Genome-Wide Association Study of Daughter Pregnancy Rate in Crossbred Dairy Cows
Abstract
1. Introduction
2. Results and Discussion
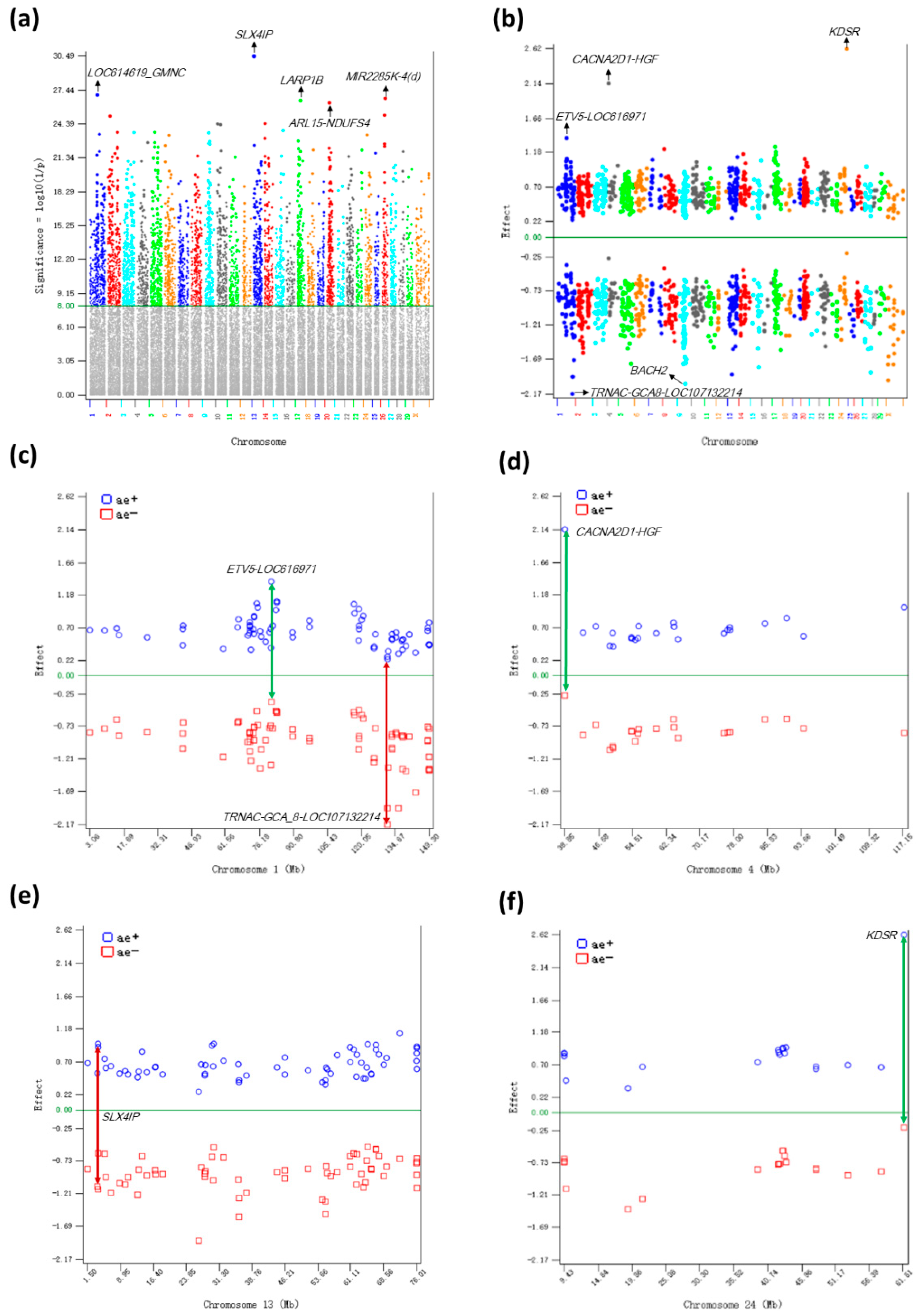
2.1. Additive Effects
2.2. Dominance Effects
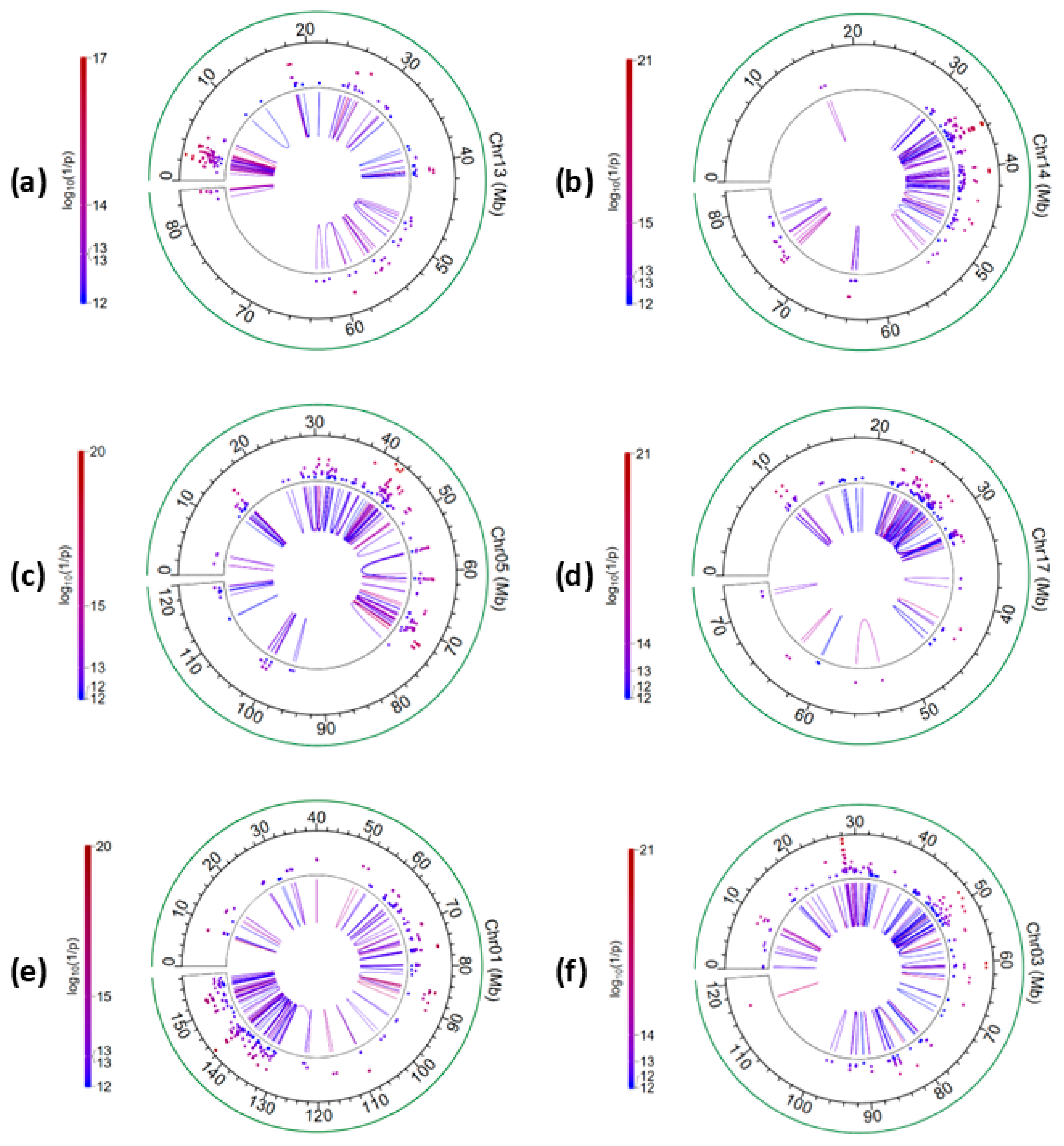
2.3. Additive × Additive (A × A) Epistasis Effects
2.4. Additive × Dominance (A × D and D × A) Epistasis Effects
2.5. Dominance × Dominance (D × D) Epistasis Effects
2.6. Genetic Mechanism of DPR Heterosis in Dairy Cattle
2.6.1. Complementary Additive Effects from Different Breeds and New Additive Effects Due to Cross Breeding
2.6.2. Two-Locus Allelic Interactions Between Loci and Between Breeds
2.6.3. Within-Locus Allelic Interaction Between Different Breeds Underlying Homozygous Advantage and Heterozygous Disadvantage
2.6.4. Genotype × Genotype Interactions Enabled by Allelic Interactions Between Breeds
2.7. Candidate Genes with Fertility and Reproductive Functions
3. Materials and Methods
3.1. Dairy Crossbred Population and SNP Data
3.2. GWAS Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Candidate Genes with Fertility and Reproductive Functions
| SNP | Chr | Position | Candidate Gene | Effect Type | Function |
| rs110391667 | 1 | 68682461 | KALRN | AAintra | Embryo development [31], Holstein cow conception rate, and DPR [16] |
| rs133450760 | 1 | 68949628 | KALRN | A | Embryo development [31], Holstein cow conception rate, and DPR [16] |
| rs110468756 | 1 | 75346472 | FGF12 | A | Progesterone synthesis [32] |
| rs136701022 | 1 | 75755758 | FGF12 | AAintra | Progesterone synthesis [32] |
| rs110789800 | 1 | 76375060 | GMNC | A | Growth and fertility [33] |
| rs41640538 | 1 | 81200460 | ETV5 | A | Spermatogenesis and folliculogenesis [34,35] |
