Genome-Wide Association Studies of Growth and Carcass Traits in Charolais Cattle Based on High-Coverage Whole-Genome Resequencing
Abstract
1. Introduction
2. Results
2.1. Phenotypic Statistics Analysis
2.2. WGS, SNPs Distribution and Principal Component Analysis (PCA)
2.3. Genome-Wide Association Study and Candidate Gene Identification
2.4. GO and KEGG Enrichment Analyses of Candidate Genes
2.5. Functional SNPs Identification and PCR-RFLP Assays
2.6. ACOX1 Gene Polymorphism Identification and Trait Association Analysis
3. Discussion
3.1. Candidate Genes Identified for Growth Traits According to the GWAS Results
3.2. Candidate Genes Identified for BFT According to the GWAS Results
3.3. PCR-RFLP for Quick Identification of SNPs
3.4. ACOX1 Polymorphism and Association Analysis
3.5. Application of SNPs and Candidate Genes in Future Genomic Breeding of Beef Cattle
4. Materials and Methods
4.1. Animals and Phenotype Data
4.2. Whole-Genome Resequencing
4.3. Alignment, Variant Identification and PCA
4.4. Genome-Wide Association Study
4.5. Candidate Gene Identification and Functional Enrichment Analysis
4.6. SNP Effect Analysis and Genotyping by PCR-RFLP
4.7. ACOX1-SNP Association Study
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GWAS | genome-wide association study |
| WGS | whole-genome sequencing |
| hcWGS | high-coverage whole-genome sequencing |
| NB | newborn |
| BW | body weight |
| BH | body height |
| CH | cross height |
| BL | body length |
| CM | chest measurement |
| BFT | back fat thickness |
| EM | eye muscle area |
| PCR-RFLP | polymerase chain reaction–restriction fragment length polymorphism |
| SNP | single nucleotide polymorphism |
| ACOX1 | Acyl-CoA oxidase 1 |
References
- de Rezende, M.P.G.; Malhado, C.H.M.; Biffani, S.; Carneiro, P.L.S.; Bozzi, R. Genetic diversity derived from pedigree information and estimation of genetic parameters for reproductive traits of Limousine and Charolais cattle raised in Italy. Ital. J. Anim. Sci. 2020, 19, 762–771. [Google Scholar] [CrossRef]
- Keogh, K.; Carthy, T.R.; McClure, M.C.; Waters, S.M.; Kenny, D.A. Genome-wide association study of economically important traits in Charolais and Limousin beef cows. Animal 2021, 15, 100011. [Google Scholar] [CrossRef] [PubMed]
