Pepino Mosaic Virus in Tomato: Challenges, Control Strategies, and Future Prospects for Resistance Breeding
Abstract
1. Introduction
2. Biology and Diversity of PepMV
2.1. General Characteristics, Host Range, and Transmission
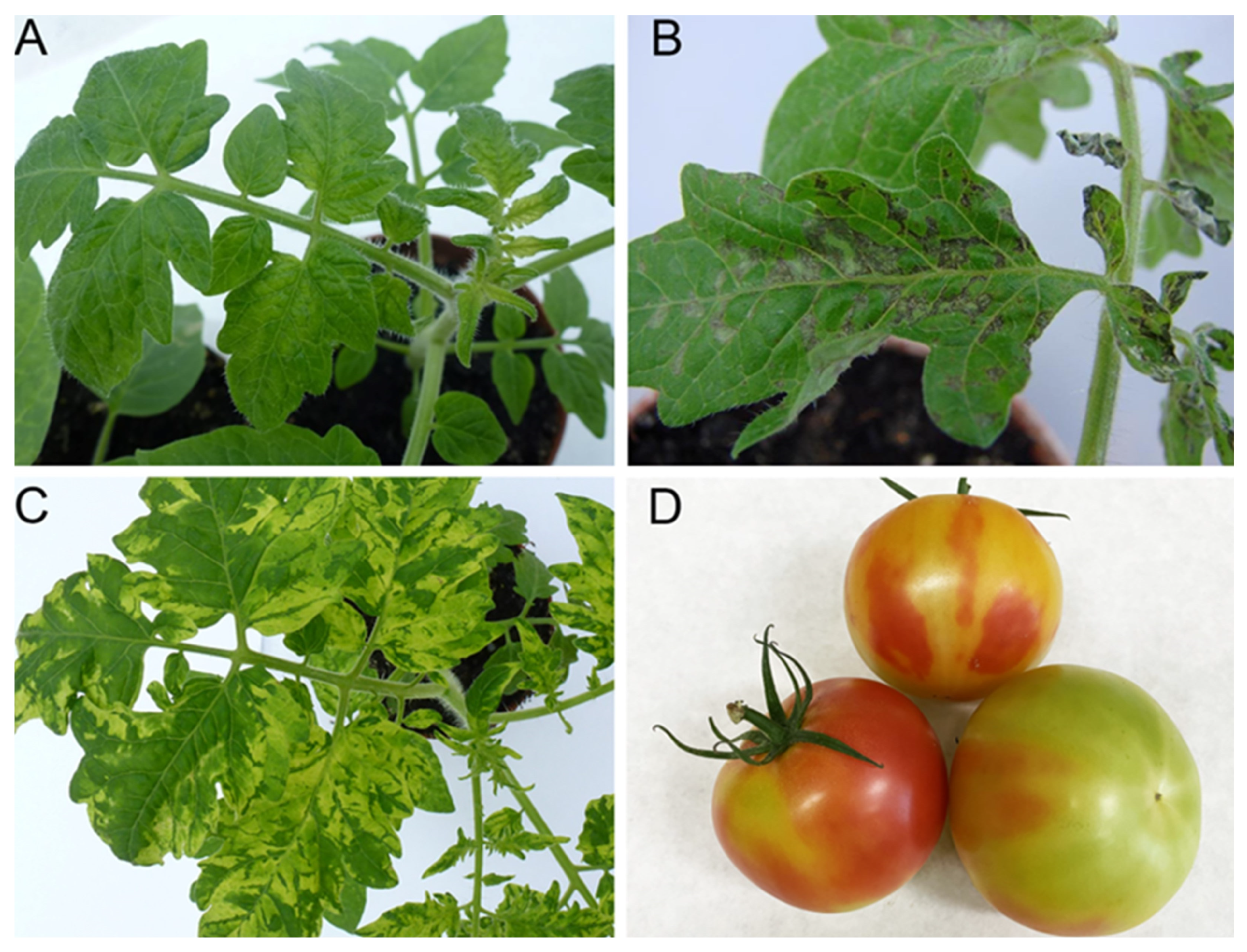
2.2. Symptomatology
2.3. Genome Organization and Functional Modules
2.4. Genetic Diversity and Evolutionary Dynamics
2.5. Genetic Determinants of Symptom Expression
3. Molecular Mechanisms of PepMV–Tomato Interactions
3.1. Codon Adaptation and Recombination as Evolutionary Drivers
3.2. Interactions with Host Chaperones
3.3. Redox Regulation and Host Oxidative Enzymes in PepMV Infection
3.4. Epitranscriptomic Manipulation: m6A Methylation and Autophagy
3.5. Host Susceptibility Factors: The Role of SlOSCA4.1
3.6. Integrative Perspective
4. Host Responses and Transcriptomic Reprogramming
4.1. Early Host Reprogramming and Metabolic–Defense Integration During PepMV Infection
4.2. Environmental Modulation of Hormonal and Redox Crosstalk
4.3. Integrative Overview
5. Sources of Resistance and Breeding Challenges
5.1. Screening for Resistance in Wild Solanum Species
5.2. Limitations and Perspectives for Durable Resistance
6. Current Control Methods
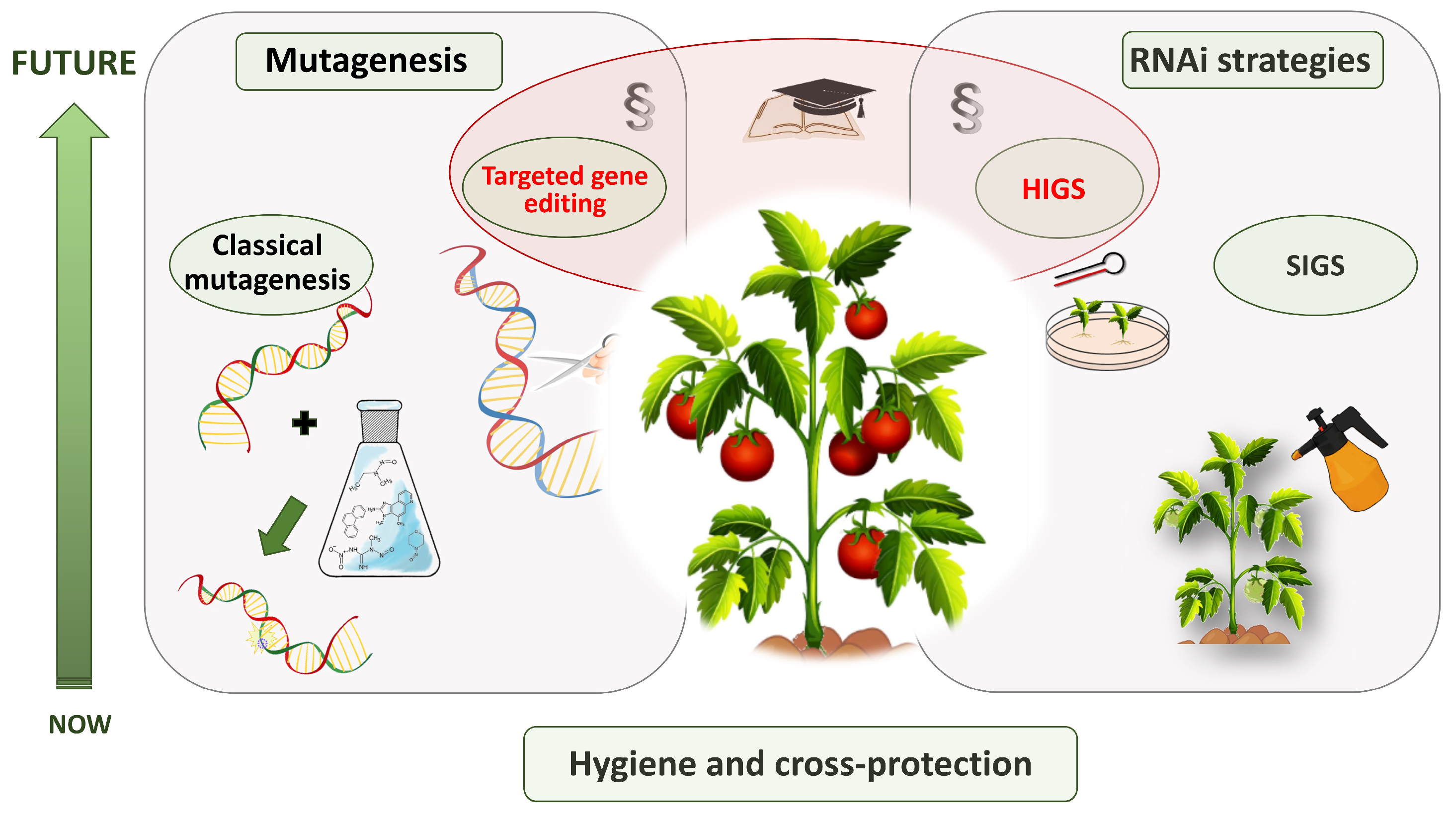
7. Emerging Strategies and Durable Resistance for PepMV in Tomato
7.1. Lessons from Other Tomato Viruses
7.2. Classical Mutagenesis and Targeted Genome Editing Approaches
7.3. RNA-Based Strategies: Transgenic and Non-Transgenic
7.4. Regulatory and Practical Considerations
7.5. Outlook and Knowledge Gaps
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| APHIS | Animal and Plant Health Inspection Service |
| CAS | CRISPR-associated system |
| CH2 | Chilean 2 genotype (of PepMV) |
| CMV | Cucumber mosaic virus |
| CP | Coat protein |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| DNA | Deoxyribonucleic acid |
| dsRNA | Double-stranded RNA |
| EAP | Europe–Asia–Pacific lineage (of PepMV) |
| ECT | Evolutionarily Conserved C-Terminal region |
| ELISA | Enzyme-linked immunosorbent assay |
| EMS | Ethyl methanesulfonate |
| EU | European genotype (of PepMV) |
| GMO | Genetically modified organism |
| HIGS | Host-induced gene silencing |
| LDH | Layered double hydroxide |
| LOS | Loss of susceptibility |
| LP | Peruvian-like genotype (of PepMV) |
| MAS | Marker-assisted selection |
| PepMoV | Pepper mottle virus |
| PepMV | Pepino mosaic virus |
| PES | Southern Peruvian genotype (of PepMV) |
| PVY | Potato virus Y |
| QTL | Quantitative trait locus |
| RDR | Host-encoded RNA-dependent RNA polymerase |
| RNA | Ribonucleic acid |
| RNAi | RNA interference |
| RT-qPCR | Reverse transcription quantitative polymerase chain reaction |
| SIGS | Spray-induced gene silencing |
| siRNA | Small interfering RNA |
| SNP | Single-nucleotide polymorphism |
| ToBRFV | Tomato brown rugose fruit virus |
| TSWV | Tomato spotted wilt virus |
| TILLING | Targeting induced local lesions in genomes |
| TYLCV | Tomato yellow leaf curl virus |
| US1 | North American genotype (of PepMV) |
References
- Hanssen, I.M.; Thomma, B.P. Pepino mosaic virus: A successful pathogen that rapidly evolved from emerging to endemic in tomato crops. Mol. Plant Pathol. 2010, 11, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.A.C.; Koenig, R.; Lesemann, D.E. Pepino mosaic virus, a new potexvirus from pepino (Solanum muricatum). Ann. Appl. Biol. 1980, 94, 61–68. [Google Scholar] [CrossRef]
- Gómez, P.; Sempere, R.N.; Aranda, M.A.; Elena, S.F. Phylodynamics of pepino mosaic virus in Spain. Eur. J. Plant Pathol. 2012, 134, 445–449. [Google Scholar] [CrossRef]
- Agüero, J.; Gómez-Aix, C.; Sempere, R.N.; García-Villalba, J.; García-Núñez, J.; Hernando, Y.; Aranda, M.A. Stable and broad spectrum cross-protection against pepino mosaic virus attained by mixed infection. Front. Plant Sci. 2018, 9, 1810. [Google Scholar] [CrossRef]
- Alcaide, C.; Aranda, M.A. Determinants of persistent patterns of pepino mosaic virus mixed infections. Front. Microbiol. 2021, 12, 694492. [Google Scholar] [CrossRef]
- Gómez, P.; Sempere, R.N.; Elena, S.F.; Aranda, M.A. Mixed infections of pepino mosaic virus strains modulate the evolutionary dynamics of this emergent virus. J. Virol. 2009, 83, 12378–12387. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Jackowiak, P.; Borodynko, N.; Figlerowicz, M.; Pospieszny, H. Quasispecies nature of pepino mosaic virus and its evolutionary dynamics. Virus Genes 2010, 41, 260–267. [Google Scholar] [CrossRef]