| rs43278453 | 1 | 142383844 | UMODL1 | DDintra | Oocyte quality [36] |
| rs41583697 | 1 | 146007587 | PCNT | D | Female fertility [37] |
| rs110783898 | 1 | 153664085 | DAZL | AAintra | Germ cells development in embryos [38] |
| rs110991306 | 2 | 94592808 | GPR1 | AAintra | Steroidogenesis in ovaries [39] |
| rs110027085 | 2 | 100793281 | ERBB4 | A | Regulation of sertoli and germ cell adhesion [40] |
| rs43315219 | 2 | 106815376 | WNT6 | AAintra | Stromal proliferation during decidualization [41] |
| rs110099308 | 2 | 122454418 | PUM1 | D | Establishment of the primordial follicle pool [42] |
| rs134335964 | 2 | 128057688 | RUNX3 | AAintra | Regulates ovarian functions and ovulation [43] |
| rs110808151 | 3 | 19209742 | TUFT1 | AAintra | Male fertility [44] |
| rs111002920 | 3 | 27938935 | NGF | AAintra | Folliculogenesis and spermatogenesis [45,46] |
| rs43712201 | 3 | 97857026 | SPATA6 | A | Essential for sperm formation [47] |
| BTB-01885061 | 4 | 38854540 | HGF | A | Male and female fertility [48] |
| rs43406699 | 4 | 76069283 | IGFBP3 | AAintra | Follicle and pregnancy maintenance [49,50] |
| rs42664831 | 4 | 77659486 | HECW1 | AAintra | Oogenesis [51] |
| rs110544309 | 4 | 77811545 | HECW1 | DAinter | Oogenesis [51] |
| BTB-01477571 | 5 | 43254214 | CNOT2 | A | Oocyte maturation and female fertility [52] |
| rs43438147 | 5 | 43898744 | FRS2 | A | Epididymal development [53] |
| rs136525396 | 5 | 45641750 | IFNG | AAintra | Success of fertilization and embryo development [54] |
| rs134371136 | 5 | 47933174 | HMGA2 | A | Male and female infertility [55] |
| rs42516642 | 5 | 75694606 | RAC2 | A | Remodeling of the endometrial stroma [56] |
| rs110601510 | 6 | 70246689 | KIT | A | Male infertility [57] |