- Santiago, G.G.; Siqueira, F.; Cardoso, F.F.; Regitano, L.C.A.; Ventura, R.; Sollero, B.P.; Souza, M.D.; Mokry, F.B.; Ferreira, A.B.R.; Torres, R.A.A. Genomewide association study for production and meat quality traits in Canchim beef cattle. J. Anim. Sci. 2017, 95, 3381–3390. [Google Scholar] [CrossRef] [PubMed]
- Jahuey-Martínez, F.J.; Parra-Bracamonte, G.M.; Sifuentes-Rincón, A.M.; Martínez-González, J.C.; Gondro, C.; García-Pérez, C.A.; López-Bustamante, L.A. Genomewide association analysis of growth traits in Charolais beef cattle. J. Anim. Sci. 2016, 94, 4570–4582. [Google Scholar] [CrossRef]
- Braz, C.U.; Rowan, T.N.; Schnabel, R.D.; Decker, J.E. Genome-wide association analyses identify genotype-by-environment interactions of growth traits in Simmental cattle. Sci. Rep. 2021, 11, 13335. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, L.; Wang, Z.; Xu, L.; Chen, Y.; Zhang, L.; Xu, L.; Gao, X.; Gao, H.; Zhu, B.; et al. Genomic prediction and association analysis with models including dominance effects for important traits in Chinese simmental beef cattle. Animals 2019, 9, 1055. [Google Scholar] [CrossRef]
- Pegolo, S.; Cecchinato, A.; Savoia, S.; Di Stasio, L.; Pauciullo, A.; Brugiapaglia, A.; Bittante, G.; Albera, A. Genome-wide association and pathway analysis of carcass and meat quality traits in Piemontese young bulls. Animal 2020, 14, 243–252. [Google Scholar] [CrossRef]
- Smith, J.L.; Wilson, M.L.; Nilson, S.M.; Rowan, T.N.; Schnabel, R.D.; Decker, J.E.; Seabury, C.M. Genome-wide association and genotype by environment interactions for growth traits in U.S. Red Angus cattle. BMC Genom. 2022, 23, 517. [Google Scholar] [CrossRef]
- Bila, L.; Putra, W.P.B.; Malatji, D.P.; Sanarana, Y.P.; Tyasi, T.L. Genome-wide association study (GWAS) for body weights of sussex cattle (Bos taurus) in South Africa. Heliyon 2024, 10, e39540. [Google Scholar] [CrossRef]
- Adhikari, M.; Kantar, M.B.; Longman, R.J.; Lee, C.N.; Oshiro, M.; Caires, K.; He, Y. Genome-wide association study for carcass weight in pasture-finished beef cattle in Hawai'i. Front. Genet. 2023, 14, 1168150. [Google Scholar] [CrossRef]
- Grigoletto, L.; Ferraz, J.B.S.; Oliveira, H.R.; Eler, J.P.; Bussiman, F.O.; Abreu Silva, B.C.; Baldi, F.; Brito, L.F. Genetic Architecture of Carcass and Meat Quality Traits in Montana Tropical® Composite Beef Cattle. Front. Genet. 2020, 11, 123. [Google Scholar] [CrossRef] [PubMed]