- Minicka, J.; Rymelska, N.; Elena, S.F.; Czerwoniec, A.; Hasiów-Jaroszewska, B. Molecular evolution of pepino mosaic virus during long-term passaging in different hosts and its impact on virus virulence. Ann. Appl. Biol. 2015, 166, 389–401. [Google Scholar] [CrossRef]
- He, M.; He, C.-Q.; Ding, N.-Z. Pepino mosaic virus: Recombination, spatiotemporal divergence and codon usage. Plant Pathol. 2024, 73, 2574–2583. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Gutiérrez-Aguirre, I.; Paeleman, A.; Goen, K.; Wittemans, L.; Lievens, B.; Vanachter, A.C.R.C.; Ravnikar, M.; Thomma, B.P.H.J. Cross-protection or enhanced symptom display in greenhouse tomato co-infected with different pepino mosaic virus isolates. Plant Pathol. 2010, 59, 13–21. [Google Scholar] [CrossRef]
- De Nayer, F.; Goen, K.; Paeleman, A.; Parra, N.; Vanachter, A.; Hanssen, I.; Wittemans, L.; Vandewoestijne, E. Cross-protection as a control strategy for pepino mosaic virus (PepMV) in greenhouse tomato. Acta Hortic. Sin. 2011, 914, 163–169. [Google Scholar] [CrossRef]
- Hernando, Y.; Aranda, M.A. Cross-protection against pepino mosaic virus, more than a decade of efficient disease control. Ann. Appl. Biol. 2024, 184, 174–182. [Google Scholar] [CrossRef]
- Moreno-Pérez, M.G.; Pagán, I.; Aragón-Caballero, L.; Cáceres, F.; Fraile, A.; García-Arenal, F. Ecological and genetic determinants of pepino mosaic virus emergence. J. Virol. 2014, 88, 3359–3368. [Google Scholar] [CrossRef]
- Van der Vlugt, R.A.A.; Stijger, C.C.M.M.; Verhoeven, J.T.J. First report of pepino mosaic virus on tomato. Plant Dis. 2000, 84, 103. [Google Scholar] [CrossRef]
- Gómez, P.; Sempere, R.; Aranda, M.A. Pepino mosaic virus and tomato torrado virus: Two emerging viruses affecting tomato crops in the Mediterranean Basin. In Advances in Virus Research; Loebenstein, G., Lecoq, H., Eds.; Elsevier Academic Press Inc.: San Diego, CA, USA, 2012; Volume 84, pp. 505–532. [Google Scholar]
- Aguilar, J.M.; Hernández-Gallardo, M.D.; Cenis, J.L.; Lacasa, A.; Aranda, M.A. Complete sequence of the pepino mosaic virus RNA genome. Arch. Virol. 2002, 147, 2009–2015. [Google Scholar] [CrossRef] [PubMed]
- Mumford, R.A.; Metcalfe, E.J. The partial sequencing of the genomic RNA of a UK isolate of pepino mosaic virus and the comparison of the coat protein sequence with other isolates from Europe and Peru. Arch. Virol. 2001, 146, 2455–2460. [Google Scholar] [CrossRef]
- Cotillon, A.C.; Girard, M.; Ducouret, S. Complete nucleotide sequence of the genomic RNA of a French isolate of pepino mosaic virus (PepMV). Arch. Virol. 2002, 147, 2231–2238. [Google Scholar] [CrossRef]
- French, C.J.; Bouthillier, M.; Bernardy, M.; Ferguson, G.; Sabourin, M.; Johnson, R.C.; Masters, C.; Godkin, S.; Mumford, R. First report of pepino mosaic virus in Canada and the United States. Plant Dis. 2001, 85, 1121. [Google Scholar] [CrossRef] [PubMed]
- Maroon-Lango, C.J.; Guaragna, M.A.; Jordan, R.L.; Hammond, J.; Bandla, M.; Marquardt, S.K. Two unique US isolates of pepino mosaic virus from a limited source of pooled tomato tissue are distinct from a third (European-like) US isolate. Arch. Virol. 2005, 150, 1187–1201. [Google Scholar] [CrossRef]
- Pospieszny, H.; Borodynko, N.; Palczewska, M. Occurrence of pepino mosaic virus in Poland. Phytopathol. Pol. 2002, 26, 91–94. [Google Scholar]
- Soler, S.; Prohens, J.; Diez, M.J.; Nuez, F. Natural occurrence of pepino mosaic virus in Lycopersicon Species in Central and Southern Peru. J. Phytopathol. 2002, 150, 49–53. [Google Scholar] [CrossRef]
- Soler, S.; López, C.; Nuez, F. Natural occurrence of viruses in Lycopersicon spp. in Ecuador. Plant Dis. 2005, 89, 1244. [Google Scholar] [CrossRef]
- Blystad, D.-R.; van der Vlugt, R.; Alfaro-Fernández, A.; del Carmen Córdoba, M.; Bese, G.; Hristova, D.; Pospieszny, H.; Mehle, N.; Ravnikar, M.; Tomassoli, L.; et al. Host range and symptomatology of pepino mosaic virus strains occurring in Europe. Eur. J. Plant Pathol. 2015, 143, 43–56. [Google Scholar] [CrossRef]
