Structure-Guided Design of Cyclic Peptide: A Potent Inhibitor Targeting PD-1/PD-L1 Axis with Antitumor Activity
Abstract
1. Introduction
2. Results
2.1. Identification of Hotspot Residues at the PD-1/PD-L1 Interface
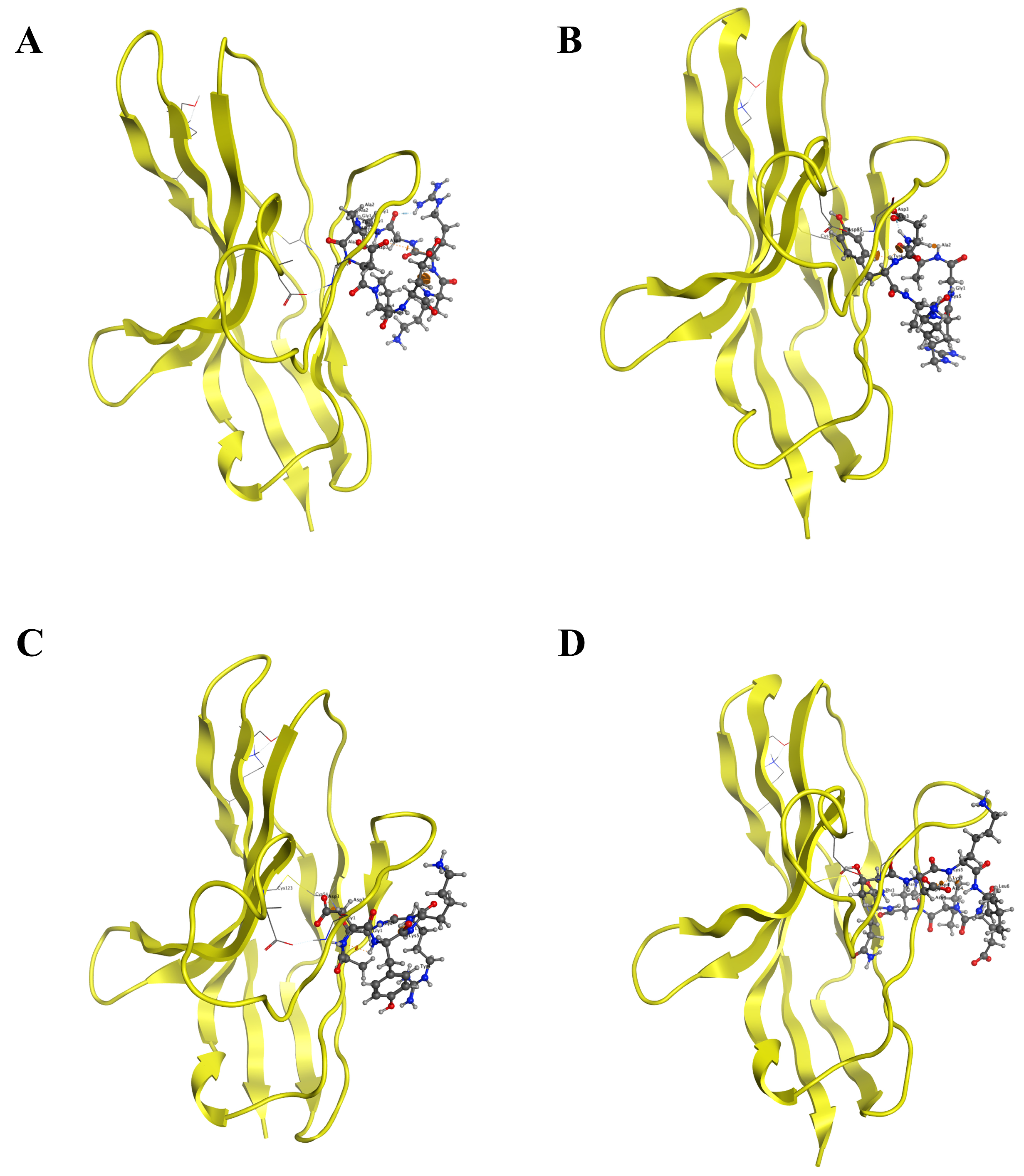
2.2. Structure-Based Design and Virtual Screening of Cyclic Peptides
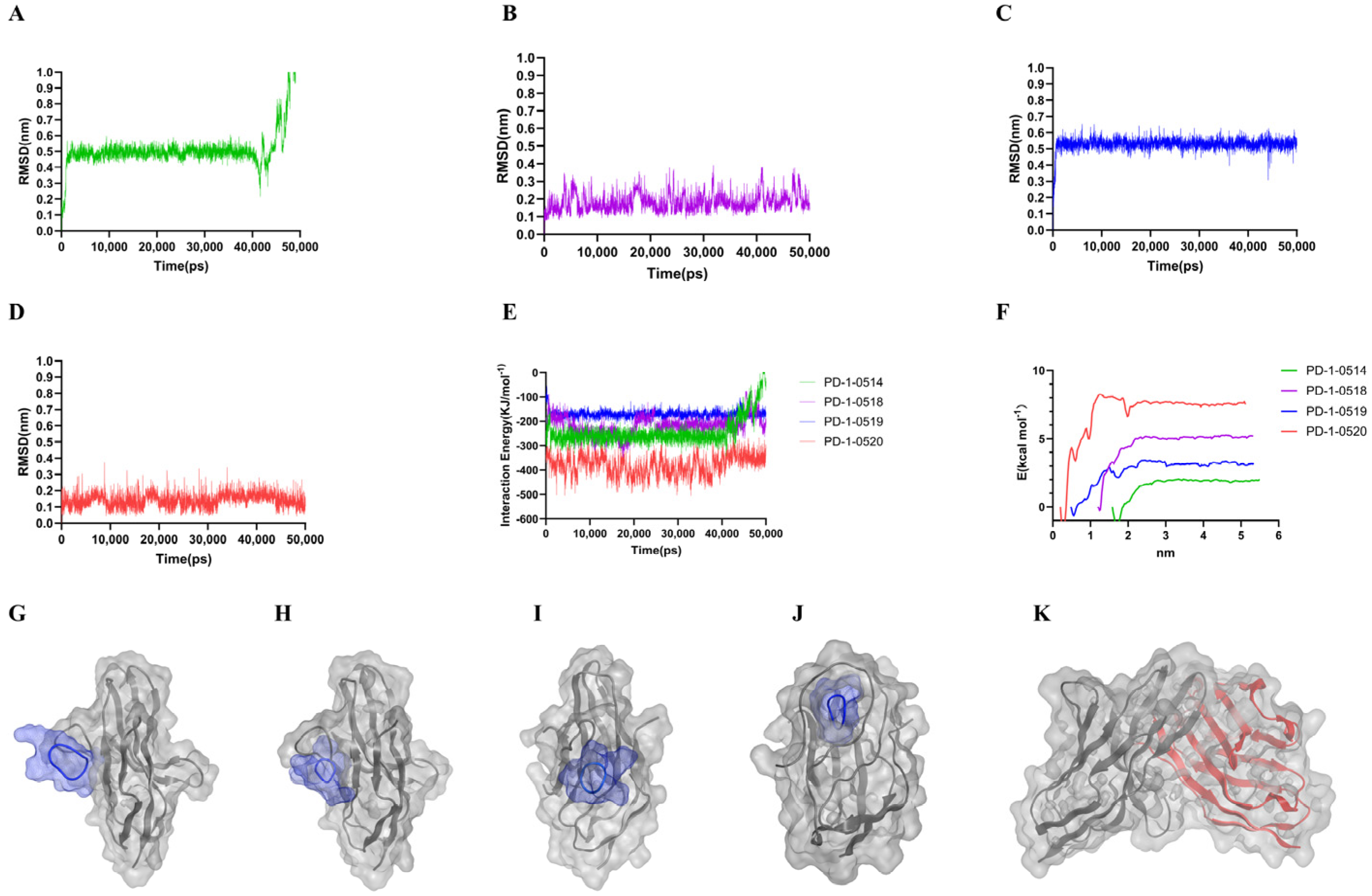
2.3. Molecular Dynamics and Free Energy Analysis
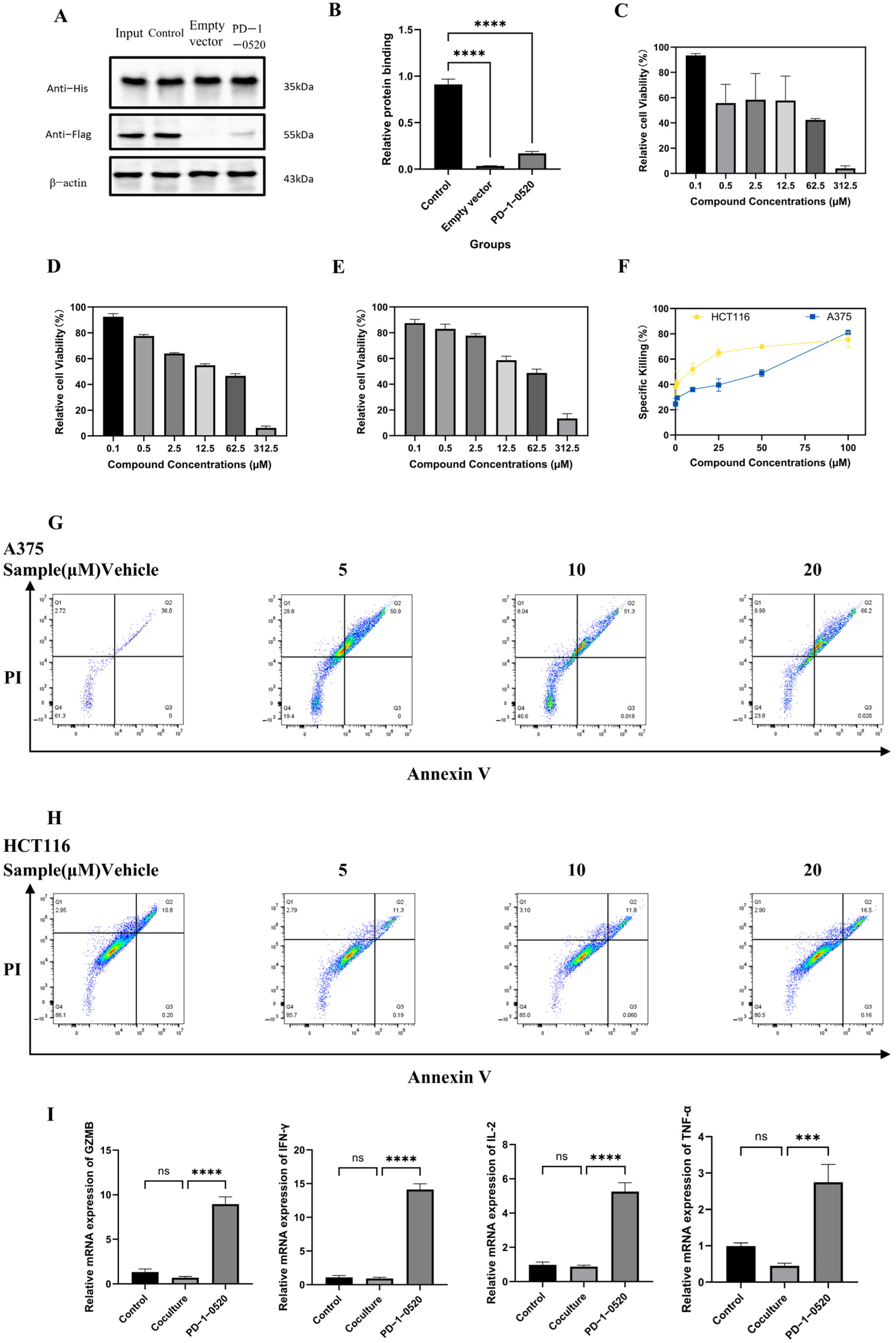
2.4. PD-1-0520 Blocks PD-1/PD-L1 Interaction In Vitro
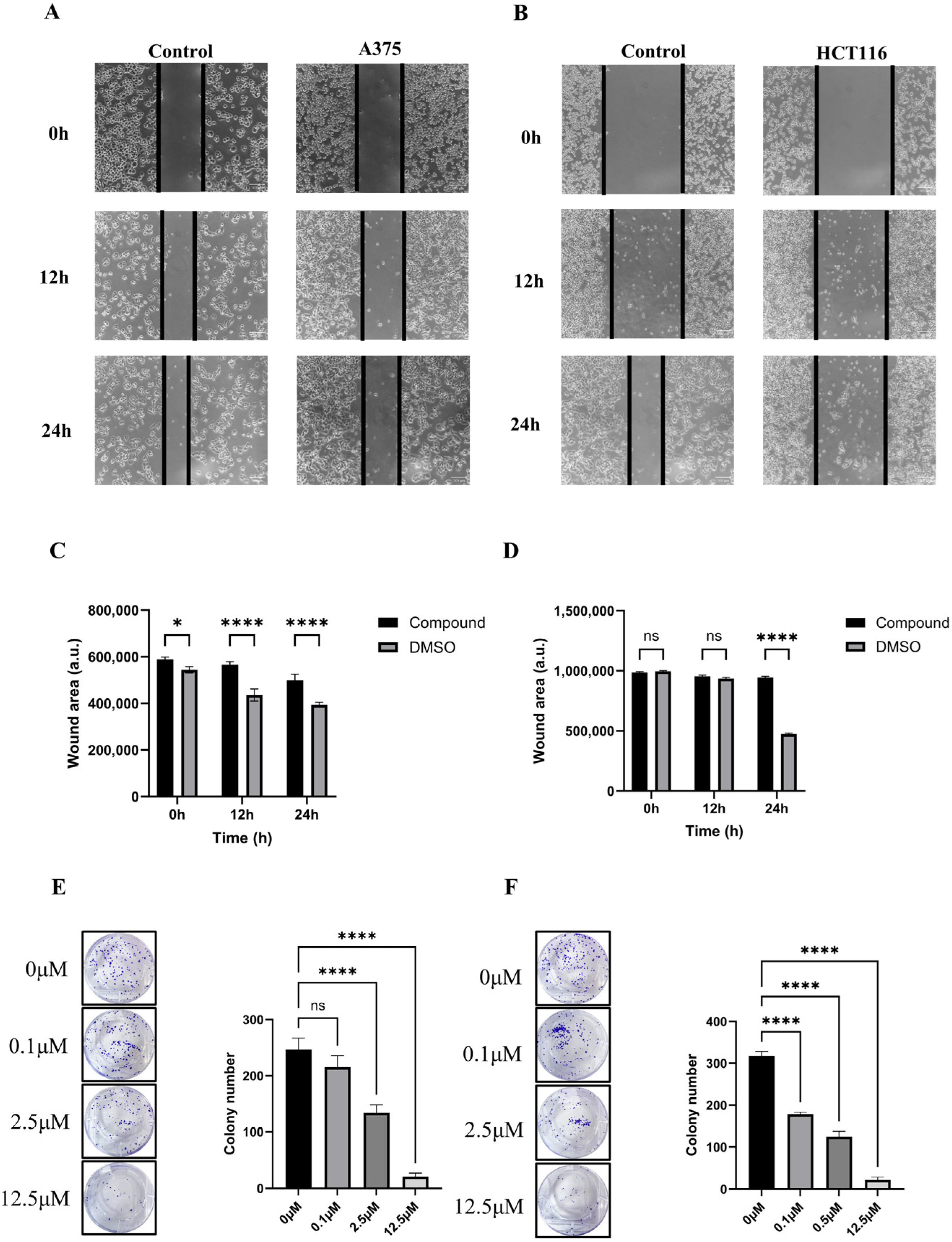
2.5. In Vitro Antitumor Activity of PD-1-0520
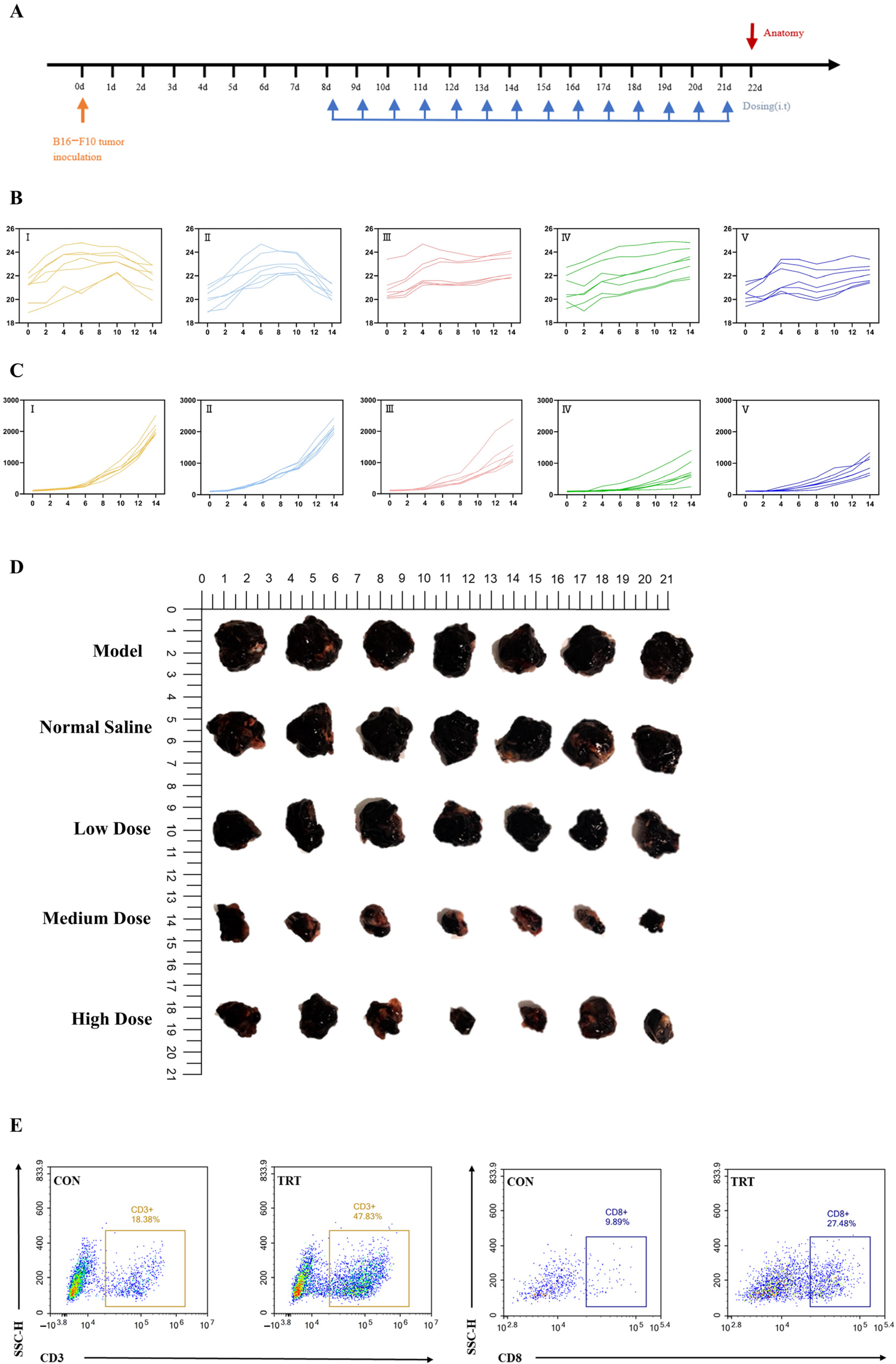
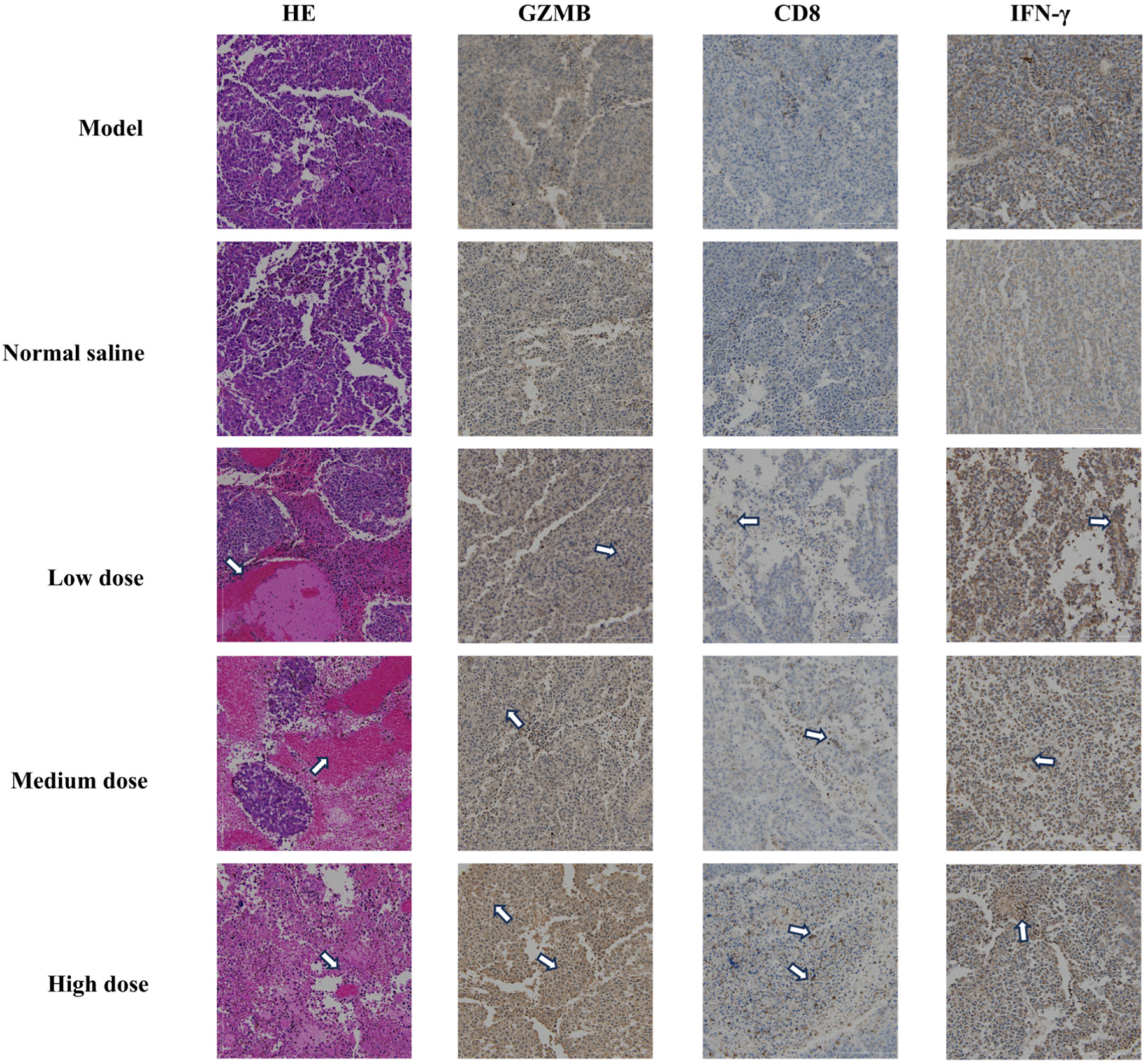
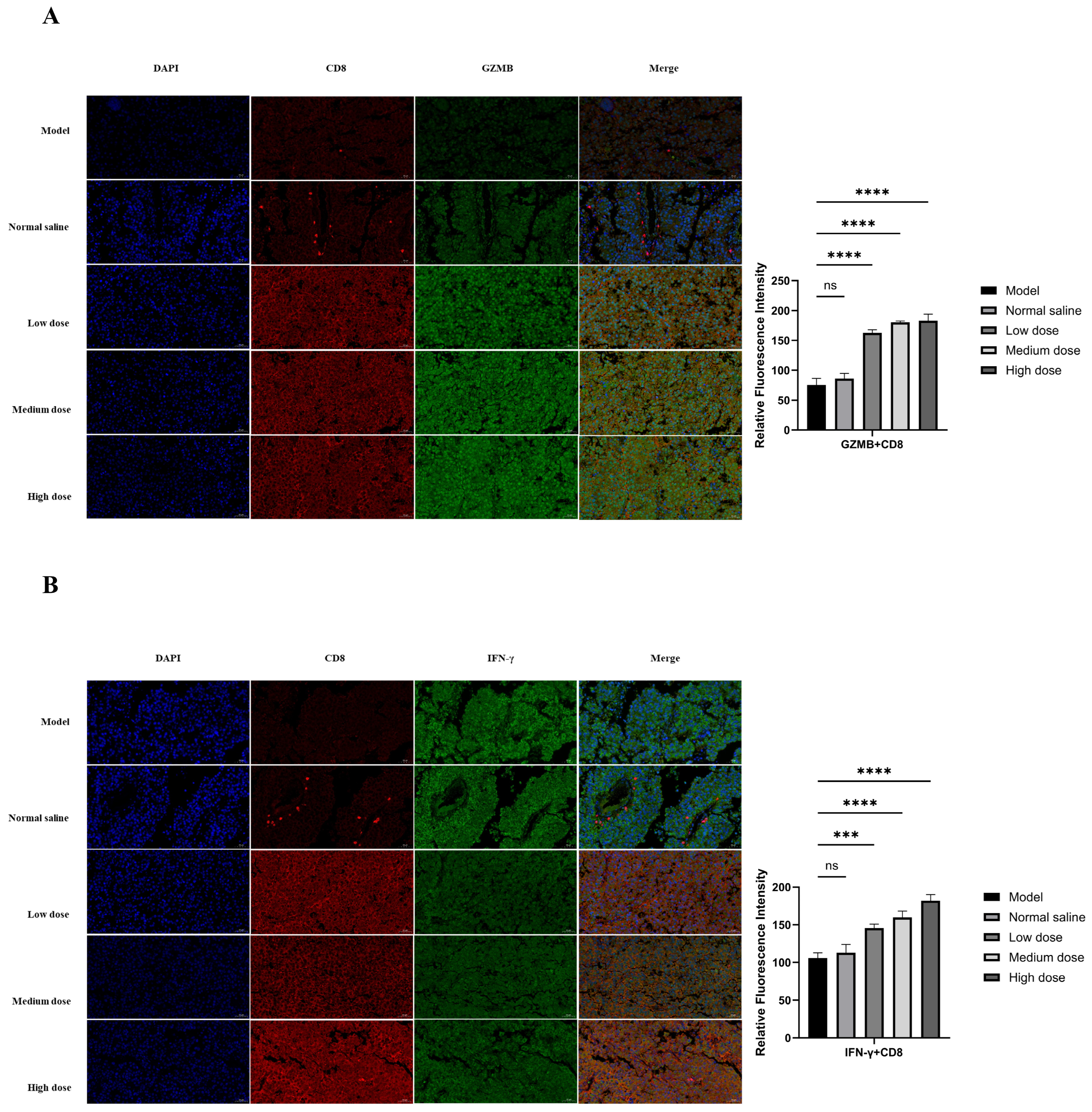
2.6. In Vivo Antitumor Activity and Immune Activation
3. Discussion
4. Materials and Methods
4.1. Experimental Materials
4.2. Molecular Docking-Based Hotspot Residue Prediction
4.3. Cyclic Peptide Design Strategy
4.4. Molecular Dynamics Simulations and Binding Free Energy Calculations
4.5. Protein Interaction Blocking Assay (Pull-Down Assay)
4.6. Cell Viability Assay
4.7. T-Cell-Mediated Tumor Cell Killing Assay
4.8. Detection of Apoptosis by Flow Cytometry
4.9. Cell Scratch Assay
4.10. Cell Clone Formation Assay
4.11. Real-Time PCR to Detect Cytokine Expression
4.12. Animal Experiments and Tissue Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PPI | protein–protein interaction |
| PD-1 | programmed death receptor-1 |
| PD-L1 | PD-1 ligand 1 |
| mAbs | monoclonal Antibodies |
| MD | Molecular dynamics |
| RMSD | root-mean-square deviation |
| MM-PBSA | molecular mechanism/Poisson–Boltzmann surface area |
| IC50 | Half-maximal inhibitory concentration |
| IFN-γ | interferon γ |
| IL-2 | interleukin 2 |
| TNF-α | Tumor Necrosis Factor-α |
| GZMB | granzyme B |
| IHC | immunohistochemistry |
| ANOVA | analysis of variance |