- Buzanskas, M.E.; Grossi, D.A.; Ventura, R.V.; Schenkel, F.S.; Sargolzaei, M.; Meirelles, S.L.; Mokry, F.B.; Higa, R.H.; Mudadu, M.A.; da Silva, M.V.; et al. Genome-wide association for growth traits in Canchim beef cattle. PLoS ONE 2014, 9, e94802. [Google Scholar] [CrossRef] [PubMed]
- Snelling, W.M.; Allan, M.F.; Keele, J.W.; Kuehn, L.A.; McDaneld, T.; Smith, T.P.; Sonstegard, T.S.; Thallman, R.M.; Bennett, G.L. Genome-wide association study of growth in crossbred beef cattle. J. Anim. Sci. 2010, 88, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, F.; Mukiibi, R.; Chen, L.; Vinsky, M.; Plastow, G.; Basarab, J.; Stothard, P.; Li, C. Genetic architecture of quantitative traits in beef cattle revealed by genome wide association studies of imputed whole genome sequence variants: II: Carcass merit traits. BMC Genom. 2020, 21, 38. [Google Scholar] [CrossRef]
- Doyle, J.L.; Berry, D.P.; Veerkamp, R.F.; Carthy, T.R.; Walsh, S.W.; Evans, R.D.; Purfield, D.C. Genomic regions associated with skeletal type traits in beef and dairy cattle are common to regions associated with carcass traits, feed intake and calving difficulty. Front. Genet. 2020, 11, 20. [Google Scholar] [CrossRef]
- Vanvanhossou, S.F.U.; Scheper, C.; Dossa, L.H.; Yin, T.; Brügemann, K.; König, S. A multi-breed GWAS for morphometric traits in four Beninese indigenous cattle breeds reveals loci associated with conformation, carcass and adaptive traits. BMC Genom. 2020, 21, 783. [Google Scholar] [CrossRef]
- Martin, P.; Taussat, S.; Vinet, A.; Krauss, D.; Maupetit, D.; Renand, G. Genetic parameters and genome-wide association study regarding feed efficiency and slaughter traits in Charolais cows. J. Anim. Sci. 2019, 97, 3684–3698. [Google Scholar] [CrossRef]
- Song, X.; Yao, Z.; Zhang, Z.; Lyu, S.; Chen, N.; Qi, X.; Liu, X.; Ma, W.; Wang, W.; Lei, C.; et al. Whole-genome sequencing reveals genomic diversity and selection signatures in Xia'nan cattle. BMC Genom. 2024, 25, 559. [Google Scholar] [CrossRef]
- Harish, A.; Lopes Pinto, F.A.; Eriksson, S.; Johansson, A.M. Genetic diversity and recent ancestry based on whole-genome sequencing of endangered Swedish cattle breeds. BMC Genom. 2024, 25, 89. [Google Scholar] [CrossRef]
- Saleh, A.A.; Xue, L.; Zhao, Y. Screening Indels from the whole genome to identify the candidates and their association with economic traits in several goat breeds. Funct. Integr. Genom. 2023, 23, 58. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, W.; Jiang, W.; Yao, J.; Zhang, D. An investigation of obesity susceptibility genes in Northern Han Chinese by targeted resequencing. Medicine 2017, 96, e6117. [Google Scholar] [CrossRef]