- Fakhro, A.; von Bargen, S.; Bandte, M.; Büttner, C.; Franken, P.; Schwarz, D. Susceptibility of different plant species and tomato cultivars to two isolates of pepino mosaic virus. Eur. J. Plant Pathol. 2011, 129, 579–590. [Google Scholar] [CrossRef]
- Papayiannis, L.C.; Kokkinos, C.D.; Alfaro-Fernández, A. Detection, characterization and host range studies of pepino mosaic virus in Cyprus. Eur. J. Plant Pathol. 2012, 132, 1–7. [Google Scholar] [CrossRef]
- Córdoba, M.C.; Martínez-Priego, L.; Jordá, C. New natural hosts of pepino mosaic virus in Spain. Plant Dis. 2004, 88, 906. [Google Scholar] [CrossRef]
- Jordá, C.; Pérez, A.L.; Martínez Culebras, P.V.; Lacasa, A. First report of pepino mosaic virus on natural hosts. Plant Dis. 2001, 85, 1292. [Google Scholar] [CrossRef]
- Just, K.; Arif, U.; Kõrve, E.-L.; Paats, P.; Kvarnheden, A. Detection of pepino mosaic virus in imported tomato fruit in Northern Europe. Plant Pathol. 2025, 74, 2427–2438. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Mumford, R.; Blystad, D.-R.; Cortez, I.; Hasiów-Jaroszewska, B.; Hristova, D.; Pagán, I.; Pereira, A.-M.; Peters, J.; Pospieszny, H.; et al. Seed transmission of pepino mosaic virus in tomato. Eur. J. Plant Pathol. 2010, 126, 145–152. [Google Scholar] [CrossRef]
- Córdoba-Sellés, M.d.C.; García-Rández, A.; Alfaro-Fernández, A.; Jordá-Gutiérrez, C. Seed transmission of pepino mosaic virus and efficacy of tomato seed disinfection treatments. Plant Dis. 2007, 91, 1250–1254. [Google Scholar] [CrossRef]
- Ling, K.S. Pepino mosaic virus on tomato seed: Virus location and mechanical transmission. Plant Dis. 2008, 92, 1701–1705. [Google Scholar] [CrossRef]
- Shipp, J.L.; Buitenhuis, R.; Stobbs, L.; Wang, K.; Kim, W.S.; Ferguson, G. Vectoring of pepino mosaic virus by bumble-bees in tomato greenhouses. Ann. Appl. Biol. 2008, 153, 149–155. [Google Scholar] [CrossRef]
- Lacasa, A.; Guerrero, M.; Hita, I.; Martínez, M.; Jordá, C.; Bielza, P.; Contreras, J.; Alcázar, A.; Cano, A. Implication of bumble bees (Bombus spp.) on pepino mosaic virus (PepMV) spread on tomato crops. Plagas 2003, 29, 393–403. [Google Scholar]
- Stobbs, L.W.; Greig, N.; Weaver, S.; Shipp, L.; Ferguson, G. The potential role of native weed species and bumble bees (Bombus impatiens) on the epidemiology of pepino mosaic virus. Can. J. Plant Pathol. 2009, 31, 254–261. [Google Scholar] [CrossRef]
- Stobbs, L.W.; Greig, N. First report of bumblebee (Bombus impatiens Cresson) transmission of pepino mosaic virus between tomato (Solanum lycopersicum L.) and perennial climbing nightshade (Solanum dulcamara L.). Can. J. Plant Pathol. 2014, 36, 529–533. [Google Scholar] [CrossRef]
- Alfaro-Fernández, A.; Del Carmen Córdoba-Sellés, M.; Herrera-Vásquez, J.Á.; Cebrián, M.d.C.; Jordá, C. Transmission of pepino mosaic virus by the fungal vector Olpidium virulentus. J. Phytopathol. 2010, 158, 217–226. [Google Scholar] [CrossRef]
- Noël, P.; Hance, T.; Bragard, C. Transmission of the pepino mosaic virus by whitefly. Eur. J. Plant Pathol. 2014, 138, 23–27. [Google Scholar] [CrossRef]
- Noël, P.; Hance, T.; Muratori, F.; Bragard, C. Biological control by parasitoids does not enhance pepino mosaic virus transmission. Eur. J. Plant Pathol. 2016, 145, 493–499. [Google Scholar] [CrossRef]
- Moerkens, R.; Pekas, A.; Bellinkx, S.; Hanssen, I.; Huysmans, M.; Bosmans, L.; Wäckers, F. Nesidiocoris tenuis as a pest in Northwest Europe: Intervention threshold and influence of pepino mosaic virus. J. Appl. Entomol. 2020, 144, 566–577. [Google Scholar] [CrossRef]
- Anastassiadou, M.; Arena, M.; Auteri, D.; Brancato, A.; Bura, L.; Carrasco Cabrera, L.; Chaideftou, E.; Chiusolo, A.; Crivellente, F.; De Lentdecker, C.; et al. Peer review of the pesticide risk assessment of the active substances pepino mosaic virus, EU strain, mild isolate Abp1 and pepino mosaic virus, CH2 strain, mild isolate Abp2. EFSA J. 2021, 19, e06388. [Google Scholar] [CrossRef]
- Klap, C.; Luria, N.; Smith, E.; Hadad, L.; Bakelman, E.; Sela, N.; Belausov, E.; Lachman, O.; Leibman, D.; Dombrovsky, A. Tomato brown rugose fruit virus contributes to enhanced pepino mosaic virus titers in tomato plants. Viruses 2020, 12, 879. [Google Scholar] [CrossRef]
