AI-Integrated Omics Analysis Reveals Cultivar-Specific Resistance Mechanisms to Powdery Mildew in Cucurbita pepo
Abstract
1. Introduction
2. Results
2.1. Podosphaera xanthii Inoculation and Disease Progression
2.2. Transcriptomic Analysis and Key Classification Features
2.3. Classification of Gene Expression Data Using Random Forest Models Random Forest Classification of Variables
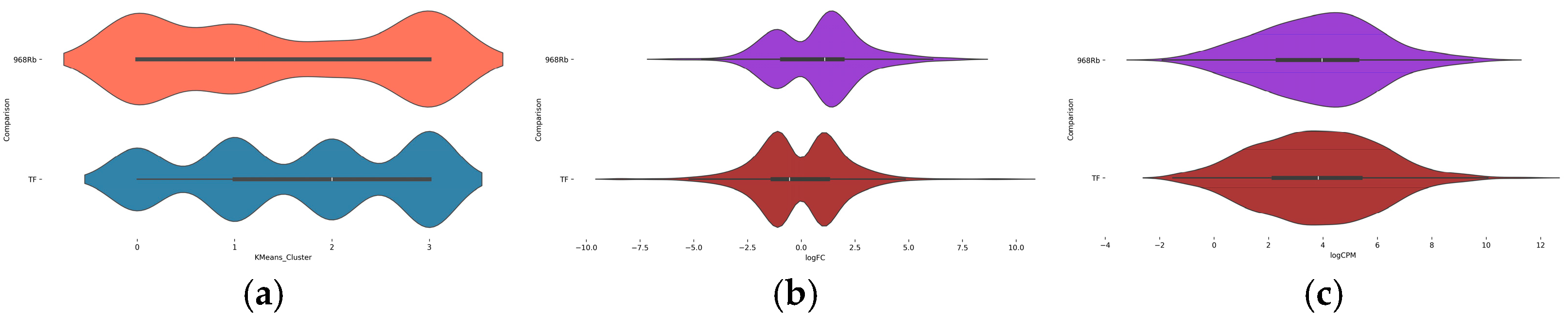
2.4. AI-Based Clustering and Interpretation of DEG Patterns
2.5. Cluster Distribution and Expression Dynamics in Contrasting Genotypes
2.6. Global Functional Interpretation of Transcriptomic Clusters via GPT-4
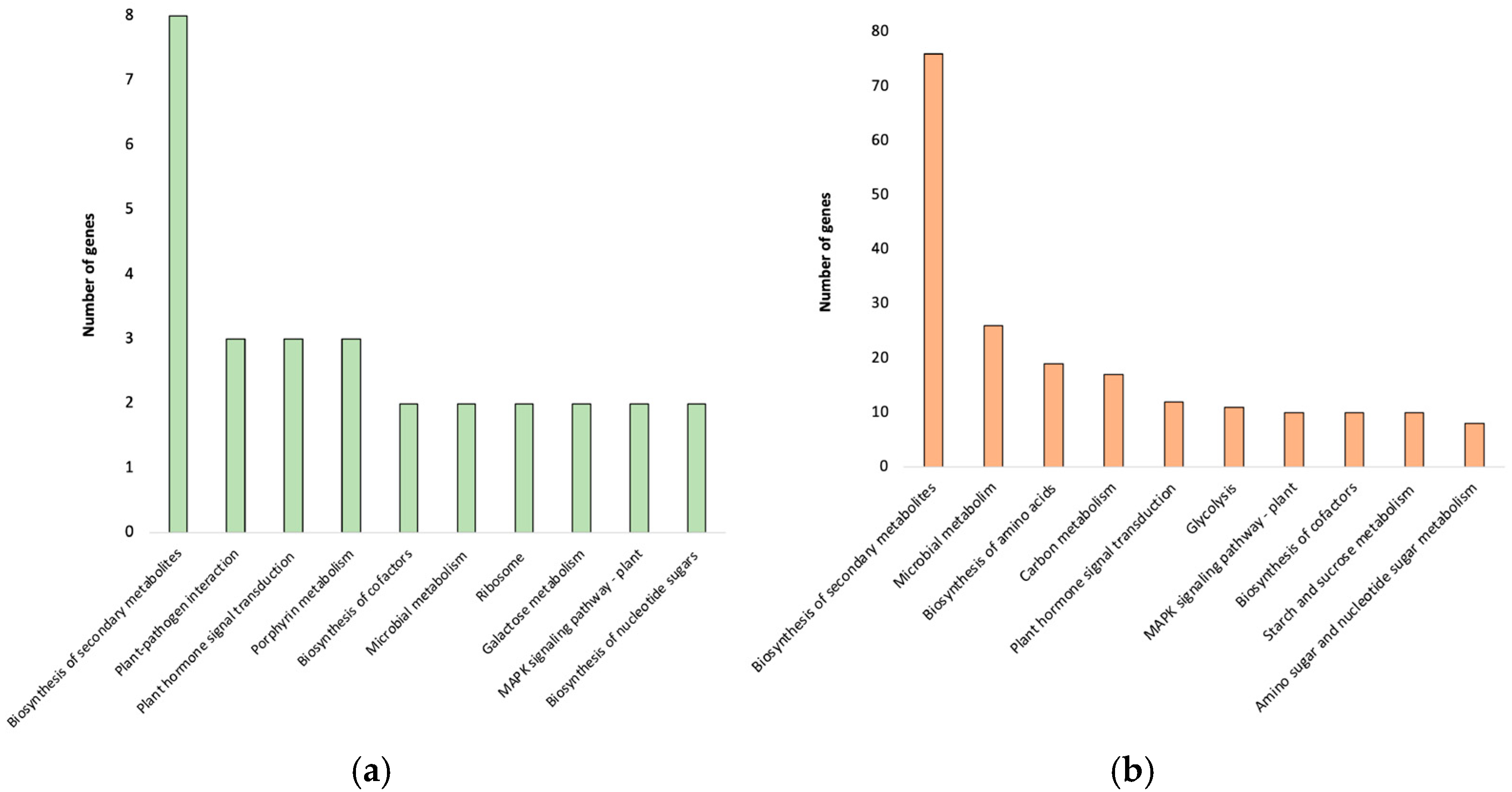
2.7. MapMan Analysis to Refine DEGs Pathway Associations
2.8. Functional Categorization of DEGs Through GO and KEGG Analysis
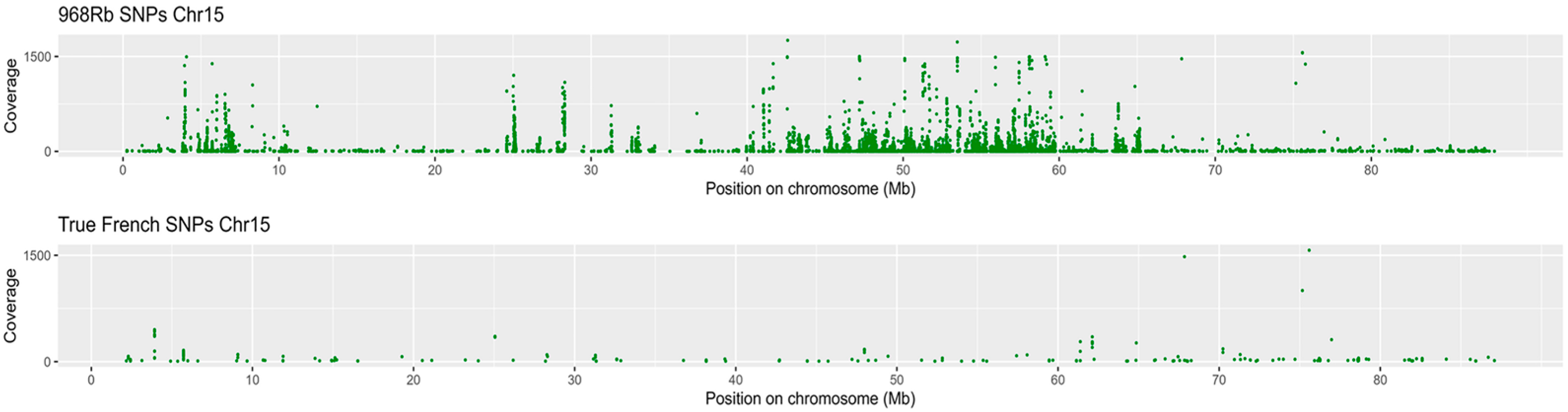
2.9. Genomic Variation Profiling
2.9.1. Detection, Classification, and Functional Relevance of SNPs and InDels
2.9.2. Genetic Variation Within K-Means Expression Clusters
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Podosphaera xanthii Inoculation
4.2. Total RNA Extraction
4.3. Transcriptomic Sequencing, Mapping and Differential Expression Analysis
4.4. Random Forest Classification of Variables
4.5. Clustering Algorithms for DEGs Classification
4.5.1. K-Means
4.5.2. Self-Organizing Map
4.5.3. Agglomerative Clustering
4.6. Conventional Functional and Orthology Annotation
4.7. AI-Based Biological Interpretation via GPT-4
4.8. Variant Calling: SNPs and InDel Investigation
4.9. Genetic Variant Analysis Within DEG Clusters
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abscisic Acid |
| AF | Allele Frequency |
| AI | Artificial Intelligence |
| ARI | Adjusted Rand Index |
| CHI | Calinski–Harabasz Index |
| CSL | Cellulose Synthase-Like |
| DBI | Davies–Bouldin Index |
| DEG(s) | Differentially Expressed Gene(s) |
| DL | Deep Learning |
| dpi | Days Post-Inoculation |
| DP | Read Depth (coverage) |
| ERF | Ethylene Response Factor |
| F | F-value |
| FBA | Fructose-Bisphosphate Aldolase |
| FDR | False Discovery Rate |
| GAE | UDP-D-Glucuronate 4-Epimerase |
| GATA | GATA Transcription Factor |
| GO | Gene Ontology |
| GQ | Genotype Quality |
| JA | Jasmonic Acid |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KO | KEGG Orthology |
| logCPM | Log Counts Per Million |
| logFC | Log Fold Change |
| MAPK | Mitogen-Activated Protein Kinase |
| ML | Machine Learning |
| MQ | Mapping Quality |
| NLR | Nucleotide-binding Leucine-rich Repeat protein |
| PCA | Principal Component Analysis |
| PGIP | Polygalacturonase-Inhibiting Protein |
| PM | Powdery Mildew |
| PPI | Protein–Protein Interaction |
| PR1 | Pathogenesis-Related Protein 1 |
| PRR(s) | Pattern Recognition Receptor(s) |