- Yu, F.; Zhu, A.C.; Liu, S.; Gao, B.; Wang, Y.; Khudaverdyan, N.; Yu, C.; Wu, Q.; Jiang, Y.; Song, J.; et al. RBM33 is a unique m(6)A RNA-binding protein that regulates ALKBH5 demethylase activity and substrate selectivity. Mol. Cell 2023, 83, 2003–2019.e2006. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, Y.; Han, J.; Yang, Y.; Chen, Y.; Tang, Z.; Gao, F. Longitudinal epitranscriptome profiling reveals the crucial role of N(6)-methyladenosine methylation in porcine prenatal skeletal muscle development. J. Genet. Genomics 2020, 47, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.C.; Jung, Y.H.; Jeong, Y.; Oh, A.R.; Lee, S.B.; Kim, K. Kctd17-mediated Chop degradation promotes adipogenic differentiation. Biochem. Biophys. Res. Commun. 2023, 653, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Skoblov, M.; Marakhonov, A.; Marakasova, E.; Guskova, A.; Chandhoke, V.; Birerdinc, A.; Baranova, A. Protein partners of KCTD proteins provide insights about their functional roles in cell differentiation and vertebrate development. BioEssays 2013, 35, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Rizk, R.; Devost, D. KCTD Proteins Have Redundant Functions in Controlling Cellular Growth. Int. J. Mol. Sci. 2024, 25, 4993. [Google Scholar] [CrossRef]
- Bai, M.; Yin, H.; Zhao, J.; Li, Y.; Wu, Y. miR-182-5p overexpression inhibits chondrogenesis by down-regulating PTHLH. Cell Biol. Int. 2019, 43, 222–232. [Google Scholar] [CrossRef]
- Elli, F.M.; Mattinzoli, D.; Lucca, C.; Piu, M.; Maffini, M.A.; Costanza, J.; Fontana, L.; Santaniello, C.; Forino, C.; Milani, D.; et al. Novel pathogenetic variants in PTHLH and TRPS1 genes causing syndromic brachydactyly. J. Bone Miner. Res. 2022, 37, 465–474. [Google Scholar] [CrossRef]
- Scheffer-Rath, M.E.A.; Veenstra-Knol, H.E.; Boot, A.M. A novel mutation in PTHLH in a family with a variable phenotype with brachydactyly, short stature, oligodontia and developmental delay. Bone Rep. 2023, 19, 101699. [Google Scholar] [CrossRef]
- Goldberg, S.R.; Georgiou, J.; Glogauer, M.; Grynpas, M.D. A 3D scanning confocal imaging method measures pit volume and captures the role of Rac in osteoclast function. Bone 2012, 51, 145–152. [Google Scholar] [CrossRef]