- Miranda-Campaña, O.A.; Diaz-Lara, A.; García-Estrada, R.S.; Carrillo-Fasio, J.A.; Tovar-Pedraza, J.M. Molecular and biological characterization of pepino mosaic virus isolates occurring in the main tomato-producing areas in Mexico. Physiol. Mol. Plant Pathol. 2024, 131, 102269. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Borodynko, N.; Jackowiak, P.; Figlerowicz, M.; Pospieszny, H. Single mutation converts mild pathotype of the pepino mosaic virus into necrotic one. Virus Res. 2011, 159, 57–61. [Google Scholar] [CrossRef]
- Sempere, R.N.; Gómez-Aix, C.; Ruíz-Ramón, F.; Gómez, P.; Hasiów-Jaroszewska, B.; Sánchez-Pina, M.A.; Aranda, M.A. Pepino mosaic virus RNA-dependent RNA polymerase POL domain Is a hypersensitive response-like elicitor shared by necrotic and mild isolates. Phytopathology 2016, 106, 395–406. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Paeleman, A.; Vandewoestijne, E.; Van Bergen, L.; Bragard, C.; Lievens, B.; Vanachter, A.C.R.C.; Thomma, B.P.H.J. Pepino mosaic virus isolates and differential symptomatology in tomato. Plant Pathol. 2009, 58, 450–460. [Google Scholar] [CrossRef]
- Spence, N.J.; Basham, J.; Mumford, R.A.; Hayman, G.; Edmondson, R.; Jones, D.R. Effect of pepino mosaic virus on the yield and quality of glasshouse-grown tomatoes in the UK. Plant Pathol. 2006, 55, 595–606. [Google Scholar] [CrossRef]
- Alcaide, C.; Méndez-López, E.; Úbeda, J.R.; Gómez, P.; Aranda, M.A. Characterization of two aggressive PepMV isolates useful in breeding programs. Viruses 2023, 15, 2230. [Google Scholar] [CrossRef] [PubMed]
- Hasiów-Jaroszewska, B.; Paeleman, A.; Ortega-Parra, N.; Borodynko, N.; Minicka, J.; Czerwoniec, A.; Thomma, B.P.H.J.; Hanssen, I.M. Ratio of mutated versus wild-type coat protein sequences in pepino mosaic virus determines the nature and severity of yellowing symptoms on tomato plants. Mol. Plant Pathol. 2013, 14, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, I.M.; Paeleman, A.; Wittemans, L.; Goen, K.; Lievens, B.; Bragard, C.; Vanachter, A.C.R.C.; Thomma, B.P.H.J. Genetic characterization of pepino mosaic virus isolates from Belgian greenhouse tomatoes reveals genetic recombination. Eur. J. Plant Pathol. 2008, 121, 131–146. [Google Scholar] [CrossRef]
- Agirrezabala, X.; Méndez-López, E.; Lasso, G.; Sánchez-Pina, M.A.; Aranda, M.; Valle, M. The near-atomic cryoEM structure of a flexible filamentous plant virus shows homology of its coat protein with nucleoproteins of animal viruses. eLife 2015, 4, e11795. [Google Scholar] [CrossRef]
- Mathioudakis, M.M.; Veiga, R.S.L.; Canto, T.; Medina, V.; Mossialos, D.; Makris, A.M.; Livieratos, I. Pepino mosaic virus triple gene block protein 1 (TGBp1) interacts with and increases tomato catalase 1 activity to enhance virus accumulation. Mol. Plant Pathol. 2013, 14, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Tilsner, J.; Linnik, O.; Louveaux, M.; Roberts, I.M.; Chapman, S.N.; Oparka, K.J. Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 2013, 201, 981–995. [Google Scholar] [CrossRef]
- Morozov, S.Y.; Solovyev, A.G. Triple gene block: Modular design of a multifunctional machine for plant virus movement. J. Gen. Virol. 2003, 84, 1351–1366. [Google Scholar] [CrossRef]
- Mathioudakis, M.M.; Rodríguez-Moreno, L.; Sempere, R.N.; Aranda, M.A.; Livieratos, I. Multifaceted capsid proteins: Multiple interactions suggest multiple roles for pepino mosaic virus capsid protein. Mol. Plant-Microbe Interact. 2014, 27, 1356–1369. [Google Scholar] [CrossRef]
- Méndez-López, E.; Aranda, M.A. A regulatory role for the redox status of the pepino mosaic virus coat protein. PLoS Pathog. 2023, 19, e1011732. [Google Scholar] [CrossRef]
- Candresse, T.; Marais, A.; Faure, C.; Dubrana, M.P.; Gombert, J.; Bendahmane, A. Multiple coat protein mutations abolish recognition of pepino mosaic potexvirus (PepMV) by the potato Rx resistance gene in transgenic tomatoes. Mol. Plant-Microbe Interact. 2010, 23, 376–383. [Google Scholar] [CrossRef]
- Duff-Farrier, C.R.A.; Candresse, T.; Bailey, A.M.; Boonham, N.; Foster, G.D. Evidence for different, host-dependent functioning of Rx against both wild-type and recombinant Pepino mosaic virus. Mol. Plant Pathol. 2016, 17, 120–126. [Google Scholar] [CrossRef]
- Duff-Farrier, C.R.A.; Bailey, A.M.; Boonham, N.; Foster, G.D. A pathogenicity determinant maps to the N-terminal coat protein region of the Pepino mosaic virus genome. Mol. Plant Pathol. 2015, 16, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Stankovic, I.; Vucurovic, A.; Zecevic, K.; Petrovic, B.; Ristic, D.; Vucurovic, I.; Krstic, B. Short communication: Pepino mosaic virus, a new threat for Serbia’s tomatoes. Span. J. Agric. Res. 2020, 18, e10SC05. [Google Scholar] [CrossRef]