| PTI | Pattern-Triggered Immunity |
| RF | Random Forest |
| SA | Salicylic Acid |
| SNP(s) | Single Nucleotide Polymorphism(s) |
| SOM | Self-Organizing Map |
| Tre6P | Trehalose-6-Phosphate |
| TF | True French |
| UGE | UDP-D-Glucuronate 4-Epimerase |
| UXS | UDP-Glucuronic Acid Decarboxylase |
| XTH(s) | Xyloglucan Endotransglucosylase/Hydrolase(s) |
References
- FAO. FAOSTAT Database. Food and Agriculture Organization of the United Nations. 2022. Available online: https://www.fao.org/faostat/en/#home (accessed on 24 November 2025).
- Andolfo, G.; Sánchez, C.S.; Cañizares, J.; Pico, M.B.; Ercolano, M.R. Large-scale gene gains and losses molded the NLR defense arsenal during the Cucurbita evolution. Planta 2021, 254, 82. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.-P.; Yang, D.; Yu, H.-Y.; Chen, F.-Y.; Wang, K.-X.; Ding, W.-Q.; Xu, W.-L.; Wang, Y.-L. QTL analysis of early flowering of female flowers in zucchini (Cucurbita pepo L.). J. Integr. Agric. 2022, 22, 3321–3330. [Google Scholar] [CrossRef]
- Xu, X.; Liu, X.; Yan, Y.; Wang, W.; Gebretsadik, K.; Qi, X.; Xu, Q.; Chen, X. Comparative proteomic analysis of cucumber powdery mildew resistance between a single-segment substitution line and its recurrent parent. Hortic. Res. 2019, 6, 115. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Burger, Y.; Katzir, N. Monitoring physiological races of Podosphaera xanthii (syn. Sphaerotheca fuliginea), the causal agent of powdery mildew in cucurbits: Factors affecting race identification and the importance for research and commerce. Phytoparasitica 2004, 32, 174–183. [Google Scholar] [CrossRef]
- Miazzi, M.; Laguardia, C.; Faretra, F. Variation in Podosphaera xanthii on cucurbits in Southern Italy. J. Phytopathol. 2011, 159, 538–545. [Google Scholar] [CrossRef]
- Zhu, Q.; Wu, C.; Amanullah, S.; Liu, S.; Gao, P.; Wang, X.; Ma, H.; Zhu, Z.; Luan, F. Genome-Wide Association Study of Powdery Mildew Resistance in a Worldwide Collection of Melon (Cucumis melo L.) Germplasm. In Cucurbitaceae 2016, XIth Eucarpia Meeting on Cucurbit Genetics & Breeding; Kozik, E.U., Paris, H.S., Eds.; Cucurbitaceae 2016 Organizing Committee: Skierniewice, Poland, 2016; pp. 50–53. [Google Scholar]
- Alavilli, H.; Lee, J.-J.; You, C.-R.; Poli, Y.; Kim, H.-J.; Jain, A.; Song, K. GWAS Reveals a Novel Candidate Gene CmoAP2/ERF in Pumpkin (Cucurbita moschata) Involved in Resistance to Powdery Mildew. Int. J. Mol. Sci. 2022, 23, 6524. [Google Scholar] [CrossRef]
- Zhang, P.; Zhu, Y.; Zhou, S. Comparative analysis of powdery mildew resistant and susceptible cultivated cucumber (Cucumis sativus L.) varieties to reveal the metabolic responses to Sphaerotheca fuliginea infection. BMC Plant Biol. 2021, 21, 24. [Google Scholar] [CrossRef]
