Data-Independent Acquisition (DIA)-Based Proteomics for the Identification of Biomarkers in Tissue Washings of Endometrial Cancer
Abstract
1. Introduction
2. Results
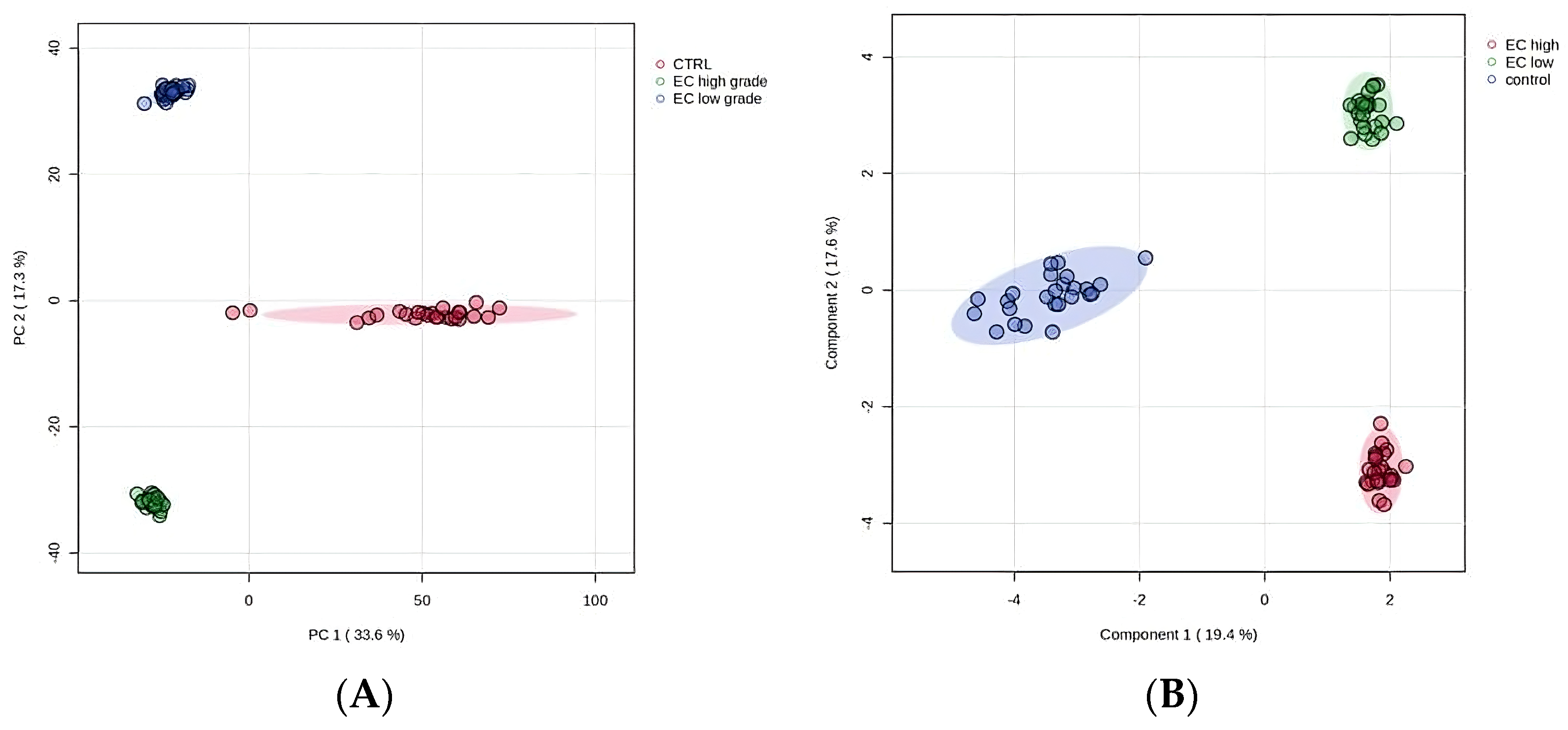
