; Agargar, Z.; Abdellaoui Maane, I.; Nassik, S. A Dual-Gene Colorimetric LAMP Assay for Genus-Level Detection of Salmonella and Specific Identification of the Non-Motile Serovar S. gallinarum Gallinarum. Int. J. Mol. Sci. 2025, 26, 12083. https://doi.org/10.3390/ijms262412083
Skenndri S, Lahkak FE, El Kamli T, Agargar Z, Abdellaoui Maane I, Nassik S. A Dual-Gene Colorimetric LAMP Assay for Genus-Level Detection of Salmonella and Specific Identification of the Non-Motile Serovar S. gallinarum Gallinarum. International Journal of Molecular Sciences. 2025; 26(24):12083. https://doi.org/10.3390/ijms262412083
Chicago/Turabian StyleSkenndri, Safae, Fatima Ezzahra Lahkak, Taha El Kamli, Zineb Agargar, Imane Abdellaoui Maane, and Saâdia Nassik. 2025. "A Dual-Gene Colorimetric LAMP Assay for Genus-Level Detection of Salmonella and Specific Identification of the Non-Motile Serovar S. gallinarum Gallinarum" International Journal of Molecular Sciences 26, no. 24: 12083. https://doi.org/10.3390/ijms262412083
APA StyleSkenndri, S., Lahkak, F. E., El Kamli, T., Agargar, Z., Abdellaoui Maane, I., & Nassik, S. (2025). A Dual-Gene Colorimetric LAMP Assay for Genus-Level Detection of Salmonella and Specific Identification of the Non-Motile Serovar S. gallinarum Gallinarum. International Journal of Molecular Sciences, 26(24), 12083. https://doi.org/10.3390/ijms262412083