- Cao, Y.; Diao, Q.; Lu, S.; Zhang, Y.; Yao, D. Comparative transcriptomic analysis of powdery mildew resistant and susceptible melon inbred lines to identify the genes involved in the response to Podosphaera xanthii infection. Sci. Hortic. 2022, 304, 111305. [Google Scholar] [CrossRef]
- Meng, X.; Yu, Y.; Song, T.; Yu, Y.; Cui, N.; Ma, Z.; Chen, L.; Fan, H. Transcriptome Sequence Analysis of the Defense Responses of Resistant and Susceptible Cucumber Strains to Podosphaera xanthii. Front. Plant Sci. 2022, 13, 872218. [Google Scholar] [CrossRef]
- Lin, E.; Lane, H.-Y. Machine learning and systems genomics approaches for multi-omics data. Biomark. Res. 2017, 5, 2. [Google Scholar] [CrossRef]
- Guo, X.; Han, J.; Song, Y.; Yin, Z.; Liu, S.; Shang, X. Using expression quantitative trait loci data and graph-embedded neural networks to uncover genotype–phenotype interactions. Front. Genet. 2022, 13, 921775. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Jackson, S.A. Machine learning and complex biological data. Genome Biol. 2019, 20, 76. [Google Scholar] [CrossRef] [PubMed]
- Sharieff, N.A.A.; Sameer, N.R. Artificial intelligence Techniques in Bioinformatics: Unravelling complex biological systems. Int. J. Adv. Res. Sci. Commun. Technol. 2023, 3, 269–275. [Google Scholar] [CrossRef]
- Tong, K.; Chen, X.; Yan, S.; Dai, L.; Liao, Y.; Li, Z.; Wang, T. PlantMine: A Machine-Learning Framework to Detect Core SNPs in Rice Genomics. Genes 2024, 15, 603. [Google Scholar] [CrossRef]
- Rezayi, S.; Kalhori, S.R.N.; Saeedi, S. Effectiveness of Artificial intelligence for Personalized Medicine in Neoplasms: A Systematic review. BioMed Res. Int. 2022, 2022, 7842566. [Google Scholar] [CrossRef]
- Singh, D.; Sachdeva, D.; Singh, L. Advancing breast cancer drug delivery: The transformative potential of bioinformatics and artificial intelligence. Curr. Cancer Ther. Rev. 2025, 21, 254–264. [Google Scholar] [CrossRef]
- Vidanagamachchi, S.M.; Waidyarathna, K.M.G.T.R. Opportunities, challenges and future perspectives of using bioinformatics and artificial intelligence techniques on tropical disease identification using omics data. Front. Digit. Health 2024, 6, 1471200. [Google Scholar] [CrossRef]
- Cohen, R.; Hanan, A.; Paris, H. Single-gene resistance to powdery mildew in zucchini squash (Cucurbita pepo). Euphytica 2003, 130, 433–441. [Google Scholar] [CrossRef]
- Luo, J.; Xia, W.; Cao, P.; Xiao, Z.; Zhang, Y.; Liu, M.; Zhan, C.; Wang, N. Integrated Transcriptome Analysis Reveals Plant Hormones Jasmonic Acid and Salicylic Acid Coordinate Growth and Defense Responses upon Fungal Infection in Poplar. Biomolecules 2019, 9, 12. [Google Scholar] [CrossRef]
- Kiani, M.; Szczepaniec, A. Effects of sugarcane aphid herbivory on transcriptional responses of resistant and susceptible sorghum. BMC Genom. 2018, 19, 774. [Google Scholar] [CrossRef]