- Gu, Y.; Williams, D.A. RAC2 GTPase deficiency and myeloid cell dysfunction in human and mouse. J. Pediatr. Hematol. Oncol. 2002, 24, 791–794. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cai, S.; Chen, L.; Yuan, R.; Nie, Y.; Ding, S.; Fang, Y.; Zhu, Q.; Chen, K.; Wei, H.; et al. Integrated miRNA-mRNA transcriptomic analysis reveals epigenetic-mediated embryonic muscle growth differences between Wuzhishan and Landrace pigs1. J. Anim. Sci. 2019, 97, 1967–1978. [Google Scholar] [CrossRef] [PubMed]
- Serão, N.V.; Veroneze, R.; Ribeiro, A.M.; Verardo, L.L.; Braccini Neto, J.; Gasparino, E.; Campos, C.F.; Lopes, P.S.; Guimarães, S.E. Candidate gene expression and intramuscular fat content in pigs. J. Anim. Breed. Genet. 2011, 128, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Alradi, M.; Askari, H.; Shaw, M.; Bhavsar, J.D.; Kingham, B.F.; Polson, S.W.; Fancher, I.S. A long-term high-fat diet induces differential gene expression changes in spatially distinct adipose tissue of male mice. Physiol. Genom. 2024, 56, 819–832. [Google Scholar] [CrossRef]
- Xia, J.; Qi, X.; Wu, Y.; Zhu, B.; Xu, L.; Zhang, L.; Gao, X.; Chen, Y.; Li, J.; Gao, H. Genome-wide association study identifies loci and candidate genes for meat quality traits in Simmental beef cattle. Mamm. Genome 2016, 27, 246–255. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, J.; Zhao, F.; Ren, H.; Xu, L.; Lu, J.; Zhang, S.; Zhang, X.; Wei, C.; Lu, G.; et al. Genome-wide association studies for growth and meat production traits in sheep. PLoS ONE 2013, 8, e66569. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Z.; Liu, X.; Chai, Y.; Yang, P.; Li, J.; Huang, Y.; Li, L.; Huang, W.; Yang, G.; et al. Copy number variation of WBP1L gene revealed its association with growth traits across Chinese cattle populations. J. Agr. Sci. 2022, 160, 528–534. [Google Scholar] [CrossRef]
- Jiang, J.; Huang, J.; Gu, J.; Cai, X.; Zhao, H.; Lu, H. Genomic analysis of a spinal muscular atrophy (SMA) discordant family identifies a novel mutation in TLL2, an activator of growth differentiation factor 8 (myostatin): A case report. Case Rep. 2019, 20, 204. [Google Scholar] [CrossRef]
- Shen, C.; Zhou, J.; Wang, X.; Yu, X.Y.; Liang, C.; Liu, B.; Pan, X.; Zhao, Q.; Song, J.L.; Wang, J.; et al. Angiotensin-II-induced muscle wasting is mediated by 25-Hydroxycholesterol via GSK3β signaling pathway. EBioMedicine 2017, 16, 238–250. [Google Scholar] [CrossRef]