| rs41654346 | 6 | 72581115 | IGFBP7 | A, D | Maintenance of pregnancy [58] |
| rs134948894 | 6 | 81156919 | EPHA5 | AAintra | Female fertility [59] |
| rs3423226650 | 6 | 85656834 | CSN3 | A | Holstein DPR [16] |
| rs43474151 | 6 | 86508590 | SLC4A4 | A | Holstein DPR [16] |
| rs110434046 | 6 | 87184768 | GC | A | Holstein cow conception rate and DPR [16] |
| rs109034709 | 6 | 87316810 | NPFFR2 | A | Holstein cow conception rate and DPR [16] |
| rs43475934 | 6 | 92430962 | CNOT6L | A | Female fertility [60] |
| rs110103910 | 7 | 52664571 | DIAPH1 | AAinter | Sex hormone production [61] |
| rs136410320 | 7 | 61491583 | PDGFRB | AAintra | Embryonic testis development [62] |
| rs43549012 | 8 | 36818848 | PTPRD | AAinter | Gestational diabetes risk in pregnant women [63] |
| rs109347873 | 8 | 82976354 | HSD17B3 | A | Ovary morphological traits of cows [64] |
| rs135050343 | 8 | 105307551 | PAPPA | A | Ovarian steroidogenesis and female fertility [65] |
| UA-IFASA-5710 | 9 | 15743940 | IMPG1 | D | Male fertility [66] |
| rs29019953 | 10 | 10520078 | HOMER1 | AAintra | Gonadotropin [67] |
| rs134257923 | 10 | 79265033 | EIF2S1 | D | Placentation and pregnancy maintenance [68] |
| rs110147881 | 12 | 20815304 | SERPINE3 | D | Follicle count and embryo development [69] |
| rs42391346 | 13 | 1641651 | PCLB1 | AAintra | Embryonic implantation failure [70] |
| rs108997231 | 13 | 3906114 | SLX4IP | A | DNA repair and maintenance [71] and embryogenesis [18] |
| rs41605729 | 13 | 3710010 | SLX4IP | AAintra | DNA repair and maintenance [71] and embryogenesis [18] |
| rs132697751 | 13 | 25821584 | ENKUR | DDintra | Sperm motility [72] |
| rs41704549 | 13 | 58979512 | BMP7 | AAintra | Embryo development [73] |
| rs41587252 | 13 | 76013752 | NCOA3 | A | Germ cell and fertility-related phenotypes [74] |