- Tiberini, A.; Davino, S.; Davino, M.; Tomassoli, L. Complete sequence, genotyping and comparative analysis of pepino mosaic virus isolates from Italy. J. Plant Pathol. 2011, 96, 437–442. [Google Scholar]
- Gómez-Aix, C.; Alcaide, C.; Gómez, P.; Aranda, M.A.; Sánchez-Pina, M.A. In situ hybridization for the localization of two pepino mosaic virus isolates in mixed infections. J. Virol. Methods 2019, 267, 42–47. [Google Scholar] [CrossRef]
- Alcaide, C.; Rabadán, M.P.; Juárez, M.; Gómez, P. Long-term cocirculation of two strains of pepino mosaic virus in tomato crops and its effect on population genetic variability. Phytopathology 2020, 110, 49–57. [Google Scholar] [CrossRef]
- Pagán, I.; del Carmen Córdoba-Sellés, M.; Martínez-Priego, L.; Fraile, A.; Malpica, J.M.; Jordá, C.; García-Arenal, F. Genetic structure of the population of pepino mosaic virus infecting tomato crops in Spain. Phytopathology 2006, 96, 274–279. [Google Scholar] [CrossRef]
- Alcaide, C.; Sardanyés, J.; Elena, S.F.; Gómez, P. Increasing temperature alters the within-host competition of viral strains and influences virus genetic variability. Virus Evol. 2021, 7, veab017. [Google Scholar] [CrossRef]
- Minicka, J.; Elena, S.F.; Borodynko-Filas, N.; Rubiś, B.; Hasiów-Jaroszewska, B. Strain-dependent mutational effects for pepino mosaic virus in a natural host. BMC Evol. Biol. 2017, 17, 67. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, S.; Batuman, O. Co-Infection of tomato brown rugose fruit virus and pepino mosaic virus in grocery tomatoes in South Florida: Prevalence and genomic diversity. Viruses 2023, 15, 2305. [Google Scholar] [CrossRef] [PubMed]
- Klap, C.; Luria, N.; Smith, E.; Bakelman, E.; Belausov, E.; Laskar, O.; Lachman, O.; Gal-On, A.; Dombrovsky, A. The potential risk of plant-virus disease initiation by infected tomatoes. Plants 2020, 9, 623. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Borodynko, N. Characterization of the necrosis determinant of the European genotype of pepino mosaic virus by site-specific mutagenesis of an infectious cDNA clone. Arch. Virol. 2012, 157, 337–341. [Google Scholar] [CrossRef]
- Cockerham, G. Genetical studies on resistance to potato viruses X and Y. Heredity 1970, 25, 309–348. [Google Scholar] [CrossRef]
- Bendahmane, A.; Kanyuka, K.; Baulcombe, D.C. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 1999, 11, 781–791. [Google Scholar] [CrossRef]
- Mathioudakis, M.M.; Veiga, R.; Ghita, M.; Tsikou, D.; Medina, V.; Canto, T.; Makris, A.M.; Livieratos, I.C. Pepino mosaic virus capsid protein interacts with a tomato heat shock protein cognate 70. Virus Res. 2012, 163, 28–39. [Google Scholar] [CrossRef]
- Méndez-López, E.; Donaire, L.; Gosálvez, B.; Díaz-Vivancos, P.; Sánchez-Pina, M.A.; Tilsner, J.; Aranda, M.A. Tomato SlGSTU38 interacts with the PepMV coat protein and promotes viral infection. New Phytol. 2023, 238, 332–348. [Google Scholar] [CrossRef]
- Mathioudakis, M.M.; Khechmar, S.; Owen, C.A.; Medina, V.; Ben Mansour, K.; Tomaszewska, W.; Spanos, T.; Sarris, P.F.; Livieratos, I.C. A Thioredoxin domain-containing protein interacts with pepino mosaic virus triple gene block protein 1. Int. J. Mol. Sci. 2018, 19, 3747. [Google Scholar] [CrossRef]
- He, M.; Li, Z.; Xie, X. The roles of N6-Methyladenosine modification in plant–RNA virus interactions. Int. J. Mol. Sci. 2023, 24, 15608. [Google Scholar] [CrossRef]
- He, H.; Ge, L.; Chen, Y.; Zhao, S.; Li, Z.; Zhou, X.; Li, F. m6A modification of plant virus enables host recognition by NMD factors in plants. Sci. China Life Sci. 2024, 67, 161–174. [Google Scholar] [CrossRef]
- He, H.; Ge, L.; Li, Z.; Zhou, X.; Li, F. Pepino mosaic virus antagonizes plant m6A modification by promoting the autophagic degradation of the m6A writer HAKAI. aBIOTECH 2023, 4, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ramón, F.; Rodríguez-Sepúlveda, P.; Bretó, P.; Donaire, L.; Hernando, Y.; Aranda, M.A. The tomato calcium-permeable channel 4.1 (SlOSCA4.1) is a susceptibility factor for pepino mosaic virus. Plant Biotechnol. J. 2023, 21, 2140–2154. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Peter van Esse, H.; Ballester, A.-R.; Hogewoning, S.W.; Parra, N.O.; Paeleman, A.; Lievens, B.; Bovy, A.G.; Thomma, B.P.H.J. Differential tomato transcriptomic responses induced by pepino mosaic virus isolates with differential aggressiveness. Plant Physiol. 2011, 156, 301–318. [Google Scholar] [CrossRef] [PubMed]