- Wang, R.; Li, Y.; Lin, Y.; Chen, D.; Sheng, X.; Zhao, N.; Liu, W. Cloning and expression characteristic analysis of goat ST13 gene. Sheng Wu Gong Cheng Xue Bao 2022, 38, 2959–2973. [Google Scholar] [CrossRef]
- Silva-Vignato, B.; Coutinho, L.L.; Cesar, A.S.M.; Poleti, M.D.; Regitano, L.C.A.; Balieiro, J.C.C. Comparative muscle transcriptome associated with carcass traits of Nellore cattle. BMC Genom. 2017, 18, 506. [Google Scholar] [CrossRef]
- Pan, Y.; Sun, G.; Li, G.; Chen, S.; Liu, H.; Li, H.; Mei, C.; Yang, W.; Zan, L. Sex-specific microbiota associations with backfat thickness, eye muscle area, and rumen fermentation in Qinchuan cattle. BMC Microbiology 2025, 25, 277. [Google Scholar] [CrossRef]
- Tasdelen, I.; Berger, R.; Kalkhoven, E. PPARγ regulates expression of carbohydrate sulfotransferase 11 (CHST11/C4ST1), a regulator of LPL cell surface binding. PLoS ONE 2013, 8, e64284. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Peng, Y.J.; Lin, Y.Y. LRRK2 Regulates CPT1A to Promote β-Oxidation in HepG2 Cells. Molecules 2020, 25, 4122. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Arshad, M.; Wang, W.; Zhao, D.; Xu, L.; Zhou, L. LRRK2 mediated Rab8a phosphorylation promotes lipid storage. Lipids Health Dis. 2018, 17, 34. [Google Scholar] [CrossRef] [PubMed]
- Messling, J.E.; Agger, K.; Andersen, K.L. Targeting RIOK2 ATPase activity leads to decreased protein synthesis and cell death in acute myeloid leukemia. Blood 2022, 139, 245–255. [Google Scholar] [CrossRef]
- Gao, Z.; Xu, C.; Fan, H. Analysis of RIOK2 Functions in Mediating the Toxic Effects of Deoxynivalenol in Porcine Intestinal Epithelial Cells. Int. J. Mol. Sci. 2022, 23, 12712. [Google Scholar] [CrossRef]
- Hebbar, P.; Abubaker, J.A.; Abu-Farha, M.; Alsmadi, O.; Elkum, N.; Alkayal, F.; John, S.E.; Channanath, A.; Iqbal, R.; Pitkaniemi, J.; et al. Genome-wide landscape establishes novel association signals for metabolic traits in the Arab population. Hum. Genet. 2021, 140, 505–528. [Google Scholar] [CrossRef]
- Nagdy, H.; Mahmoud, K.G.M.; Kandiel, M.M.M.; Helmy, N.A.; Ibrahim, S.S.; Nawito, M.F.; Othman, O.E. PCR-RFLP of bone morphogenetic protein 15 (BMP15/FecX) gene as a candidate for prolificacy in sheep. Int. J. Vet. Sci. Med. 2018, 6, S68–S72. [Google Scholar] [CrossRef]
- Paek, H.J.; Li, Z.Y.; Quan, B.H.; Yin, X.J. Application of PCR-RFLP for quick identification of MSTN mutants in MSTN mutant pig breeding. Anim. Biotechnol. 2023, 34, 2231–2239. [Google Scholar] [CrossRef]