- Kiani, M.; Bryan, B.; Rush, C.; Szczepaniec, A. Transcriptional responses of resistant and susceptible wheat exposed to wheat curl mite. Int. J. Mol. Sci. 2021, 22, 2703. [Google Scholar] [CrossRef] [PubMed]
- De Mello, U.S.; Vidigal, P.M.P.; Vital, C.E.; Tomaz, A.C.; De Figueiredo, M.; Peternelli, L.A.; Barbosa, M.H.P. An overview of the transcriptional responses of two tolerant and susceptible sugarcane cultivars to borer (Diatraea saccharalis) infestation. Funct. Integr. Genom. 2020, 20, 839–855. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Zhang, X.; Yao, W.; Gao, Y.; Zhao, K.; Guo, Q.; Zhou, B.; Jiang, T. Genome-wide identification and expression analysis of the xyloglucan endotransglucosylase/hydrolase gene family in poplar. BMC Genom. 2021, 22, 804. [Google Scholar] [CrossRef] [PubMed]
- De Caroli, M.; Manno, E.; Piro, G.; Lenucci, M.S. Ride to cell wall: Arabidopsis XTH11, XTH29 and XTH33 exhibit different secretion pathways and responses to heat and drought stress. Plant J. 2021, 107, 448–466. [Google Scholar] [CrossRef]
- Han, J.; Liu, Y.; Shen, Y.; Li, W. A surprising diversity of Xyloglucan Endotransglucosylase/Hydrolase in wheat: New in sight to the roles in drought tolerance. Int. J. Mol. Sci. 2023, 24, 9886. [Google Scholar] [CrossRef]
- Qiao, T.; Zhang, L.; Yu, Y.; Pang, Y.; Tang, X.; Wang, X.; Li, L.; Li, B.; Sun, Q. Identification and expression analysis of xyloglucan endotransglucosylase/hydrolase (XTH) family in grapevine (Vitis vinifera L.). PeerJ 2022, 10, e13546. [Google Scholar] [CrossRef]
- Liu, T.; Shen, C.; Wang, Y.; Huang, C.; Shi, J. New Insights into Regulation of Proteome and Polysaccharide in Cell Wall of Elsholtzia splendens in Response to Copper Stress. PLoS ONE 2014, 9, e109573. [Google Scholar] [CrossRef]
- Sun, Q. Structural variation and polysaccharide profiling of intervessel pit membranes. bioRxiv 2021. [Google Scholar] [CrossRef]
- Chen, L.; Xu, S.; Liu, Y.; Zu, Y.; Zhang, F.; Du, L.; Chen, J.; Li, L.; Wang, K.; Wang, Y.; et al. Identification of key gene networks controlling polysaccharide accumulation in different tissues of Polygonatum cyrtonema Hua by integrating metabolic phenotypes and gene expression profiles. Front. Plant Sci. 2022, 13, 1012231. [Google Scholar] [CrossRef]
- Cheng, S.; Li, R.; Lin, L.; Shi, H.; Liu, X.; Yu, C. Recent Advances in Understanding the Function of the PGIP Gene and the Research of Its Proteins for the Disease Resistance of Plants. Appl. Sci. 2021, 11, 11123. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, X.; Sun, Y.; Wang, P.; Li, X.; Pei, Y.; Li, F.; Hou, Y. Molecular evidence for the involvement of a polygalacturonase-inhibiting protein, GhPGIP1, in enhanced resistance to Verticillium and Fusarium wilts in cotton. Sci. Rep. 2017, 7, 39840. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Lu, L.; Pan, X.; Hu, Z.; Ling, F.; Yan, Y.; Liu, Y.; Lin, Y. Functional analysis of OsPGIP1 in rice sheath blight resistance. Plant Mol. Biol. 2014, 87, 181–191. [Google Scholar] [CrossRef]