References
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2015, 348, 56–61. [Google Scholar] [CrossRef]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, Activity, and Immune Correlates of Anti-PD-1 Antibody in Cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Beck, A.; Wurch, T.; Bailly, C.; Corvaia, N. Strategies and challenges for the next generation of therapeutic antibodies. Nat. Rev. Immunol. 2010, 10, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Postow, M.A.; Sidlow, R.; Hellmann, M.D. Immune-Related Adverse Events Associated with Immune Checkpoint Blockade. N. Engl. J. Med. 2018, 378, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Craik, D.J.; Fairlie, D.P.; Liras, S.; Price, D. The Future of Peptide-based Drugs. Chem. Biol. Drug Des. 2013, 81, 136–147. [Google Scholar] [CrossRef]
- Craik, D.J.; Du, J.Q. Cyclotides as drug design scaffolds. Curr. Opin. Chem. Biol. 2017, 38, 8–16. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Kim, S.-H.; Decker, C.G.; Wong, D.Y.; Loo, J.A.; Maynard, H.D. A heparin-mimicking polymer conjugate stabilizes basic fibroblast growth factor. Nat. Chem. 2013, 5, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, Y.; Guo, Y.; Pan, Z.; Zhong, S.; Jin, X.; Zhuang, W.; Chen, S.; Gao, J.; Huang, W.; et al. Discovery of phenyl-linked symmetric small molecules as inhibitors of the programmed cell death-1/programmed cell death-ligand 1 interaction. Eur. J. Med. Chem. 2021, 223, 113637. [Google Scholar] [CrossRef]
- Lin, D.Y.-W.; Tanaka, Y.; Iwasaki, M.; Gittis, A.G.; Su, H.-P.; Mikami, B.; Okazaki, T.; Honjo, T.; Minato, N.; Garboczi, D.N. The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc. Natl. Acad. Sci. USA 2008, 105, 3011–3016. [Google Scholar] [CrossRef]