2.1. Proteomics Study from EC Tissue Washes
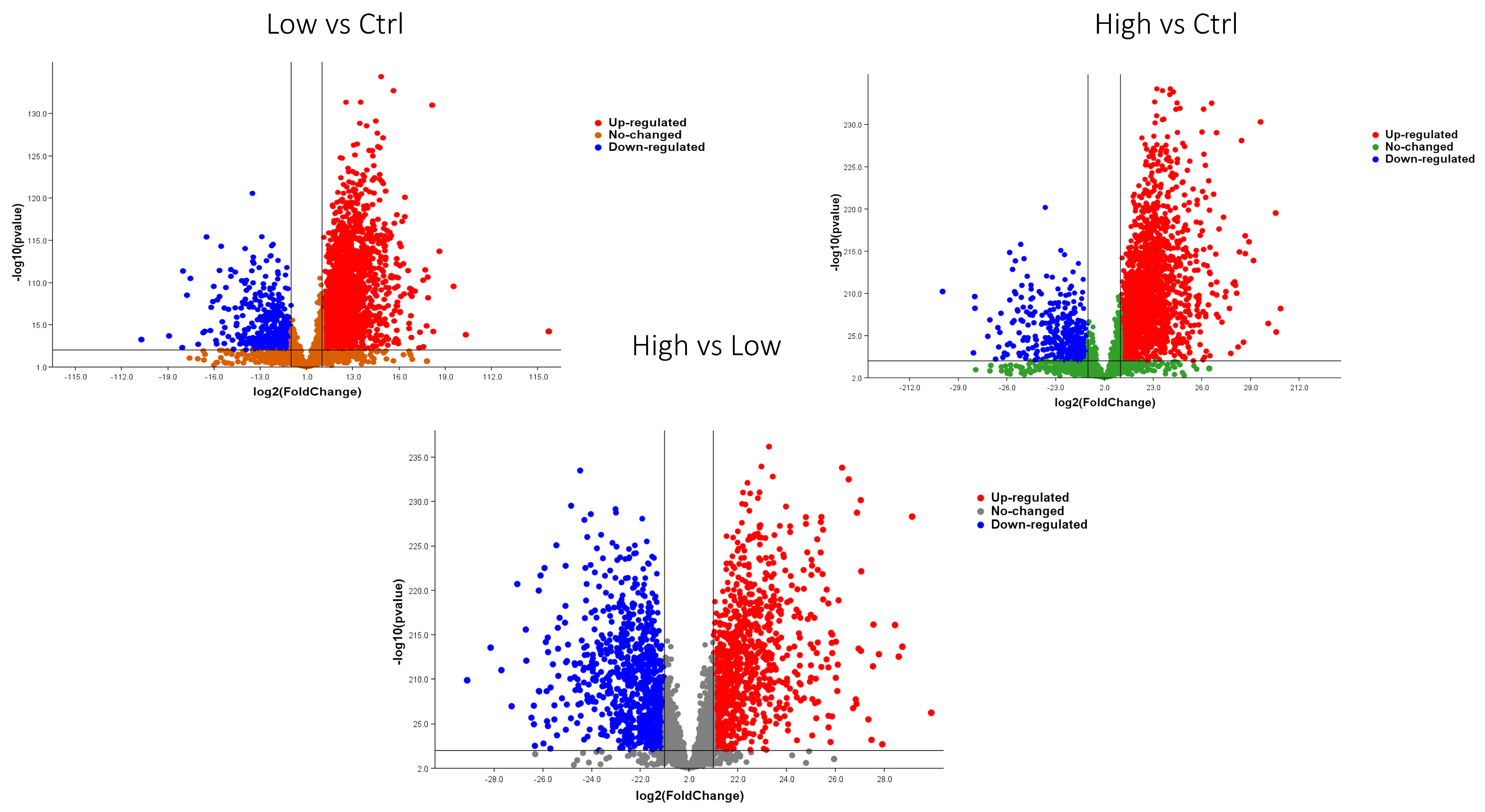
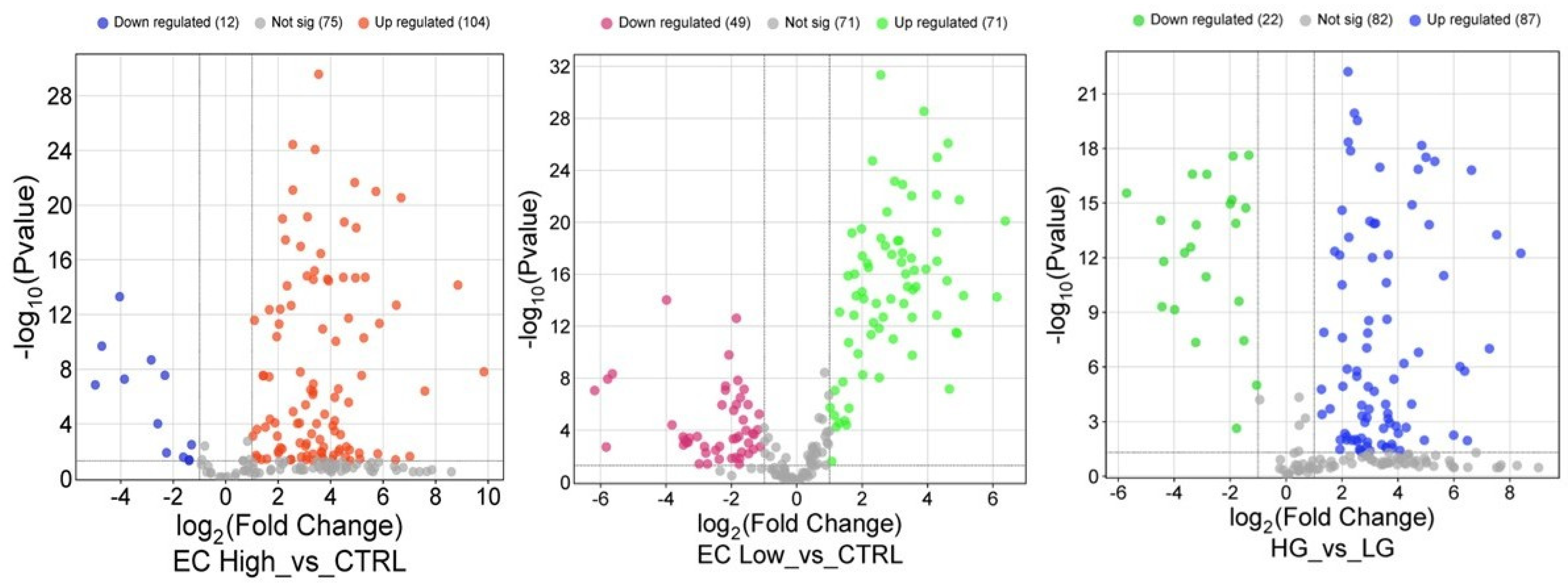
2.2. Statistical Analysis
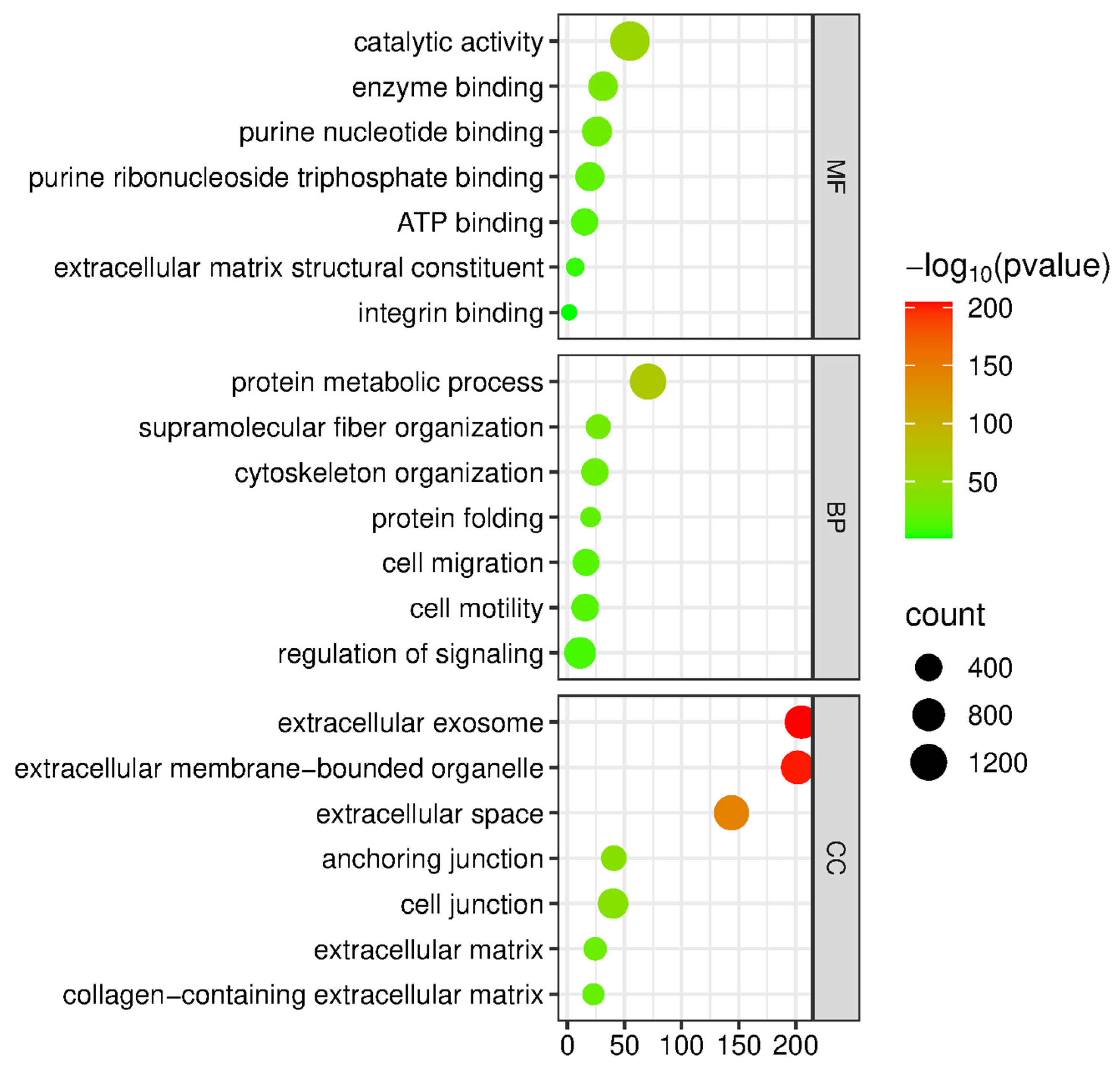
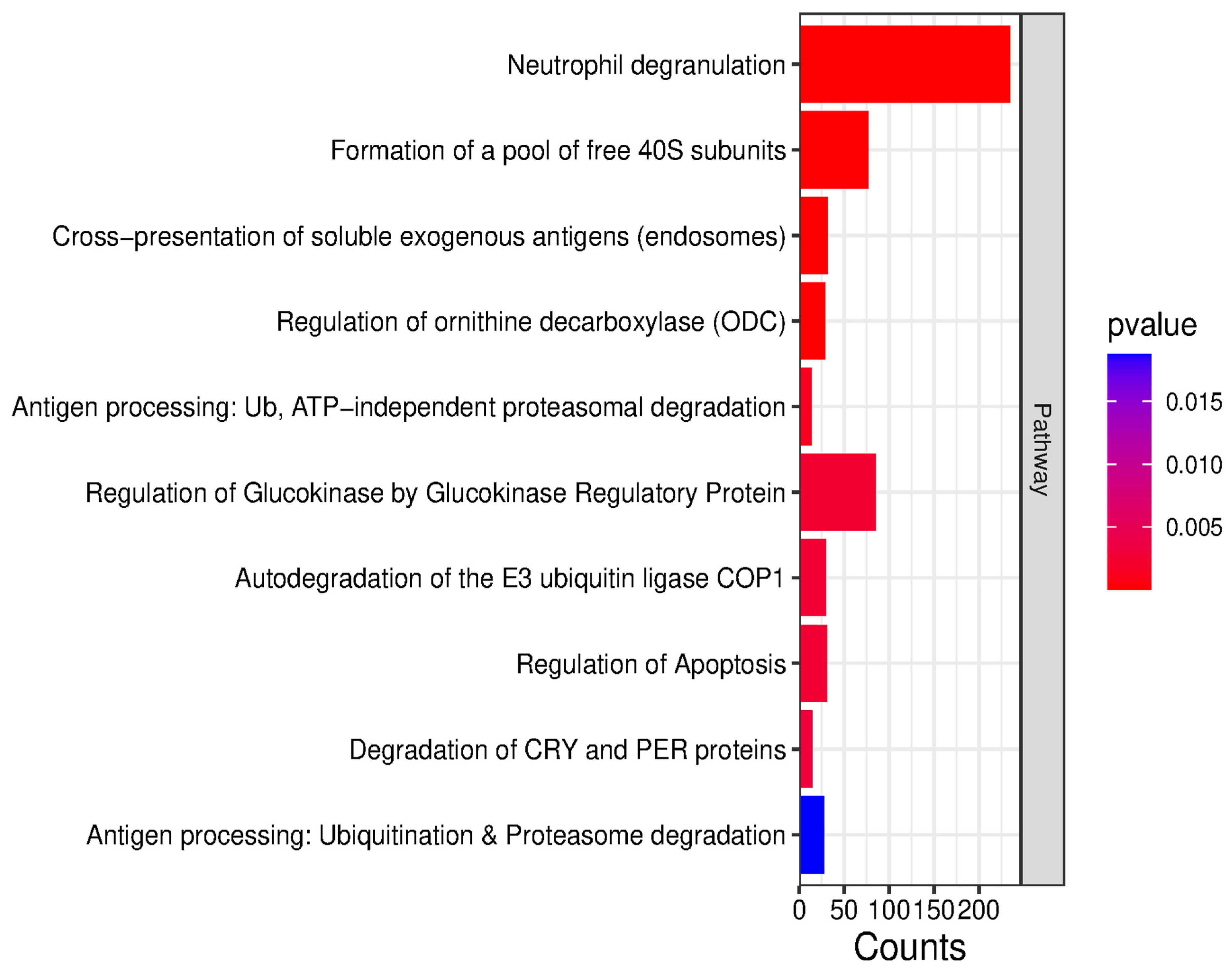