- Parasar, P.; Bhushan, B. Characterization of BoLA class II DQA and DQB by PCR-RFLP, cloning, and sequencing reveals sequence diversity in crossbred cattle. Anim. Biotechnol. 2023, 34, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Sundaramahalingam, M.A.; Amrutha, C.; Rajeshbanu, J.; Thirukumaran, K.; Manibalan, S.; Ashokkumar, M.; Sivashanmugam, P. In silico approach for enhancing innate lipid content of Yarrowia lipolytica, by blocking the acyl-CoA oxidase-1 enzyme, using various analogous compounds of lipids. J. Biomol. Struct. Dyn. 2023, 41, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Xiong, Q.; Tao, H.; Liu, Y.; Zhang, N.; Li, X.F.; Suo, X.J.; Yang, Q.P.; Chen, M.X. ACOX1, regulated by C/EBPα and miR-25-3p, promotes bovine preadipocyte adipogenesis. J. Mol. Endocrinol. 2021, 66, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Zan, L.S.; Liu, Y.F.; Wang, H.B. Molecular characterization, polymorphism of the ACOX1 gene and association with ultrasound traits in Bos taurus. Genet. Mol. Res. 2011, 10, 1948–1957. [Google Scholar] [CrossRef]
- Lee, J.K.; Cho, Y.M.; Lee, J.H. Association of Bovine CSRP3 and ACOX1 Genes with Carcass and Meat Quality Traits. Korean J. Agr. Sci. 2010, 37, 231–238. [Google Scholar]
- Dominguez-Castaño, P.; Marchi Maiorano, A.; Silva Lopes, J.E.; Vargas de Oliveira, M.H.; Michel Castilhos, A.; Vasconcelos Silva, J.A.I. Genetic parameters for mouth size and their influence on growth traits in pasture-raised Nelore cattle. J. Anim. Sci. 2023, 101, skad150. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
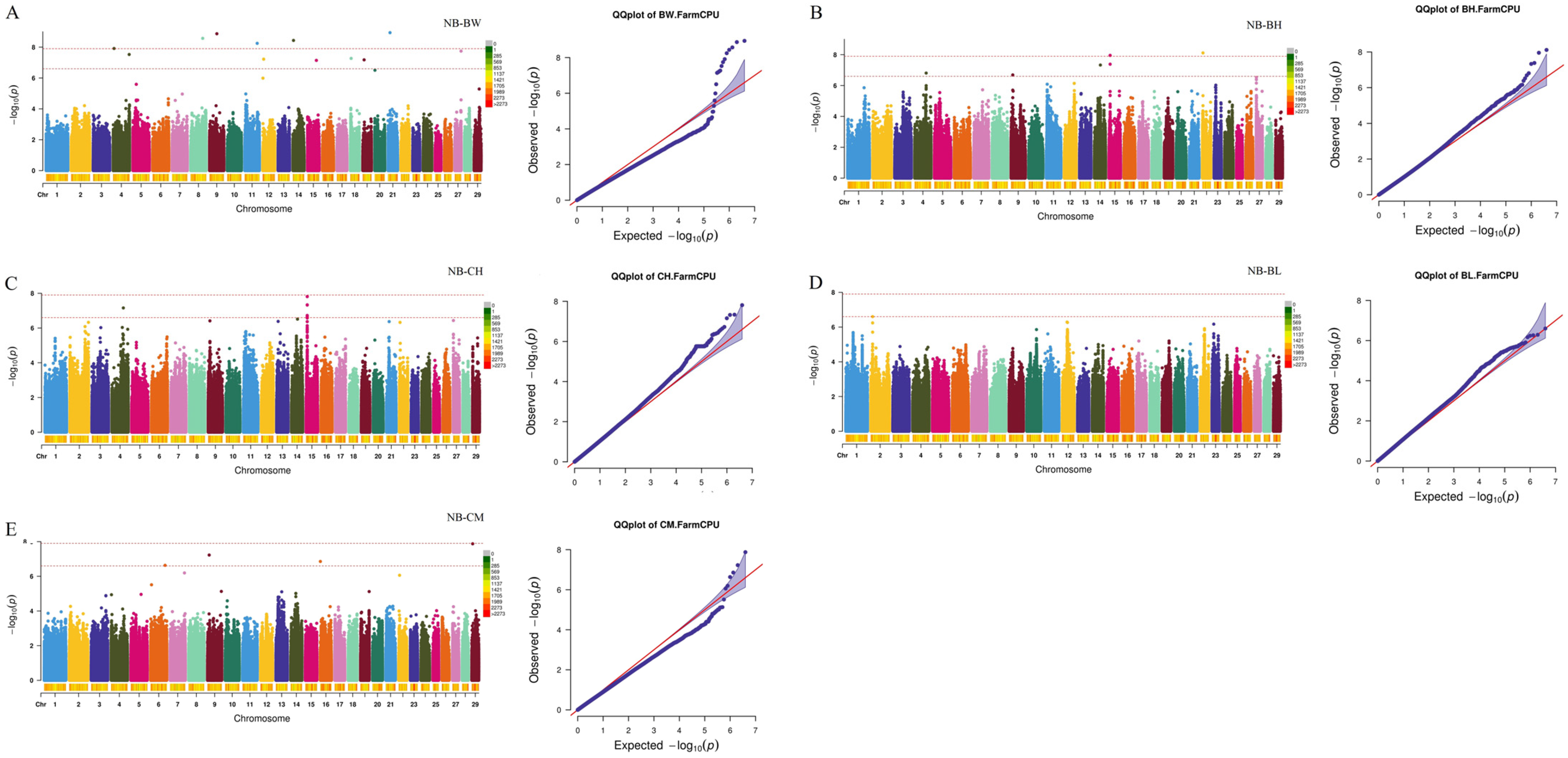
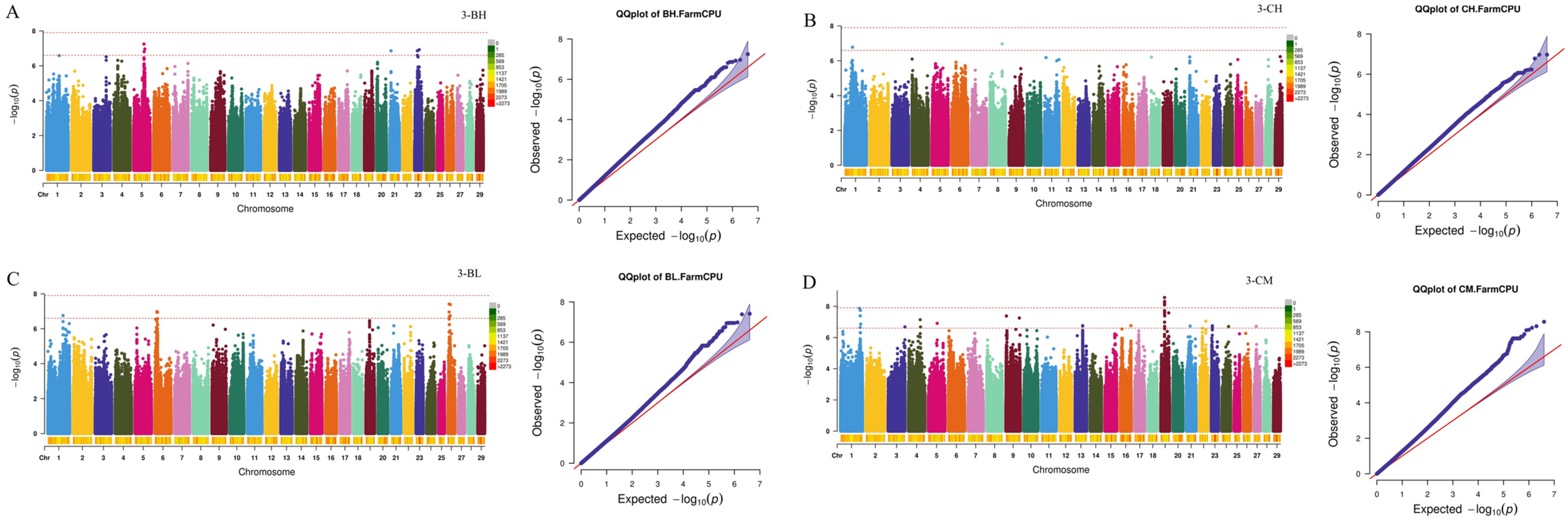


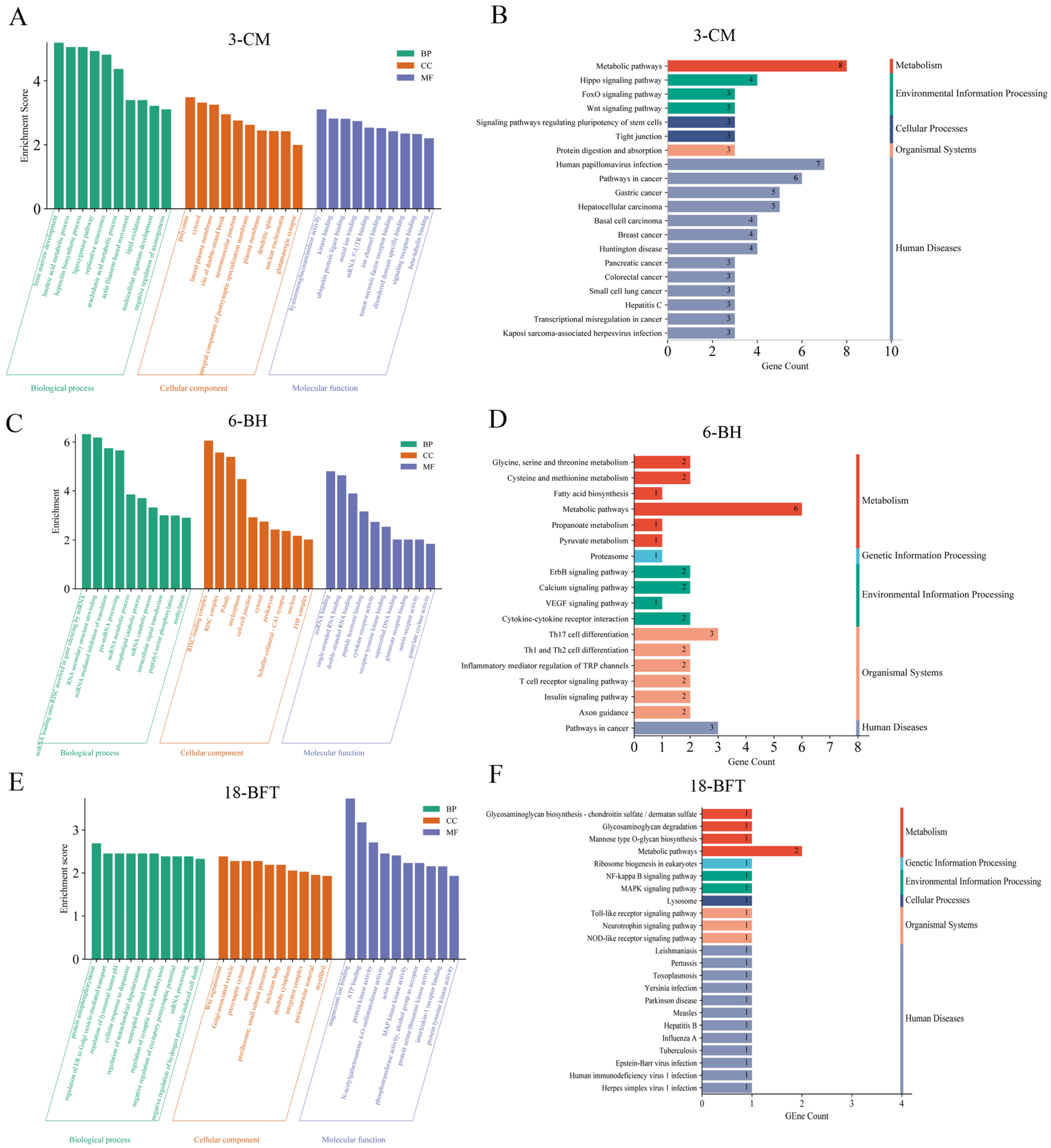

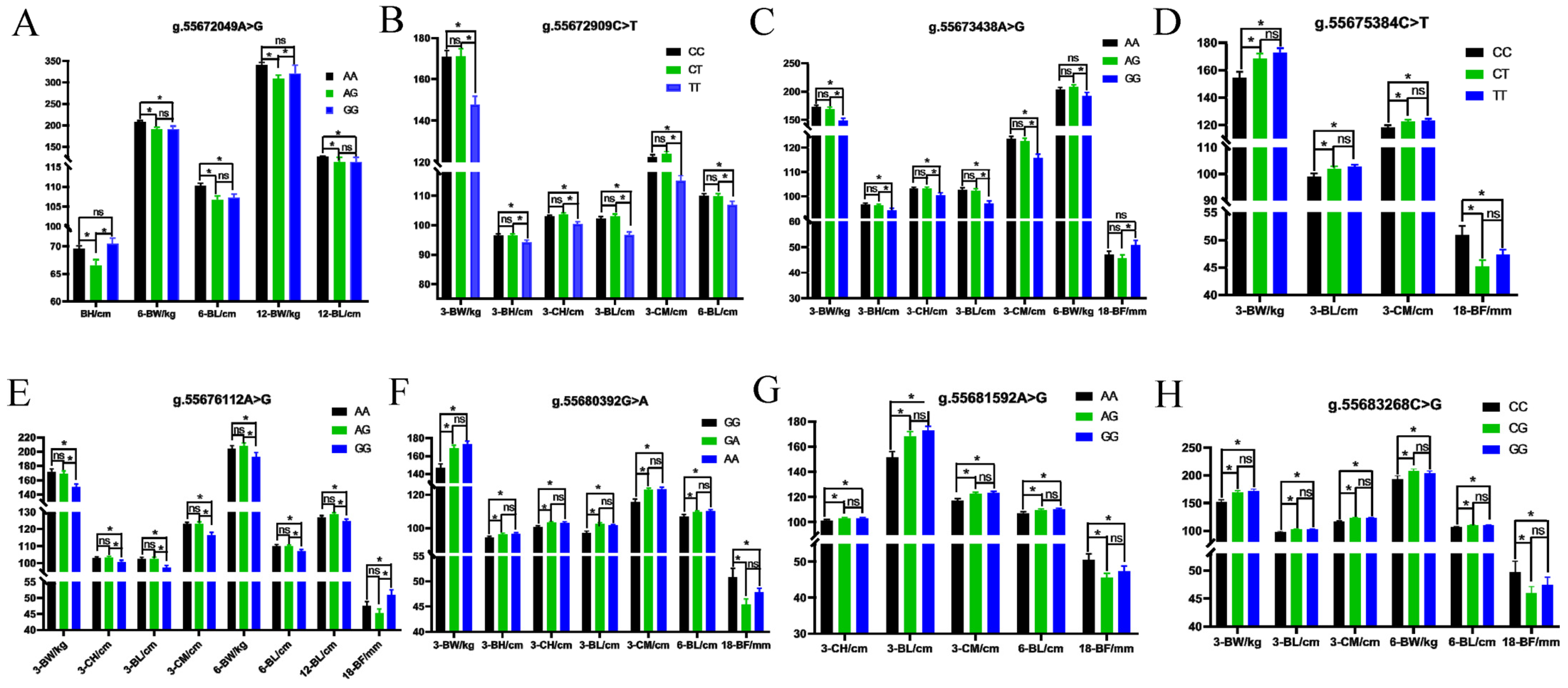
| Traits a | Individuals | Mean ± SD b | Maximum | Minimum | CV c |
|---|---|---|---|---|---|
| NB-BW | 142 | 39.99 ± 7.77 | 57.00 | 15.00 | 0.19 |
| NB-BH | 142 | 68.90 ± 5.49 | 80.00 | 50.00 | 0.08 |
| NB-CH | 142 | 74.46 ± 5.88 | 87.00 | 54.00 | 0.08 |
| NB-BL | 142 | 64.25 ± 6.01 | 78.00 | 50.00 | 0.09 |
| NB-CM | 142 | 73.94 ± 6.06 | 87.00 | 55.00 | 0.08 |
| 3-BW | 139 | 166.92 ± 26.59 | 254.00 | 96.00 | 0.16 |
| 3-BH | 139 | 96.03 ± 4.30 | 105.00 | 84.00 | 0.04 |
| 3-CH | 139 | 102.71 ± 4.55 | 115.00 | 86.00 | 0.04 |
| 3-BL | 139 | 101.45 ± 6.72 | 115.00 | 80.00 | 0.06 |
| 3-CM | 139 | 121.81 ± 9.24 | 144.00 | 92.00 | 0.07 |