- Weber, J.S. Immuno-oncology Comes of Age-Introduction. Semin. Oncol. 2014, 41, S1–S2. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Wang, Y.; Yao, Z.; Gao, X.; Liu, C.; Wang, X.; Wu, H.; Ding, X.; Hu, J.; Lin, B.; et al. Enhanced Safety and Antitumor Efficacy of Switchable Dual Chimeric Antigen Receptor-Engineered T Cells against Solid Tumors through a Synthetic Bifunctional PD-L1-Blocking Peptide. J. Am. Chem. Soc. 2020, 142, 18874–18885. [Google Scholar] [CrossRef] [PubMed]
- Rui, M.; Zhang, W.; Mi, K.; Ni, H.; Ji, W.; Yu, X.; Qin, J.; Feng, C. Design and evaluation of α-helix-based peptide inhibitors for blocking PD-1/PD-L1 interaction. Int. J. Biol. Macromol. 2023, 253, 16. [Google Scholar] [CrossRef]
- Li, Y.; Ren, Q.; Wang, X.; Luoreng, Z.; Wei, D. Bta-miR-199a-3p Inhibits LPS-Induced Inflammation in Bovine Mammary Epithelial Cells via the PI3K/AKT/NF-κB Signaling Pathway. Cells 2022, 11, 3518. [Google Scholar] [CrossRef]
- Sullivan, C.F.; Davari, A.; Kim, J.S.; Parker, B.L.; Skinner, M. Evaluation of a guardian plant system to suppress Frankliniella occidentalis (Thysanoptera: Thripidae) in greenhouse ornamentals. Pest Manag. Sci. 2023, 79, 3559–3569. [Google Scholar] [CrossRef]
- Li, X.; Tian, L.; Li, H.; Cai, W. Ultrastructural Variations of Antennae and Labia Are Associated with Feeding Habit Shifts in Stink Bugs (Heteroptera: Pentatomidae). Biology 2021, 10, 1161. [Google Scholar] [CrossRef]
- Le, H.T.T.; Hioki, Y.; Danova, A.; Nguyen, V.; Duong, T.H.; Kita, M.; Chavasiri, W. α-Glucosidase inhibition of sesquiterpenoids from the heartwood of Mansonia gagei. Phytochemistry 2023, 213, 8. [Google Scholar] [CrossRef]
- Wang, X.Y.; Cui, L.D.; Wang, Y.C.; Zeng, Z.M.; Wang, H.J.; Tian, L.Y.; Guo, J.; Chen, Y.C. Mechanistic investigation of wogonin in delaying the progression of endothelial mesenchymal transition by targeting the TGF-β1 pathway in pulmonary hypertension. Eur. J. Pharmacol. 2024, 978, 12. [Google Scholar] [CrossRef]