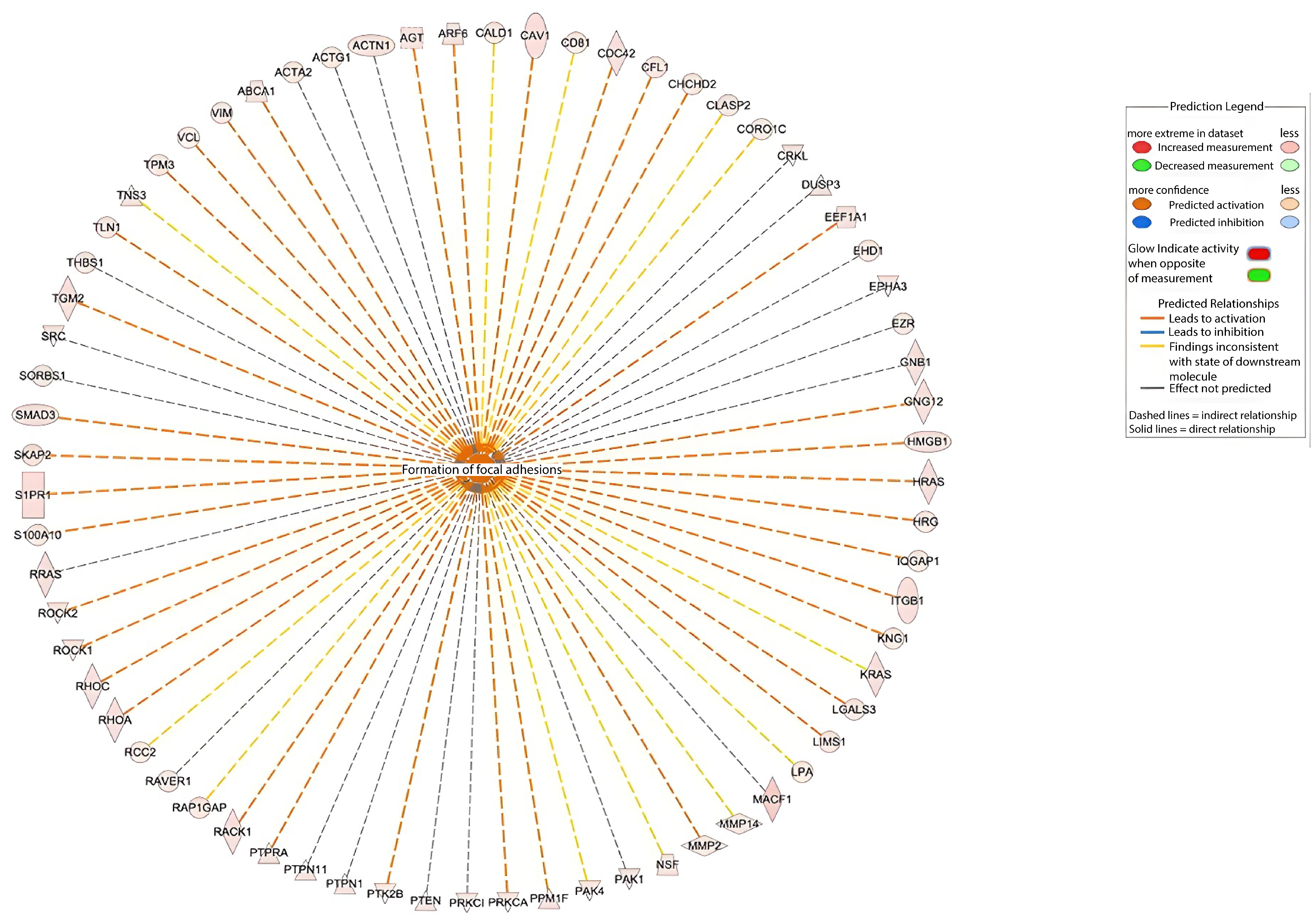
2.3. Bioinformatic Analysis of Identified Proteins
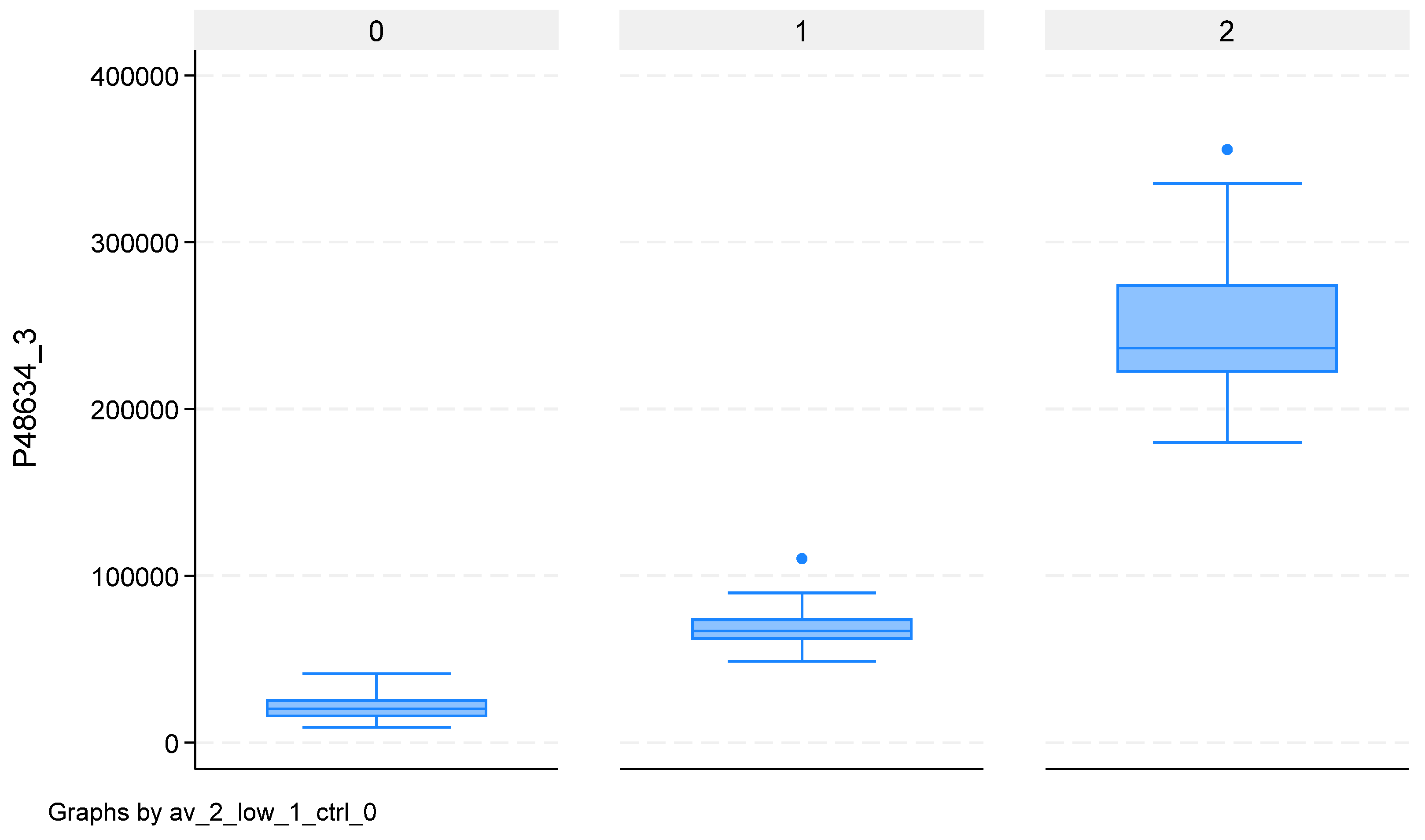
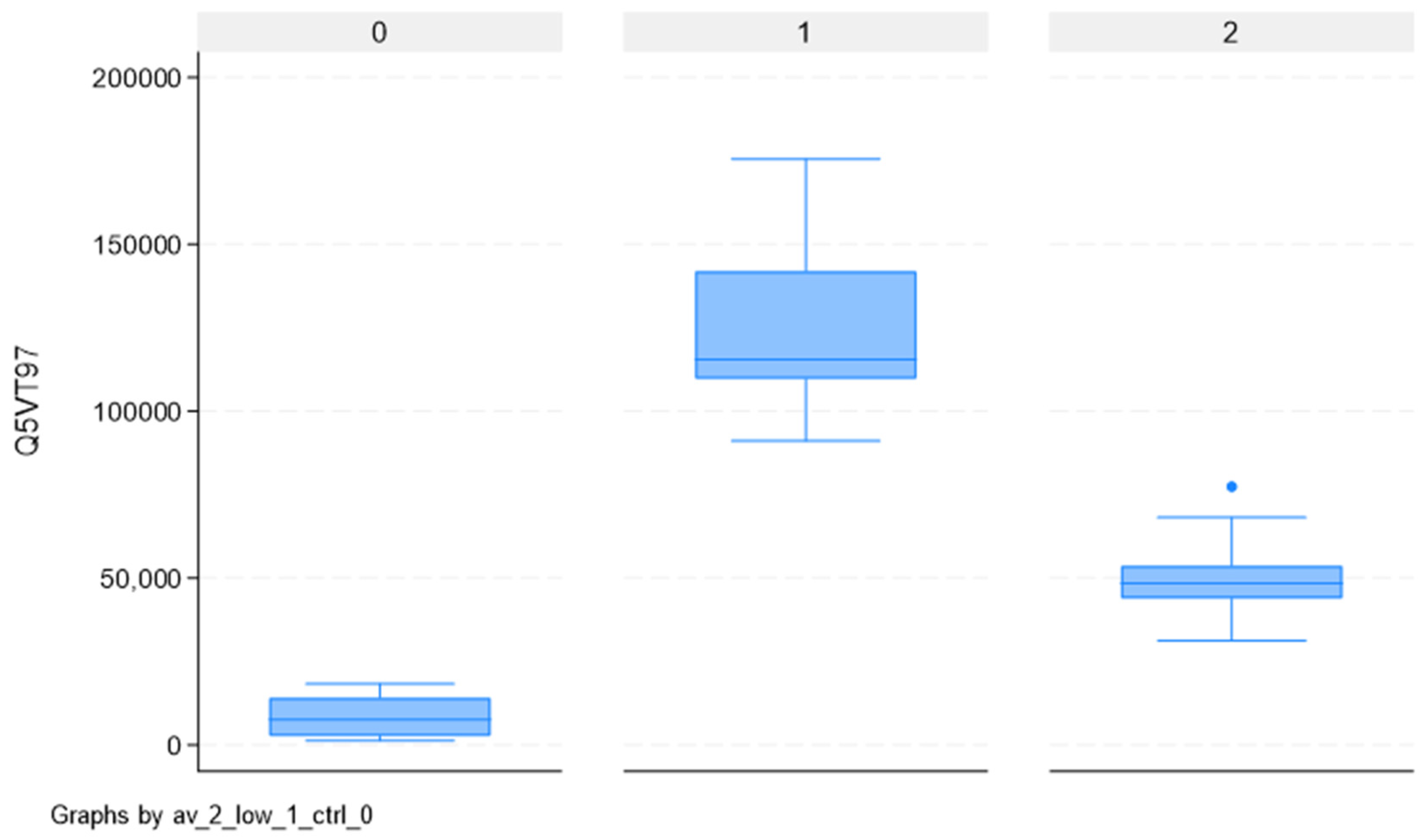
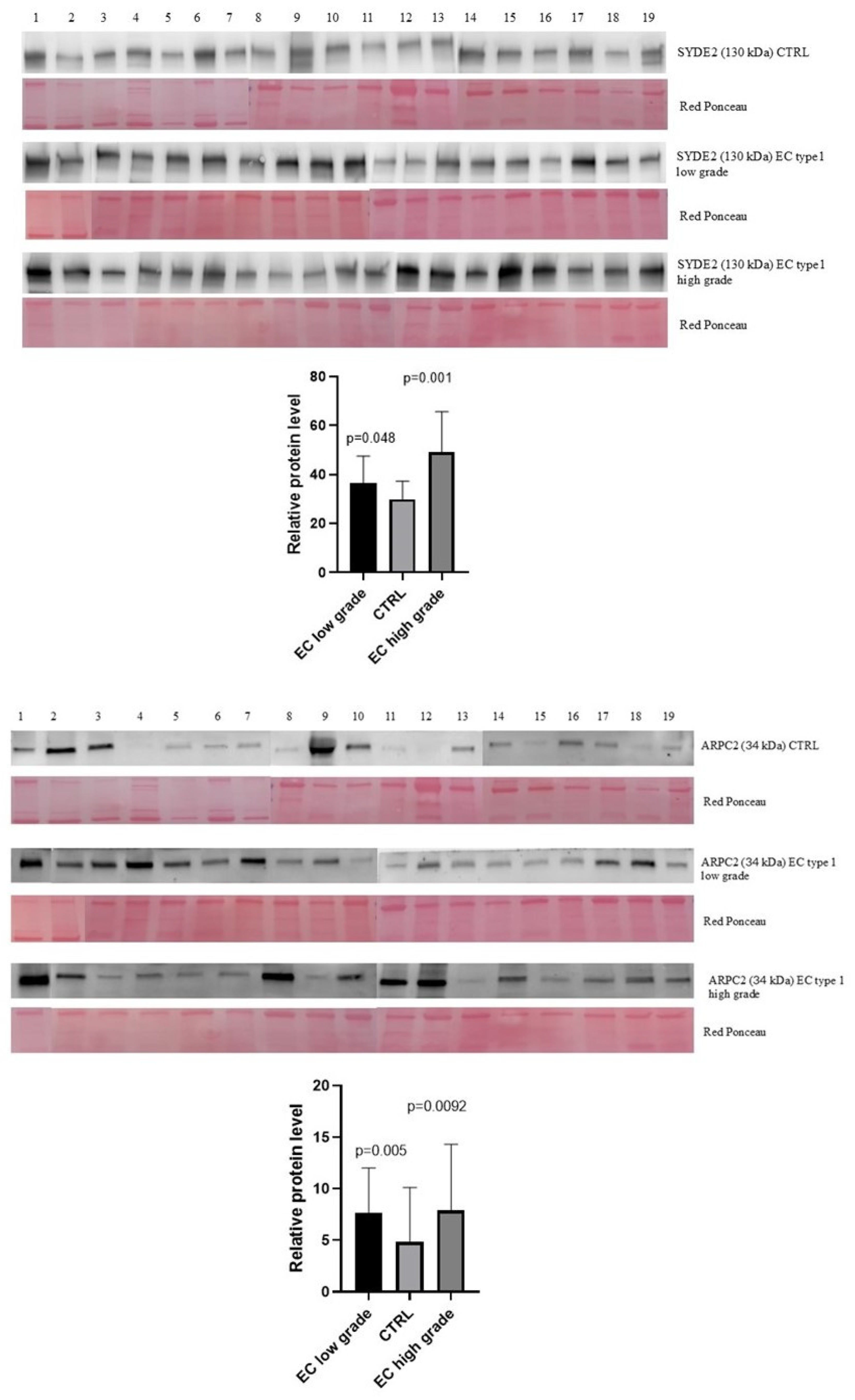
2.4. Mass-Spectrometry Validation by Western Blotting in an Independent Cohort
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Sample Collection
4.3. Protein Digestion and MS Analysis
4.4. Western Blotting
4.5. Bioinformatic Analysis
4.6. Statistical Approach
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morice, P.; Leary, A.; Creutzberg, C.; Abu-Rustum, N.; Darai, E. Endometrial cancer. Lancet 2016, 387, 1094–1108. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 7, 209–249. [Google Scholar]
- International Agency for Research on Cancer. Cancer Today [Internet]. Available online: http://gco.iarc.fr/today/home (accessed on 27 April 2024).