- Alcaide, C.; Donaire, L.; Aranda, M.A. Transcriptome analyses unveiled differential regulation of AGO and DCL genes by pepino mosaic virus strains. Mol. Plant Pathol. 2022, 23, 1592–1607. [Google Scholar] [CrossRef] [PubMed]
- de Moya-Ruiz, C.; Rivero, R.M.; Pardo-Hernández, M.; Gómez, P. Heat stress and viral infection elicit a specific and differential gene response in thermo-tolerant and thermo-susceptible tomato plants. J. Plant Interact. 2025, 20, 2554136. [Google Scholar] [CrossRef]
- Zarrougui, N.E.; Spanos, T.; Kissoudis, C.; Livieratos, I.C. The effect of soil salinity and pepino mosaic virus infection on the expression of tomato defence-related genes. Plant Pathol. 2022, 71, 1304–1312. [Google Scholar] [CrossRef]
- Tsitsekian, D.; Daras, G.; Templalexis, D.; Avgeri, F.; Lotos, L.; Orfanidou, C.G.; Ntoukakis, V.; Maliogka, V.I.; Rigas, S. A subset of highly responsive transcription factors upon tomato infection by pepino mosaic virus. Plant Biol. 2023, 25, 529–540. [Google Scholar] [CrossRef]
- Ludman, M.; Szalai, G.; Janda, T.; Fátyol, K. Hierarchical contribution of Argonaute proteins to antiviral protection. J. Exp. Bot. 2023, 74, 6760–6772. [Google Scholar] [CrossRef]
- Ling, K.-S.; Scott, J.W. Sources of resistance to pepino mosaic virus in tomato accessions. Plant Dis. 2007, 91, 749–753. [Google Scholar] [CrossRef]
- Soler-Aleixandre, S.; López, C.; Cebolla-Cornejo, J.; Nuez, F. Sources of resistance to pepino mosaic virus (PepMV) in tomato. HortScience 2007, 42, 40–45. [Google Scholar] [CrossRef]
- Soler, S.; López, C.; Prohens, J.; Nuez, F. New sources of resistance to PepMV in tomato. J. Plant Dis. Prot. 2011, 118, 149–155. [Google Scholar] [CrossRef]
- Vogelaar, A.; Rozendaal, A.; van der Vlugt, R.; Schürch, A. Detection of Pepino Mosaic Virus. U.S. Patent US9637757B2, 2 May 2017. [Google Scholar]
- Ling, K.S.; Wechter, W.P.; Jordan, R. Development of a one-step immunocapture real-time TaqMan RT-PCR assay for the broad spectrum detection of pepino mosaic virus. J. Virol. Methods 2007, 144, 65–72. [Google Scholar] [CrossRef]
- Picó, B.; Herraiz, J.; Ruiz, J.J.; Nuez, F. Widening the genetic basis of virus resistance in tomato. Sci. Hortic. 2002, 94, 73–89. [Google Scholar] [CrossRef]
- Canady, M.A.; Meglic, V.; Chetelat, R.T. A library of Solanum lycopersicoides introgression lines in cultivated tomato. Genome 2005, 48, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Hak, H.; Spiegelman, Z. The tomato brown rugose fruit virus movement protein overcomes Tm-22 resistance in tomato while attenuating viral transport. Mol. Plant-Microbe Interact. 2021, 34, 1024–1032. [Google Scholar] [CrossRef]
- Khatodia, S.; Bhatotia, K.; Tuteja, N. Development of CRISPR/Cas9 mediated virus resistance in agriculturally important crops. Bioengineered 2017, 8, 274–279. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, X.; Zhou, G.; Zhang, T. Engineering plant virus resistance: From RNA silencing to genome editing strategies. Plant Biotechnol. J. 2020, 18, 328–336. [Google Scholar] [CrossRef]
- Wang, A. Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu. Rev. Phytopathol. 2015, 53, 45–66. [Google Scholar] [CrossRef]
- Hashimoto, M.; Neriya, Y.; Yamaji, Y.; Namba, S. Recessive resistance to plant viruses: Potential resistance genes beyond translation initiation factors. Front. Microbiol. 2016, 7, 1695. [Google Scholar] [CrossRef]
- Zimny, T. Regulation of GMO field trials in the EU and new genomic techniques: Will the planned reform facilitate experimenting with gene-edited plants? BioTechnologia 2023, 104, 75–83. [Google Scholar] [CrossRef]
- Eriksson, D.; Custers, R.; Edvardsson Björnberg, K.; Hansson, S.O.; Purnhagen, K.; Qaim, M.; Romeis, J.; Schiemann, J.; Schleissing, S.; Tosun, J.; et al. Options to reform the European Union legislation on GMOs: Risk governance. Trends Biotechnol. 2020, 38, 349–351. [Google Scholar] [CrossRef] [PubMed]