- Fu, Q.; Chen, P.; Wang, Z.; Liu, B.; Zhou, Q.; Anwar, I.; Wang, Y. The function of TRIML2 on the temozolomide resistance in glioblastoma. Ann. Med. Surg. 2025, 87, 506–514. [Google Scholar] [CrossRef]
- Barone, G.; Buonomano, A.; Forzano, C.; Palombo, A. Multi-objective optimization for comparative energy and economic analyses of a novel evacuated solar collector prototype (ICSSWH) under different weather conditions. Renew. Energy 2023, 210, 701–714. [Google Scholar] [CrossRef]
- Price, R.J.; Bullock, T.N.; Sheybani, N.D. Letter to the editor regarding “Translation of focused ultrasound for blood-brain barrier opening in glioma”. J. Control. Release 2022, 349, 16–17. [Google Scholar] [CrossRef]
- Sandra, D.A.; A Olson, J.; Langer, E.J.; Roy, M. Presenting a sham treatment as personalised increases the placebo effect in a randomised controlled trial. eLife 2023, 12, 27. [Google Scholar] [CrossRef]
- He, C.; Song, Y.; He, X.; Zhang, W.; Liao, L. No association of endometriosis with galactose-1-phosphate uridyl transferase mutations in a Chinese population. Environ. Mol. Mutagen. 2006, 47, 307–309. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, R.; Cherney, M.M.; Cherney, L.T.; Garen, C.R.; Moradian, F.; James, M.N. The crystal structures of ornithine carbamoyltransferase from Mycobacterium tuberculosis and its ternary complex with carbamoyl phosphate and L-norvaline reveal the enzyme’s catalytic mechanism. J. Mol. Biol. 2008, 375, 1052–1063. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.; Vasilatos, S.N.; Yin, J.; Qin, Y.; Zhang, L.; Davidson, N.E.; Huang, Y. Restoration of TFPI2 by LSD1 inhibition suppresses tumor progression and potentiates antitumor immunity in breast cancer. Cancer Lett. 2024, 600, 13. [Google Scholar] [CrossRef]