- Lu, W.; Tang, X.; Huo, Y.; Xu, R.; Qi, S.; Huang, J.; Zheng, C.; Wu, C.-A. Identification and characterization of fructose 1,6-bisphosphate aldolase genes in Arabidopsis reveal a gene family with diverse responses to abiotic stresses. Gene 2012, 503, 65–74. [Google Scholar] [CrossRef]
- Sun, H.; Dai, H.; Wang, X.; Wang, G. Physiological and proteomic analysis of selenium-mediated tolerance to Cd stress in cucumber (Cucumis Sativus L.). Ecotoxicol. Environ. Saf. 2016, 133, 114–126. [Google Scholar] [CrossRef]
- Uçar, S.; Alım, Ş.; Kasapoğlu, A.G.; Yigider, E.; İlhan, E.; Turan, M.; Polat, A.; Dikbaş, N.; Aydın, M. Genome-Wide Analysis and Characterization of FBA (Fructose 1,6-bisphosphate aldolase) Gene Family of Phaseolus vulgaris L. J. Agric. Prod. 2024, 5, 30–40. [Google Scholar] [CrossRef]
- Shinde, S.; Abida, P.S.; Saakre, M.; Bhaskar, H.; Beena, R.; Preetha, R. Identification and comparative analysis of differential proteins expression in rice under biotic stress by protein sequencing. Cereal Res. Commun. 2024, 52, 1587–1598. [Google Scholar] [CrossRef]
- Mutuku, J.M.; Nose, A. Changes in the Contents of Metabolites and Enzyme Activities in Rice Plants Responding to Rhizoctonia solani Kuhn Infection: Activation of Glycolysis and Connection to Phenylpropanoid Pathway. Plant Cell Physiol. 2012, 53, 1017–1032. [Google Scholar] [CrossRef]
- Liu, J.; Wang, B.; Li, Y.; Huang, L.; Zhang, Q.; Zhu, H.; Wen, Q. RNA sequencing analysis of low temperature and low light intensity-responsive transcriptomes of zucchini (Cucurbita pepo L.). Sci. Hortic. 2020, 265, 109263. [Google Scholar] [CrossRef]
- Chen, B.-H.; Guo, W.-L.; Yang, H.-L.; Li, Q.-F.; Zhou, J.-G.; Li, X.-Z. Photosynthetic properties and biochemical metabolism of Cucurbita moschata genotypes following infection with powdery mildew. J. Plant Pathol. 2020, 102, 1021–1027. [Google Scholar] [CrossRef]
- Hassan, M.U.; Nawaz, M.; Shah, A.N.; Raza, A.; Barbanti, L.; Skalicky, M.; Hashem, M.; Brestic, M.; Pandey, S.; Alamri, S.; et al. Trehalose: A key player in plant growth regulation and tolerance to abiotic stresses. J. Plant Growth Regul. 2022, 42, 4935–4957. [Google Scholar] [CrossRef]