- Ling, K.-S. Effectiveness of chemo- and thermotherapeutic treatments on pepino mosaic virus in tomato seed. Plant Dis. 2010, 94, 325–328. [Google Scholar] [CrossRef]
- Schenk, M.F.; Hamelink, R.; van der Vlugt, R.A.A.; Vermunt, A.M.W.; Kaarsenmaker, R.C.; Stijger, I.C.C.M.M. The use of attenuated isolates of pepino mosaic virus for cross-protection. Eur. J. Plant Pathol. 2010, 127, 249–261. [Google Scholar] [CrossRef]
- Authority, E.F.S. Conclusion on the peer review of the pesticide risk assessment of the active substance pepino mosaic virus strain CH2 isolate 1906. EFSA J. 2015, 13, 3951. [Google Scholar]
- Authority, E.F.S. Peer review of the pesticide risk assessment of the active substance mild pepino mosaic virus isolate VX1. EFSA J. 2017, 15, e04906. [Google Scholar] [CrossRef]
- Spanos, T.; Ghodbane, A.; Rezazga, A.; Ludman, M.; Fátyol, K.; Elbeaino, T.; Livieratos, I. Pre-inoculation of ago2 and DCL2/4-deficient Nicotiana benthamiana plants with the pepino mosaic virus EU mild isolate confers complete protection against superinfection with the aggressive isolate. Front. Virol. 2025, 5, 1584535. [Google Scholar] [CrossRef]
- Imane, K.; Siham, E.; Kacem, M.; Malika, B.; Tayeb, K.; Aicha, R.L.; Lydia, F.; Stéphane, V.J.; Fouad, B.; Amal, S.; et al. Protective effect of symmetrical N-heterocyclic 1,3,4-oxadiazole and 1,3,4-thiadiazole derivatives against pepino mosaic virus of tomato. J. Phytopathol. 2022, 170, 557–573. [Google Scholar] [CrossRef]
- Ezzouine, N.; El Kaim Billah, R.; Soufiane, A.; Esserti, S.; Belfaiza, M.; Rifai, L.A.; Makroum, K.; Koussa, T.; Faize, L.; Alburquerque, N.; et al. Protection of Solanum lycopesicum induced by chitosan and chitosan nano-hydroxyapatite against pepino mosaic virus and Verticillium dahliae. Biocatal. Agric. Biotechnol. 2022, 43, 102386. [Google Scholar] [CrossRef]
- Caro, M.; Verlaan, M.G.; Julián, O.; Finkers, R.; Wolters, A.-M.A.; Hutton, S.F.; Scott, J.W.; Kormelink, R.; Visser, R.G.F.; Díez, M.J.; et al. Assessing the genetic variation of Ty-1 and Ty-3 alleles conferring resistance to tomato yellow leaf curl virus in a broad tomato germplasm. Mol. Breed. 2015, 35, 132. [Google Scholar] [CrossRef] [PubMed]
- Voorburg, C.M.; Yan, Z.; Bergua-Vidal, M.; Wolters, A.-M.A.; Bai, Y.; Kormelink, R. Ty-1, a universal resistance gene against geminiviruses that is compromised by co-replication of a betasatellite. Mol. Plant Pathol. 2020, 21, 160–172. [Google Scholar] [CrossRef]
- Cao, X.; Huang, M.; Wang, S.; Li, T.; Huang, Y. Tomato yellow leaf curl virus: Characteristics, influence, and regulation mechanism. Plant Physiol. Biochem. 2024, 213, 108812. [Google Scholar] [CrossRef] [PubMed]
- Kabas, A.; Fidan, H.; Kucukaydin, H.; Atan, H.N. Screening of wild tomato species and interspecific hybrids for resistance/tolerance to tomato brown rugose fruit virus (ToBRFV). Chil. J. Agric. Res. 2022, 82, 189–196. [Google Scholar] [CrossRef]
- Zinger, A.; Lapidot, M.; Harel, A.; Doron-Faigenboim, A.; Gelbart, D.; Levin, I. Identification and mapping of tomato genome loci controlling tolerance and resistance to tomato brown rugose fruit virus. Plants 2021, 10, 179. [Google Scholar] [CrossRef] [PubMed]
- Zinger, A.; Doron-Faigenboim, A.; Gelbart, D.; Levin, I.; Lapidot, M. Contribution of the tobamovirus resistance gene Tm-1 to control of tomato brown rugose fruit virus (ToBRFV) resistance in tomato. PLoS Genet. 2025, 21, e1011725. [Google Scholar] [CrossRef]
- Rochsar, E.; Torgeman, S.; Bandel, K.; Koren, A.; Klap, C.; Dombrovsky, A.; Zamir, D. Tissue-specific resistance and susceptibility to the tomato brown rugose fruit virus (ToBRFV) conferred by Solanum pennellii loci. BMC Plant Biol. 2025, 25, 51. [Google Scholar] [CrossRef]
- Jewehan, A.; Salem, N.; Tóth, Z.; Salamon, P.; Szabó, Z. Screening of Solanum (sections Lycopersicon and Juglandifolia) germplasm for reactions to the tomato brown rugose fruit virus (ToBRFV). J. Plant Dis. Prot. 2022, 129, 117–123. [Google Scholar] [CrossRef]
- Syngenta’s ToBRFV Information Center. Available online: https://www.syngentavegetables.com/resources/ToBRFV (accessed on 25 September 2025).
- Results from Our ToBRFV-Resistant Tomato Varieties Exceed Expectations. Available online: https://www.rijkzwaan.com/en/news/Results-from-our-ToBRFV-resistant-tomato-varieties-exceed-expectations (accessed on 25 September 2025).