- Wang, B.K.; Li, N.; Huang, S.Y.; Hu, J.H.; Wang, Q.; Tang, Y.P.; Yang, T.; Asmutola, P.; Wang, J.; Yu, Q.H. Enhanced soluble sugar content in tomato fruit using CRISPR/Cas9-mediated SlINVINH1 and SlVPE5 gene editing. PeerJ 2021, 9, 19. [Google Scholar] [CrossRef]
- Mittal, L.; Srivastava, M.; Kumari, A.; Tonk, R.K.; Awasthi, A.; Asthana, S. Interplay among Structural Stability, Plasticity, and Energetics Determined by Conformational Attuning of Flexible Loops in PD-1. J. Chem. Inf. Model 2021, 61, 358–384. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, Q.; Du, M.; Vermorken, A.; Cui, Y.; Zhang, L.; Guo, L.; Ma, L.; Chen, M. Cetuximab and Doxorubicin loaded dextran-coated Fe3O4 magnetic nanoparticles as novel targeted nanocarriers for non-small cell lung cancer. J. Magn. Magn. Mater. 2019, 481, 122–128. [Google Scholar] [CrossRef]
- Primiano, A.; Persichilli, S.; Di Giacinto, F.; Ciasca, G.; Baroni, S.; Ferraro, P.M.; De Spirito, M.; Urbani, A.; Gervasoni, J. Attenuated total reflection-Fourier transform infrared spectroscopy (ATR-FTIR) detection as a rapid and convenient screening test for cystinuria. Clin. Chim. Acta 2021, 518, 128–133. [Google Scholar] [CrossRef]
- Zhu, S.; Ehnert, S.; Rouß, M.; Häussling, V.; Aspera-Werz, R.H.; Chen, T.; Nussler, A.K. From the Clinical Problem to the Basic Research-Co-Culture Models of Osteoblasts and Osteoclasts. Int. J. Mol. Sci. 2018, 19, 2284. [Google Scholar] [CrossRef]
- Alibayov, B.; Vidal, A.G.J.; Murin, L.; Scasny, A.; Takeshita, K.; Beeton, K.; Edwards, K.S.; Punshon, T.; Jackson, B.P.; McDaniel, L.S.; et al. Oxidation of hemoglobin in the lung parenchyma facilitates the differentiation of pneumococci into encapsulated bacteria. bioRxiv 2023. [Google Scholar] [CrossRef]