- Kopczewski, T.; Kuźniak, E.; Ciereszko, I.; Kornaś, A. Alterations in Primary Carbon Metabolism in Cucumber Infected with Pseudomonas syringae pv lachrymans: Local and Systemic Responses. Int. J. Mol. Sci. 2022, 23, 12418. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Li, M.; Sun, M.; Jiang, X.; Qiao, F. Plant hormone signals regulate trehalose accumulation against osmotic stress in watermelon cells. Protoplasma 2022, 259, 1351–1369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Cui, H.; Luan, F.; Liu, H.; Ding, Z.; Amanullah, S.; Zhang, M.; Ma, T.; Gao, P. A recessive gene Cmpmr2F confers powdery mildew resistance in melon (Cucumis melo L.). Theor. Appl. Genet. 2023, 136, 4. [Google Scholar] [CrossRef]
- Dun, B.; Zhuo, D.; Shuai, W.; Yane, S.; Yahang, L.; Haonan, C. Allantoin and jasmonic acid synergistically induce resistance response to powdery mildew in melon as revealed by combined hormone and transcriptome analysis. Sci. Hortic. 2023, 327, 112797. [Google Scholar] [CrossRef]
- Li, B.; Meng, X.; Shan, L.; He, P. Transcriptional regulation of Pattern-Triggered immunity in plants. Cell Host Microbe 2016, 19, 641–650. [Google Scholar] [CrossRef]
- Yu, X.; Feng, B.; He, P.; Shan, L. From Chaos to Harmony: Responses and Signaling upon Microbial Pattern Recognition. Annu. Rev. Phytopathol. 2017, 55, 109–137. [Google Scholar] [CrossRef]
- Ma, Q.; Hu, Z.; Mao, Z.; Mei, Y.; Feng, S.; Shi, K. The novel leucine-rich repeat receptor-like kinase MRK1 regulates resistance to multiple stresses in tomato. Hortic. Res. 2022, 9, uhab088. [Google Scholar] [CrossRef]
- Hashemi, L.; Golparvar, A.R.; Nasr-Esfahani, M.; Golabadi, M. Expression analysis of defense-related genes in cucumber (Cucumis sativus L.) against Phytophthora melonis. Mol. Biol. Rep. 2020, 47, 4933–4944. [Google Scholar] [CrossRef]
- Guo, W.-L.; Yang, H.-L.; Zhao, J.-P.; Bian, S.-J.; Guo, Y.-Y.; Chen, X.-J.; Li, X.-Z. A pathogenesis-related protein 1 of Cucurbita moschata responds to powdery mildew infection. Front. Genet. 2023, 14, 1168138. [Google Scholar] [CrossRef]
- Perez-Moro, C.; D’Esposito, D.; Pérez-De-Castro, A.; Ercolano, M.; Capuozzo, C.; Guadagno, A. Comparative analysis of Cucurbita pepo genomes shed light in agronomic traits variation. Res. Sq. 2024; preprint. [Google Scholar] [CrossRef]
- Duan, X.; Yuan, Y.; Real, N.; Tang, M.; Ren, J.; Wei, J.; Liu, B.; Zhang, X. Fine mapping and identification of candidate genes associated with powdery mildew resistance in melon (Cucumis melo L.). Hortic. Res. 2024, 11, uhae222. [Google Scholar] [CrossRef]
- Zhang, K.; Jia, L.; Yang, D.; Hu, Y.; Njogu, M.K.; Wang, P.; Lu, X.; Yan, C. Genome-Wide Identification, Phylogenetic and Expression Pattern Analysis of GATA Family Genes in Cucumber (Cucumis sativus L.). Plants 2021, 10, 1626. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Tang, L.; Li, J. Genome-wide identification of the GATA gene family in melon (Cucumis melo) and analysis of their expression characteristics under biotic and abiotic stresses. Front. Plant Sci. 2024, 15, 1462924. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cruz, J.M.; Polonio, Á.; Ruiz-Jiménez, L.; Vielba-Fernández, A.; Hierrezuelo, J.; Romero, D.; De Vicente, A.; Fernández-Ortuño, D.; Pérez-García, A. Suppression of Chitin-Triggered Immunity by a New Fungal Chitin-Binding Effector Resulting from Alternative Splicing of a Chitin Deacetylase Gene. J. Fungi 2022, 8, 1022. [Google Scholar] [CrossRef] [PubMed]