- Ishikawa, M.; Yoshida, T.; Matsuyama, M.; Kouzai, Y.; Kano, A.; Ishibashi, K. Tomato brown rugose fruit virus resistance generated by quadruple knockout of homologs of tobamovirus multiplication1 in tomato. Plant Physiol. 2022, 189, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Mullins, E.; Bresson, J.-L.; Dalmay, T.; Dewhurst, I.C.; Epstein, M.M.; Firbank, L.G.; Guerche, P.; Hejatko, J.; Moreno, F.J.; Naegeli, H.; et al. In vivo and in vitro random mutagenesis techniques in plants. EFSA J. 2021, 19, e06611. [Google Scholar] [CrossRef] [PubMed]
- Comai, L.; Henikoff, S. TILLING: Practical single-nucleotide mutation discovery. Plant J. 2006, 45, 684–694. [Google Scholar] [CrossRef]
- Henikoff, S.; Till, B.J.; Comai, L. TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 2004, 135, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Piron, F.; Nicolaï, M.; Minoïa, S.; Piednoir, E.; Moretti, A.; Salgues, A.; Zamir, D.; Caranta, C.; Bendahmane, A. An induced mutation in tomato eIF4E leads to immunity to two Potyviruses. PLoS ONE 2010, 5, e11313. [Google Scholar] [CrossRef]
- Akhter, M.S.; Nakahara, K.S.; Masuta, C. Resistance induction based on the understanding of molecular interactions between plant viruses and host plants. Virol. J. 2021, 18, 176. [Google Scholar] [CrossRef]
- Leibman, D.; Ortega-Parra, N.; Wolf, D.; Shterkman, M.; Hanssen, I.; Gal-On, A. A transgenic RNAi approach for developing tomato plants immune to pepino mosaic virus. Plant Pathol. 2021, 70, 1003–1012. [Google Scholar] [CrossRef]
- Praveen, S.; Ramesh, S.V.; Mishra, A.K.; Koundal, V.; Palukaitis, P. Silencing potential of viral derived RNAi constructs in tomato leaf curl virus-AC4 gene suppression in tomato. Transgenic Res. 2010, 19, 45–55. [Google Scholar] [CrossRef]
- Fuentes, A.; Ramos, P.L.; Fiallo, E.; Callard, D.; Sánchez, Y.; Peral, R.; Rodríguez, R.; Pujol, M. Intron–hairpin RNA derived from replication associated protein C1 gene confers immunity to tomato yellow leaf curl virus infection in transgenic tomato plants. Transgenic Res. 2006, 15, 291–304. [Google Scholar] [CrossRef]
- Carbonell, A.; López, C.; Daròs, J.-A. Fast-forward identification of highly effective artificial small RNAs against different tomato spotted wilt virus isolates. Mol. Plant-Microbe Interact. 2019, 32, 142–156. [Google Scholar] [CrossRef]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Xu, Z.P.; Carroll, B.J. Induction of virus resistance by exogenous application of double-stranded RNA. Curr. Opin. Virol. 2017, 26, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, C.; Meghwanshi, K.K.; Shukla, N.; Shukla, J.N. Innate and adaptive resistance to RNAi: A major challenge and hurdle to the development of double stranded RNA-based pesticides. 3 Biotech 2021, 11, 498. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Imran, M.; Feng, X.; Shen, X.; Sun, Z. Spray-induced gene silencing for crop protection: Recent advances and emerging trends. Front. Plant Sci. 2025, 16, 1527944. [Google Scholar] [CrossRef]
- Tabein, S.; Jansen, M.; Noris, E.; Vaira, A.M.; Marian, D.; Behjatnia, S.A.A.; Accotto, G.P.; Miozzi, L. The induction of an effective dsRNA-mediated resistance against tomato spotted wilt virus by exogenous application of double-stranded RNA largely depends on the selection of the viral RNA target region. Front. Plant Sci. 2020, 11, 533338. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Martín, J.; Ruiz, L.; Janssen, D.; Velasco, L. Exogenous application of dsRNA for the control of viruses in cucurbits. Front. Plant Sci. 2022, 13, 895953. [Google Scholar] [CrossRef]
- Voigt, B. EU regulation of gene-edited plants—A reform proposal. Front. Genome Ed. 2023, 5, 1119442. [Google Scholar] [CrossRef]
- Eriksson, D.; Kershen, D.; Nepomuceno, A.; Pogson, B.J.; Prieto, H.; Purnhagen, K.; Smyth, S.; Wesseler, J.; Whelan, A. A comparison of the EU regulatory approach to directed mutagenesis with that of other jurisdictions, consequences for international trade and potential steps forward. New Phytol. 2019, 222, 1673–1684. [Google Scholar] [CrossRef]
- Fernández Ríos, D.; Quintana, S.A.; Gómez Paniagua, P.; Arrúa, A.A.; Brozón, G.R.; Bertoni Hicar, M.S.; Castro Alegría, A.; Goberna, M.F. Regulatory challenges and global trade implications of genome editing in agriculture. Front. Bioeng. Biotechnol. 2025, 13, 1609110. [Google Scholar] [CrossRef]
- Schenk, P.; Hervé, P. Letter from NexGen plants to Aphis. Available online: https://www.aphis.usda.gov/biotechnology/downloads/reg_loi/19-095-02_air_inquiry_cbidel_a1.pdf (accessed on 31 October 2025).
| Species | Accession(s) | Resistance Level | Crossability with S. lycopersicum | Reference |
|---|---|---|---|---|
| S. lycopersicoides | LA1964, LA4123, LA4126, LA4131 | Complete resistance (symptom-free, no virus detected) | Highly incompatible (introgression lines required) | [87] |
| LA1966, LA2408, LA2951 | Moderate resistance (limited systemic infection, reduced virus accumulation) | [87] | ||
| S. pseudocapsicum | AN-CA-214 | Complete resistance (symptom-free, no virus detected) | Incompatible | [86] |
| S. habrochaites | LA1731 | Broad spectrum resistance (symptom-free or reduced symptoms, virus detectable) | Partially compatible (interspecific hybrids often show reduced fertility) | [85], this study |
| LA2156, LA2167 | Variable resistance (symptom suppression or susceptible) | [85], this study | ||
| S. peruvianum | CIAPAN-15, CIAPAN-16, PI212407, PI251311, LA0107, LA1305 | Moderate resistance (reduced virus accumulation) | Partially compatible (embryo rescue often required) | [85,86] |
| NCIMB 41927–42069 | High to near-complete resistance | [88] | ||
| S. chilense | ECU-527, LA0458, LA0470, LA1963, LA1968, LA1971, LA2762, CDP03677, CDP04506, CDP06600, LA2748 | Moderate resistance (subsets symptom-free; low or undetectable virus accumulation) | Partially compatible (frequent embryo rescue) | [85,86,87] |
| S. ochranthum | ECU-335 | Moderate resistance (mild symptoms; reduced virus accumulation) | Highly incompatible | [86] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowakowska, M.; Minicka, J.; Nowicki, M.; Szczechura, W.; Hasiów-Jaroszewska, B. Pepino Mosaic Virus in Tomato: Challenges, Control Strategies, and Future Prospects for Resistance Breeding. Int. J. Mol. Sci. 2025, 26, 11749. https://doi.org/10.3390/ijms262311749
Nowakowska M, Minicka J, Nowicki M, Szczechura W, Hasiów-Jaroszewska B. Pepino Mosaic Virus in Tomato: Challenges, Control Strategies, and Future Prospects for Resistance Breeding. International Journal of Molecular Sciences. 2025; 26(23):11749. https://doi.org/10.3390/ijms262311749
Chicago/Turabian StyleNowakowska, Marzena, Julia Minicka, Marcin Nowicki, Wojciech Szczechura, and Beata Hasiów-Jaroszewska. 2025. "Pepino Mosaic Virus in Tomato: Challenges, Control Strategies, and Future Prospects for Resistance Breeding" International Journal of Molecular Sciences 26, no. 23: 11749. https://doi.org/10.3390/ijms262311749
APA StyleNowakowska, M., Minicka, J., Nowicki, M., Szczechura, W., & Hasiów-Jaroszewska, B. (2025). Pepino Mosaic Virus in Tomato: Challenges, Control Strategies, and Future Prospects for Resistance Breeding. International Journal of Molecular Sciences, 26(23), 11749. https://doi.org/10.3390/ijms262311749