- Chen, L.; Li, W.C.; Li, Z.Q.; Song, Y.R.; Zhao, J.; Chen, Z.H.; Kazobinka, G.; Li, L.; Xing, Y.; Hou, T. circNUDT21 promotes bladder cancer progression by modulating the miR-16-1-3p/MDM2/p53 axis. Mol. Ther-Nucl. Acids 2021, 26, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Deng, G.Y.; Song, Z.Y.; Zhang, K.; Deng, J.M.; Jiang, K.; Han, H.Y. Enhancing antibacterial immunotherapy for bacterial pneumonia via nanovaccines coated with outer membrane vesicles. Chem. Eng. J. 2022, 436, 12. [Google Scholar] [CrossRef]
- Guo, Q.; Yin, M.; Fan, J.; Yang, Y.; Liu, T.; Qian, H.; Dai, X.; Wang, X. Peroxidase-mimicking TA-V Ox nanobranches for enhanced photothermal/chemodynamic therapy of glioma by inhibiting the expression of HSP60. Mater. Des. 2022, 224, 17. [Google Scholar] [CrossRef]
- Zhai, W.J.; Zhou, X.M.; Zhai, M.X.; Li, W.Q.; Ran, Y.H.; Sun, Y.X.; Du, J.; Zhao, W.; Xing, L.; Qi, Y.; et al. Blocking of the PD-1/PD-L1 interaction by a novel cyclic peptide inhibitor for cancer immunotherapy. Sci. China Life Sci. 2021, 64, 548–562. [Google Scholar] [CrossRef]
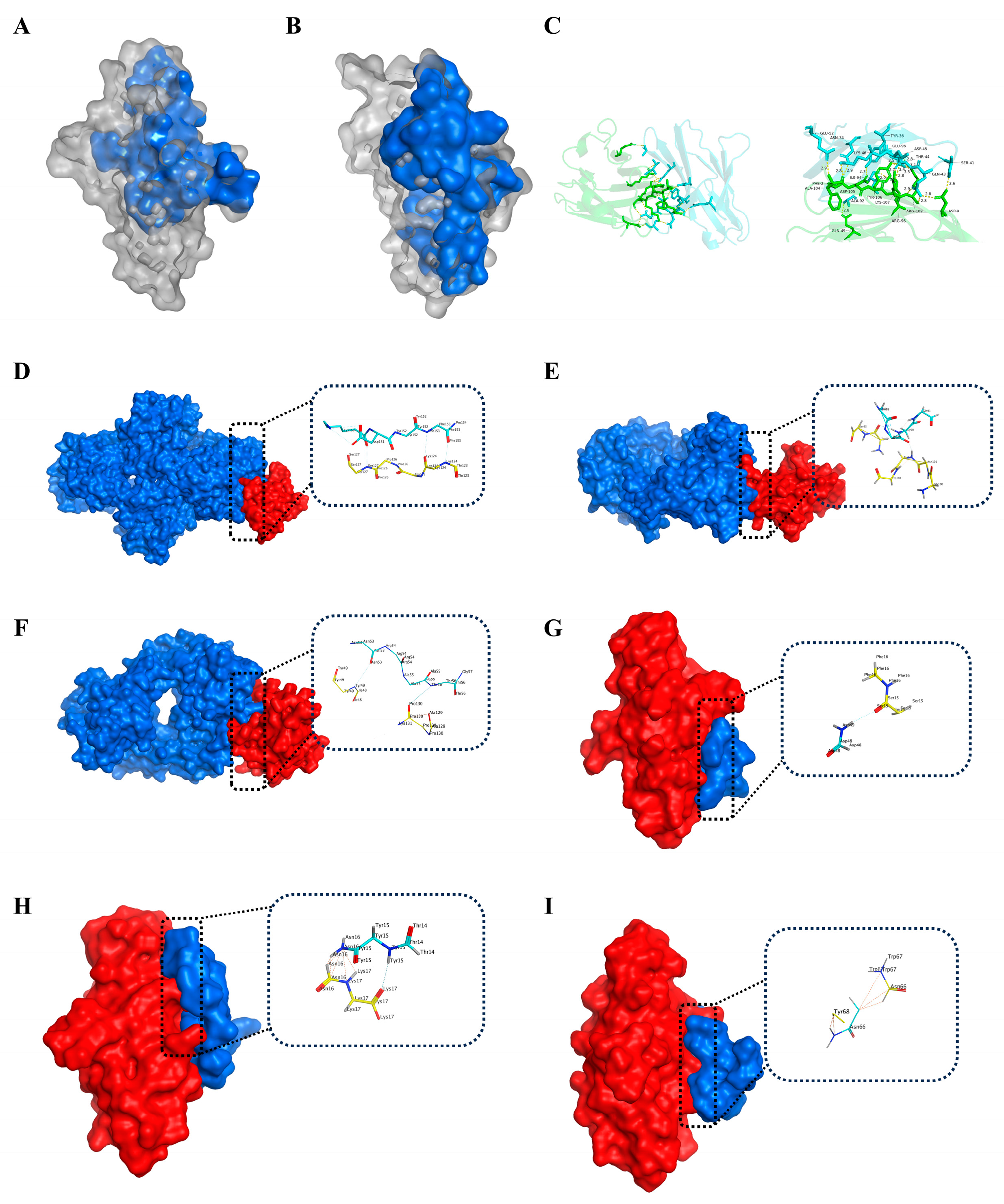
| PD-1 Residues | PD-L1 Residues | Interaction Types |
|---|---|---|
| Ile134, Glu136, Thr76 | Tyr123 | Alkyl-π hyperconjugation, H-bond |
| Asn66 | Ala121 | H-bond |
| Glu136 | Arg125, Arg113 | H-bond, salt bridge |
| Gln75 | Arg125, Asp26 | H-bond |
| Ile134 | Glu58, Glu60 | H-bond |
| Ala132 | Gln66 | H-bond |
| Thr76 | Tyr124 | H-bond |
| Lys78 | Phe19 | H-bond |
| Asn66 | Ala121 | H-bond |
| Asn34 | Phe2,Ala204 | H-bond |
| Aer41 | Asp9 | H-bond |
| Protein | Peptide | Sequence | Scores | Number of Hbonds |
|---|---|---|---|---|
| PD-1 | PD-1-0201 | D-W-F-K-A-F-W | −168.8490856 | 2 |
| PD-1-0212 | D-W-F-R-A-F-Y | −153.0645272 | 2 | |
| PD-1-0305 | D-W-F-K-P-F-Y | −165.3617216 | 1 | |
| PD-1-0310 | D-W-Y-K-A-F-Y | −150.91688 | 3 | |
| PD-1-0317 | E-W-F-K-A-F-Y | −163.098596 | 5 | |
| PD-1-0326 | D-W-F-K-G-F-Y | −159.6430304 | 1 | |
| PD-1-0329 | D-W-F-K-C-F-Y-C | −168.3935827 | 2 | |