- Polonio, Á.; Fernández-Ortuño, D.; De Vicente, A.; Pérez-García, A. A haustorial-expressed lytic polysaccharide monooxygenase from the cucurbit powdery mildew pathogen Podosphaera xanthii contributes to the suppression of chitin-triggered immunity. Mol. Plant Pathol. 2021, 22, 580–601. [Google Scholar] [CrossRef]
- Figueroa-Macías, J.P.; García, Y.C.; Núñez, M.; Díaz, K.; Olea, A.F.; Espinoza, L. Plant Growth-Defense Trade-Offs: Molecular processes leading to physiological changes. Int. J. Mol. Sci. 2021, 22, 693. [Google Scholar] [CrossRef]
- Abdelkhalek, A.; Király, L.; Al-Mansori, A.-N.A.; Younes, H.A.; Zeid, A.; Elsharkawy, M.M.; Behiry, S.I. Defense Responses and Metabolic Changes Involving Phenylpropanoid Pathway and PR Genes in Squash (Cucurbita pepo L.) following Cucumber mosaic virus Infection. Plants 2022, 11, 1908. [Google Scholar] [CrossRef]
- Dublino, R.; Ercolano, M. Artificial intelligence redefines agricultural genetics by unlocking the enigma of genomic complexity. Crop. J. 2025, 13, 1350–1362. [Google Scholar] [CrossRef]
- Heidary, M.; Pahlevan Kakhki, M. TRIzol-Based RNA Extraction: A Reliable Method for Gene Expression Studies. J. Sci. Islam. Repub. Iran 2014, 25, 13–17. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 November 2025).
- Zheng, Y.; Wu, S.; Bai, Y.; Sun, H.; Jiao, C.; Guo, S.; Zhao, K.; Blanca, J.; Zhang, Z.; Huang, S.; et al. Cucurbit Genomics Database (CuGenDB): A central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Res. 2018, 47, D1128–D1136. [Google Scholar] [CrossRef]
- Montero-Pau, J.; Blanca, J.; Bombarely, A.; Ziarsolo, P.; Esteras, C.; Martí-Gómez, C.; Ferriol, M.; Gómez, P.; Jamilena, M.; Mueller, L.; et al. De novo assembly of the zucchini genome reveals a whole-genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol. J. 2017, 16, 1161–1171. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Vettigli, G. MiniSom: Minimalistic and Numpy-Based Implementation of the Self Organizing Map [Computer Software]. GitHub. 2018. Available online: https://github.com/JustGlowing/minisom (accessed on 24 November 2025).
- Caswell, T.A.; Droettboom, M.; Lee, A.; Hunter, J.; Firing, E.; Sales De Andrade, E.; Hoffmann, T.; Stansby, D.; Klymak, J.; Varoquaux, N.; et al. Matplotlib/Matplotlib: REL: v3.3.0. 2020. Available online: https://github.com/matplotlib/matplotlib/tree/v3.3.0 (accessed on 24 November 2025).