- Cao, S.-Y.; Fan, Y.; Zhang, Y.-F.; Ruan, J.-Y.; Mu, Y.; Li, J.-K. Recurrence and survival of patients with stage III endometrial cancer after radical surgery followed by adjuvant chemo- or chemoradiotherapy: A systematic review and meta-analysis. BMC Cancer 2023, 3, 31. [Google Scholar] [CrossRef]
- Bokhman, J.V. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983, 15, 10–17. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Feng, Q.; Jia, L.; Sha, X.; Zhang, T.; Lu, L.; Wang, R.; Bai, B. Present progress in biomarker discovery of endometrial cancer by multi-omics approaches. Clin. Proteom. 2025, 22, 15. [Google Scholar] [CrossRef]
- Sommella, E.; Capaci, V.; Aloisio, M.; Salviati, E.; Campiglia, P.; Molinario, G.; Licastro, D.; Di Lorenzo, G.; Romano, F.; Ricci, G.; et al. A Label-Free Proteomic Approach for the Identification of Biomarkers in the Exosome of Endometrial Cancer Serum. Cancers 2022, 14, 6262. [Google Scholar] [CrossRef]
- Ura, B.; Monasta, L.; Arrigoni, G.; Franchin, C.; Radillo, O.; Peterlunger, I.; Ricci, G.; Scrimin, F. A proteomic approach for the identification of biomarkers in endometrial cancer uterine aspirate. Oncotarget 2017, 8, 109536–109545. [Google Scholar] [CrossRef]
- Njoku, K.; Pierce, A.; Chiasserini, D.; Geary, B.; Campbell, A.E.; Kelsall, J.; Reed, R.; Geifman, N.; Whetton, A.D.; Emma, J.; et al. Detection of endometrial cancer in cervico-vaginal fluid and blood plasma: Leveraging proteomics and machine learning for biomarker discovery. EBio-Medicine 2024, 102, 105064. [Google Scholar] [CrossRef] [PubMed]
- Njoku, K.; Pierce, A.; Bethany Geary, B.; Campbell, A.E.; Kelsall, J.; Reed, R.; Armit, A.; Da Sylva, R.; Zhang, L.; Agnew, H.; et al. Quantitative SWATH-based proteomic profiling of urine for the identification of endometrial cancer biomarkers in symptomatic women. Br. J. Cancer 2023, 128, 1723–1732. [Google Scholar] [CrossRef]
- Giusti, L.; Iacconi, P.; Da Valle, Y.; Ciregia, F.; Ventroni, T.; Donadio, E.; Giannaccini, G.; Massimo Chiarugi, M.; Liborio Torregrossa, L.; Agnese Proietti, A.; et al. A proteomic profile of washing fluid from the colorectal tract to search for potential biomarkers of colon cancer. Mol. Biosyst. 2012, 8, 1088–1099. [Google Scholar] [CrossRef]
- Tarney, C.M.; Wang, G.; Bateman, N.W.; Conrads, K.A.; Zhou, M.; Hood, B.L.; Loffredo, J.; Tian, C.; Darcy, K.M.; Hamilton, C.A.; et al. Biomarker panel for early detection of endometrial cancer in the Prostate, Lung, Colorectal, and Ovarian cancer screening trial. Am. J. Obstet. Gynecol. 2019, 22, 472.e1–472.e10. [Google Scholar] [CrossRef]
- Zhou, Y.; Tao, L.; Qiu, J.; Xu, J.; Yang, X.; Zhang, Y.; Tian, X.; Guan, X.; Cen, X.; Zhao, Y. Tumor biomarkers for diagnosis, prognosis and targeted therapy. Signal Transduct. Target. Ther. 2024, 9, 132. [Google Scholar] [CrossRef]
- Passaro, A.; Al Bakir, M.; Hamilton, E.G.; Diehn, M.; André, F.; Roy-Chowdhuri, S.; Mountzios, G.; Wistuba, I.I.; Swanton, C.; Peters, S. Cancer biomarkers: Emerging trends and clinical implications for personalized treatment. Cell 2024, 187, 1617–1635. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Li, Y.; Li, J.; Song, L.; Chen, L.; Zhao, N.; Li, X.; Chen, N.; Long, L.; Zhao, J.; et al. Comprehensive deep profiling of the plasma proteome with protein corona on zeolite, NaY. J. Pharm. Anal. 2023, 13, 503–513. [Google Scholar] [CrossRef]
- Pieper, R.; Su, Q.; Gatlin, C.L.; Huang, S.; Anderson, N.L.; Steiner, S. Multi-component immunoaffinity subtraction chromatography: An innovative step towards a comprehensive survey of the human plasma proteome. Proteomics 2003, 3, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Celsi, F.; Monasta, L.; Arrigoni, G.; Battisti, I.; Licastro, D.; Aloisio, M.; Di Lorenzo, G.; Federico Romano, F.; Ricci, G.; Ura, B. Gel-based proteomic identification of suprabasin as a potential new candidate biomarker in endometrial cancer. Int. J. Mol. Sci. 2022, 23, 2076. [Google Scholar] [CrossRef]
- Agliarulo, I.; Swann Matassa, D.; Amoroso, M.R.; Maddalena, F.; Sisinni, L.; Sepe, L.; Ferrari, M.C.; Arzeni, D.; Avolio, R.; Paolella, G.; et al. TRAP1 controls cell migration of cancer cells in metabolic stress conditions: Correlations with AKT/p70S6K pathways. Biochim. Biophys. Acta 2015, 1853 Pt A, 2570–2579. [Google Scholar] [CrossRef]
- Wei, Z.; Xiaoyi, L.; Chunming, C.; Wei, Y.; Ping, Y. Metabolism of Amino Acids in Cancer. Front. Cell Dev. Biol. 2021, 8, 603837. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, Y.; Li, X.; Wang, Y.; Zhang, Z.; Wang, Z.; Zhang, X. Participation of protein metabolism in cancer progression and its potential targeting for the management of cancer. Amino Acids 2023, 55, 1223–1246. [Google Scholar] [CrossRef]
- De Berardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef]
- Kumar, M.A.; Baba, S.K.; Sadida, H.Q.; Marzooqi, S.A.; Jerobin, J.; Altemani, F.H.; Algehainy, N.; Alanazi, M.A.; Abou-Samra, A.B.; Kumar, R.; et al. Extracellular vesicles as tools and targets in therapy for diseases. Signal Transduct. Target. Ther. 2024, 9, 27. [Google Scholar] [CrossRef]