| PD-1-0402 | D-W-F-N-A-F-Y | −141.9094456 | 2 | |
| PD-1-0411 | D-W-F-K-A-W-Y | −172.3911617 | 1 | |
| PD-1-0415 | D-W-F-K-A-F-H | −158.3387286 | 5 | |
| PD-1-0423 | D-W-F-K-S-F-Y | −138.1337358 | 3 | |
| PD-1-0503 | G-W-F-K-A-F-Y-G | −167.2862139 | 1 | |
| PD-1-0514 | G-A-D-P-E-K-R-G | −211.771068 | 7 | |
| PD-1-0518 | G-A-D-Y-K-R | −248.83294 | 8 | |
| PD-1-0519 | D-A-D-Y-K-R | −213.1262656 | 5 | |
| PD-1-0617 | N-Q-T-D-K-L-E-I | −263.3777792 | 1 | |
| PD-1-0520 | G-A-D-Y-K-G | −287.7939296 | 11 | |
| PD-1-0529 | G-A-D-P-E-K | −112.2599597 | 2 | |
| PD-1-0604 | G-K-D-F-Y-A | −129.4551328 | 2 | |
| PD-1-0609 | E-M-E-D-F-A | −157.1481112 | 2 | |
| PD-1-0619 | D-W-F-K-P-A-G | −132.4389846 | 1 | |
| PD-1-0623 | D-E-M-E-D-G | −128.5718201 | 2 |
| PD-1-0514 | PD-1-0518 | PD-1-0519 | PD-1-0520 | PD-1 | |
|---|---|---|---|---|---|
| Binding (kJ/mol) | −48.382 | −63.251 | −83.743 | −167.351 | −90.052 |
| MM (kJ/mol) | −254.604 | −327.048 | −538.087 | −627.593 | −983.159 |
| PB (kJ/mol) | 203.937 | 437.715 | 552.547 | 654.193 | 894.752 |
| SA (kJ/mol) | −23.658 | −31.926 | −40.293 | −47.076 | −49.219 |
| COU (kJ/mol) | −54.683 | −436.614 | −157.610 | −499.379 | −683.425 |
| VDW (kJ/mol) | −221.314 | −194.803 | −304.674 | −298.015 | −337.686 |
| PBcom (kJ/mol) | −5136.531 | −4792.264 | −5328.217 | −5753.738 | −6783.682 |
| PBpro (kJ/mol) | −4284.739 | −4592.976 | −4736.592 | −5231.265 | −5064.689 |
| PBlig (kJ/mol) | −1375.384 | −1683.242 | −2763.218 | −3258.629 | −4758.405 |
| SAcom (kJ/mol) | 153.352 | 167.427 | 179.969 | 198.859 | 283.371 |
| SApro (kJ/mol) | 159.527 | 158.736 | 159.037 | 161.482 | 163.648 |
| SAlig (kJ/mol) | 57.735 | 68.273 | 93.304 | 126.483 | 175.732 |
| Pre-residue of the peptide (kJ/mol) | −3.233 | −5.964 | −2.958 | −10.114 | / |
| Primer | Sequence | Number |
|---|---|---|
| IL-2 (human) | Forward: GTT GTT TCA GAT CCC TTT AGT TCC A | SEQ ID NO.1 |