| 6-BW | 135 | 203.50 ± 29.17 | 287.00 | 126.0 | 0.14 |
| 6-BH | 135 | 100.71 ± 4.10 | 110.00 | 90.00 | 0.04 |
| 6-CH | 135 | 108.70 ± 5.94 | 153.00 | 96.00 | 0.05 |
| 6-BL | 135 | 109.34 ± 5.89 | 133.00 | 95.00 | 0.05 |
| 6-CM | 135 | 131.98 ± 6.51 | 152.00 | 110.00 | 0.04 |
| 12-BW | 115 | 336.92 ± 53.49 | 463.00 | 185.00 | 0.16 |
| 12-BH | 115 | 114.10 ± 4.22 | 123.00 | 103.00 | 0.03 |
| 12-CH | 115 | 121.89 ± 4.79 | 132.00 | 111.00 | 0.03 |
| 12-BL | 115 | 127.00 ± 6.55 | 143.00 | 110.00 | 0.05 |
| 12-CM | 115 | 158.13 ± 9.33 | 184.00 | 129.00 | 0.06 |
| 18-BW | 100 | 457.25 ± 75.90 | 654.00 | 280.00 | 0.16 |
| 18-BH | 100 | 123.27 ± 4.90 | 141.00 | 112.00 | 0.03 |
| 18-CH | 100 | 132.40 ± 4.59 | 143.00 | 121.00 | 4.59 |
| 18-BL | 100 | 141.53 ± 7.25 | 159.00 | 121.00 | 0.05 |
| 18-CM | 100 | 177.67 ± 10.59 | 212.00 | 146.00 | 0.06 |
| 18-BFT | 190 | 50.72 ± 10.91 | 84.90 | 2.70 | 0.21 |
| 18-EM | 190 | 76.24 ± 10.23 | 106.70 | 50.70 | 0.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Wang, C.; Shangguan, A.; Suo, X.; Chen, M.; Tao, H.; Jiang, F.; Xu, T.; Zhang, N.; Hua, Z.; et al. Genome-Wide Association Studies of Growth and Carcass Traits in Charolais Cattle Based on High-Coverage Whole-Genome Resequencing. Int. J. Mol. Sci. 2025, 26, 11411. https://doi.org/10.3390/ijms262311411
Zhang F, Wang C, Shangguan A, Suo X, Chen M, Tao H, Jiang F, Xu T, Zhang N, Hua Z, et al. Genome-Wide Association Studies of Growth and Carcass Traits in Charolais Cattle Based on High-Coverage Whole-Genome Resequencing. International Journal of Molecular Sciences. 2025; 26(23):11411. https://doi.org/10.3390/ijms262311411
Chicago/Turabian StyleZhang, Feng, Chengmei Wang, Aishao Shangguan, Xiaojun Suo, Mengjie Chen, Hu Tao, Fan Jiang, Tian Xu, Nian Zhang, Zaidong Hua, and et al. 2025. "Genome-Wide Association Studies of Growth and Carcass Traits in Charolais Cattle Based on High-Coverage Whole-Genome Resequencing" International Journal of Molecular Sciences 26, no. 23: 11411. https://doi.org/10.3390/ijms262311411
APA StyleZhang, F., Wang, C., Shangguan, A., Suo, X., Chen, M., Tao, H., Jiang, F., Xu, T., Zhang, N., Hua, Z., Chai, J., & Xiong, Q. (2025). Genome-Wide Association Studies of Growth and Carcass Traits in Charolais Cattle Based on High-Coverage Whole-Genome Resequencing. International Journal of Molecular Sciences, 26(23), 11411. https://doi.org/10.3390/ijms262311411