- Yuan, Z.; Li, Y.; Zhang, S.; Wang, X.; Dou, H.; Yu, X.; Zhang, Z.; Yang, S.; Xiao, M. Extracellular matrix remodeling in tumor progression and immune escape: From mechanisms to treatments. Mol. Cancer 2023, 22, 48. [Google Scholar] [CrossRef]
- Tong, D.; Zhao, L.; He, K.; Sun, H.; Cai, D.; Ni, L.; Sun, R.; Chang, S.; Song, T.; Huang, C. MECP2 promotes the growth of gastric cancer cells by suppressing miR-338-mediated antiproliferative effect. Oncotarget 2016, 7, 34845–34859. [Google Scholar] [CrossRef] [PubMed]
- Castro-Piedras, I.; Vartak, D.; Sharma, M.; Pandey, S.; Casas, L.; Molehin, D.; Rasha, F.; Fokar, M.; Nichols, J.; Almodovar, S.; et al. Identification of Novel MeCP2 Cancer-Associated Target Genes and Post-Translational Modifications. Front. Oncol. 2020, 10, 576362. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Liu, J.; Wang, F.; Li, Y. Integrated analysis of Solute carrier family-2 members reveals SLC2A4 as an independent favorable prognostic biomarker for breast cancer. Channels 2021, 15, 555–568. [Google Scholar] [CrossRef]
- Bohlen, J.; Roiuk, M.; Neff, M.; Aurelio, A.; Teleman, A.A. PRRC2 proteins impact translation initiation by promoting leaky scanning. Nucleic Acids Res. 2023, 51, 3391–3409. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, S.; Pan, Y.; Li, M.; Song, M.; Zhang, H.; Deng, M.; Yang, X.; Xu, J.; Zhang, S.; et al. m6A Reader PRRC2A Promotes Colorectal Cancer Progression via CK1ε-Mediated Activation of WNT and YAP Signaling Pathways. Adv. Sci. 2025, 12, e2406935. [Google Scholar] [CrossRef]
- Havelková, L.; Nanda, G.; Martinek, J.; Bellinvia, E.; Sikorová, L.; Šlajcherová, K.; Seifertová, D.; Fischer, L.; Fišerová, J.; Petrášek, J.; et al. Arp2/3 complex subunit ARPC2 binds to microtubules. Plant Sci. 2015, 241, 96–108. [Google Scholar] [CrossRef]
- Jang, H.J.; Yoon, Y.J.; Choi, J.; Lee, Y.J.; Lee, S.; Cho, W.; Byun, W.G.; Park, S.B.; Han, D.C.; Kwon, B.M. S-Benproperine, an Active Stereoisomer of Benproperine, Suppresses Cancer Migration and Tumor Metastasis by Targeting ARPC2. Pharmaceuticals 2022, 15, 1462. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Yu, C.J.; Dai, F.; Xiong, J.; Li, H.J.; Wu, Z.S.; Ding, R.; Wang, H. Role of ARPC2 in Human Gastric Cancer. Mediators Inflamm. 2017, 2017, 5432818. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Wu, J.; Zhou, Z.; Ma, J.; Dong, L. Two novel lncRNAs AF111167.2 and AL162377.1 targeting miR-21-5p mediated down expression of SYDE2 correlates with poor prognosis and tumor immune infiltration of ccRCC. Heliyon 2022, 8, e11079. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Wang, J.; Yang, M.; Li, H. Transcriptome analysis reveals the effect of oral contraceptive use on cervical cancer. Mol. Med. Rep. 2014, 10, 1703–1708. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Garcia, E.; Lesur, A.; Devis, L.; Campos, A.; Cabrera, S.; van Oostrum, J.; Matias-Guiu, X.; Gil-Moreno, A.; Reventos, J.; Colas, E.; et al. Development of a sequential workflow based on LC-PRM for the verification of endometrial cancer protein biomarkers in uterine aspirate samples. Oncotarget 2016, 7, 53102–53115. [Google Scholar] [CrossRef]
- Ura, B.; Scrimin, F.; Arrigoni, G.; Athanasakis, E.; Aloisio, M.; Monasta, L.; Ricci, G. Abnormal expression of leiomyoma cytoskeletal proteins involved in cell migration. Oncol. Rep. 2016, 35, 3094–3100. [Google Scholar] [CrossRef]
- Ura, B.; Scrimin, F.; Franchin, C.; Arrigoni, G.; Licastro, D.; Monasta, L.; Ricci, G. Identification of proteins with different abundance associated with cell migration and proliferation in leiomyoma interstitial fluid by proteomics. Oncol. Lett. 2017, 13, 3912–3920. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monasta, L.; Capaci, V.; Kharrat, F.; Ciampechini, M.; Balasan, N.; Conti, A.; Golino, V.; Campiglia, P.; Aloisio, M.; Licastro, D.; et al. Data-Independent Acquisition (DIA)-Based Proteomics for the Identification of Biomarkers in Tissue Washings of Endometrial Cancer. Int. J. Mol. Sci. 2025, 26, 11498. https://doi.org/10.3390/ijms262311498
Monasta L, Capaci V, Kharrat F, Ciampechini M, Balasan N, Conti A, Golino V, Campiglia P, Aloisio M, Licastro D, et al. Data-Independent Acquisition (DIA)-Based Proteomics for the Identification of Biomarkers in Tissue Washings of Endometrial Cancer. International Journal of Molecular Sciences. 2025; 26(23):11498. https://doi.org/10.3390/ijms262311498
Chicago/Turabian StyleMonasta, Lorenzo, Valeria Capaci, Feras Kharrat, Milena Ciampechini, Nour Balasan, Andrea Conti, Valentina Golino, Pietro Campiglia, Michelangelo Aloisio, Danilo Licastro, and et al. 2025. "Data-Independent Acquisition (DIA)-Based Proteomics for the Identification of Biomarkers in Tissue Washings of Endometrial Cancer" International Journal of Molecular Sciences 26, no. 23: 11498. https://doi.org/10.3390/ijms262311498
APA StyleMonasta, L., Capaci, V., Kharrat, F., Ciampechini, M., Balasan, N., Conti, A., Golino, V., Campiglia, P., Aloisio, M., Licastro, D., Di Lorenzo, G., Romano, F., Ricci, G., & Ura, B. (2025). Data-Independent Acquisition (DIA)-Based Proteomics for the Identification of Biomarkers in Tissue Washings of Endometrial Cancer. International Journal of Molecular Sciences, 26(23), 11498. https://doi.org/10.3390/ijms262311498