| Reverse: ACA GAA CTG AAA CAT CTT CAG TGT C | SEQ ID NO.2 | |
| GZMB (human) | Forward: GCA GGA AGA TCG AAA GTG CG | SEQ ID NO.3 |
| Reverse: TAC AGC GGG GGC TTA GTT TG | SEQ ID NO.4 | |
| TNF-α (human) | Forward: AGC CCA TGT TGT AGC AAA CC | SEQ ID NO.5 |
| Reverse: GGA AGA CCC CTC CCA GAT AG | SEQ ID NO.6 | |
| IFN-γ (human) | Forward: GAA AAG CTG ACT AAT TAT TCG GTA ACT G | SEQ ID NO.7 |
| Reverse: GTT CAG CCA TCA CTT GGA TGA G | SEQ ID NO.8 | |
| Actin-β (human) | Forward: ATT CCT TCA GCA GCA TCC | SEQ ID NO.9 |
| Reverse: CAA TGC CAG GGT ACA TGG TG | SEQ ID NO.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, W.; Gu, W.; Chen, W.; Zhao, J.; Che Ajuyo, N.-M.; Pei, Y.; Min, Y.; Wang, D. Structure-Guided Design of Cyclic Peptide: A Potent Inhibitor Targeting PD-1/PD-L1 Axis with Antitumor Activity. Int. J. Mol. Sci. 2025, 26, 11308. https://doi.org/10.3390/ijms262311308
Peng W, Gu W, Chen W, Zhao J, Che Ajuyo N-M, Pei Y, Min Y, Wang D. Structure-Guided Design of Cyclic Peptide: A Potent Inhibitor Targeting PD-1/PD-L1 Axis with Antitumor Activity. International Journal of Molecular Sciences. 2025; 26(23):11308. https://doi.org/10.3390/ijms262311308
Chicago/Turabian StylePeng, Wenyu, Wenyu Gu, Wujuan Chen, Jiazheng Zhao, Nuela-Manka’a Che Ajuyo, Yechun Pei, Yi Min, and Dayong Wang. 2025. "Structure-Guided Design of Cyclic Peptide: A Potent Inhibitor Targeting PD-1/PD-L1 Axis with Antitumor Activity" International Journal of Molecular Sciences 26, no. 23: 11308. https://doi.org/10.3390/ijms262311308
APA StylePeng, W., Gu, W., Chen, W., Zhao, J., Che Ajuyo, N.-M., Pei, Y., Min, Y., & Wang, D. (2025). Structure-Guided Design of Cyclic Peptide: A Potent Inhibitor Targeting PD-1/PD-L1 Axis with Antitumor Activity. International Journal of Molecular Sciences, 26(23), 11308. https://doi.org/10.3390/ijms262311308