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 2013, 37, 1250–1258. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wu, S.; Sun, H.; Wang, X.; Tang, X.; Guo, S.; Zhang, Z.; Huang, S.; Xu, Y.; Weng, Y.; et al. CuGenDBv2: An updated database for cucurbit genomics. Nucleic Acids Res. 2022, 51, D1457–D1464. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2019, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2022, 51, D587–D592. [Google Scholar] [CrossRef]
- TAIR. The Arabidopsis Information Resource. 2000. Available online: https://www.arabidopsis.org/aboutarabidopsis.html (accessed on 24 November 2025).
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2022, 51, D638–D646. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Hu, M.; Alkhairy, S.; Lee, I.; Pillich, R.T.; Fong, D.; Smith, K.; Bachelder, R.; Ideker, T.; Pratt, D. Evaluation of large language models for discovery of gene set function. Nat. Methods 2025, 22, 82–91. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Cingolani, P. Variant annotation and Functional Prediction: SNPEFF. In Methods in Molecular Biology; Springer: New York, NY, USA, 2012; pp. 289–314. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed]







| Algorithms | Adjusted Rand Index | Calinski–Harabasz Index | Davies–Bouldin Index |
|---|---|---|---|
| agglomerative clustering | 0.5037 | 1060.4 | 0.9368 |
| k-means | 0.4142 | 1171.0 | 0.8430 |
| self-organizing maps | 0.5491 | 1126.4 | 0.9082 |
| Cluster | Expression Pattern | Cultivar | GPT-4 Title | GPT-4 Biological Interpretation |
|---|---|---|---|---|
| 1 | Highly upregulated | 968Rb | Early immune activation through pattern recognition signaling | Genes involved in early immune perception and signal transduction, activating membrane-bound PRRs, kinases, and WRKY/MYB transcription factors typical of PTI. |
| True French | Transcriptional and hormonal regulation in stress signaling | Genes involved in general stress-responsive regulation, including hormonal crosstalk and transcriptional activation under abiotic and biotic stimuli; less immune-specific. | ||
| 2 | Highly downregulated | 968Rb | Repression of growth- and homeostasis-related pathways | Strong suppression of developmental, transport, and hormonal response genes, indicating metabolic shift prioritizing defense overgrowth and nutrient flow. |
| True French | Oxidative stress response and redox metabolism | Enrichment in redox and detoxification pathways, including ROS scavenging; general basal stress response without activation of immune-specific transcription factors. | ||
| 3 | Moderately upregulated | 968Rb | Cell wall reinforcement and metabolic adjustment | Genes involved in structural defense via cell wall remodeling and protein synthesis; also linked to chloroplast activity and metabolic adaptation. |
| True French | Membrane-linked signaling and transcriptional regulation | General stress-adaptive signaling and transcriptional activation, likely associated with hormonal adjustment and metabolic regulation. | ||
| 4 | Moderately downregulated | 968Rb | Late-stage modulation of immune response | Fine-tuning of immune signaling and transport activity during late immune response phase; includes proteases and hormone modulators. |
| True French | Signal transduction and basal metabolic activity | Metabolic and signaling adjustment under stress, lacking strong immune-specific functional signatures. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dublino, R.; D’Esposito, D.; Guadagno, A.; Capuozzo, C.; Crinò, P.; Formisano, G.; Ercolano, M.R. AI-Integrated Omics Analysis Reveals Cultivar-Specific Resistance Mechanisms to Powdery Mildew in Cucurbita pepo. Int. J. Mol. Sci. 2025, 26, 11488. https://doi.org/10.3390/ijms262311488
Dublino R, D’Esposito D, Guadagno A, Capuozzo C, Crinò P, Formisano G, Ercolano MR. AI-Integrated Omics Analysis Reveals Cultivar-Specific Resistance Mechanisms to Powdery Mildew in Cucurbita pepo. International Journal of Molecular Sciences. 2025; 26(23):11488. https://doi.org/10.3390/ijms262311488
Chicago/Turabian StyleDublino, Rita, Daniela D’Esposito, Anna Guadagno, Claudio Capuozzo, Paola Crinò, Gelsomina Formisano, and Maria Raffaella Ercolano. 2025. "AI-Integrated Omics Analysis Reveals Cultivar-Specific Resistance Mechanisms to Powdery Mildew in Cucurbita pepo" International Journal of Molecular Sciences 26, no. 23: 11488. https://doi.org/10.3390/ijms262311488
APA StyleDublino, R., D’Esposito, D., Guadagno, A., Capuozzo, C., Crinò, P., Formisano, G., & Ercolano, M. R. (2025). AI-Integrated Omics Analysis Reveals Cultivar-Specific Resistance Mechanisms to Powdery Mildew in Cucurbita pepo. International Journal of Molecular Sciences, 26(23), 11488. https://doi.org/10.3390/ijms262311488