| rs3423093320 | 14 | 35595531 | TRPA1 | AAintra | Spermatogenesis and uterine function [75,76] |
| rs134052004 | 14 | 40390594 | PEX2 | A, AAintra | Male fertility [77] |
| rs110600324 | 15 | 75950150 | PEX16 | A | Spermatocyte maturation [78] |
| rs41576959 | 16 | 15915181 | BRINP3 | D | Influencing horse reproductive traits [79] |
| rs110348666 | 17 | 25067804 | PCDH10 | AAintra | Embryo development [80] |
| rs109778958 | 17 | 29595964 | LARP1B | A | Oogenesis [81] |
| rs109289243 | 17 | 66235451 | TPST2 | DDintra | Male infertility [82] |
| rs136993800 | 18 | 12377354 | FOXC2 | AAintra | Embryo development [83] |
| rs109688013 | 18 | 14705671 | MC1R | D | Fertility and prenatal development [84] |
| rs137810605 | 18 | 16507273 | ABCC12 | AAinter | Male fertility [85] |
| rs135072252 | 18 | 40169055 | HYDIN | AAintra | Sperm-tail movement [86] |
| rs133429914 | 18 | 43836519 | PEPD | A | Holstein cow conception rate and DPR [16] |
| rs41879557 | 18 | 44031531 | CHST8 | A | Holstein cow conception rate, DPR [16], and productive life [87] |
| rs110061700 | 18 | 47814171 | SIPA1L3 | A | Embryo development [31], Holstein cow conception rate, and DPR [16] |
| rs133706373 | 18 | 56085584 | PRMT1 | D | Male fertility and embryo development [88,89] |
| rs41600357 | 19 | 58869903 | SOX9 | AAintra | Testis development and fertility [90] |
| rs110823845 | 19 | 61080519 | MAP2K6 | A | Testis determination [91] |
| rs41638404 | 20 | 25229625 | NDUFS4 | A | Embryo development [92] |
| rs42278649 | 20 | 29647234 | HCN1 | ADintra | Functional significance in ovaries [93] |
| rs135853296 | 20 | 32077537 | GHR | A | Ovarian follicular growth [94] |
| rs109014148 | 20 | 33024411 | PLCXD3 | DDintra | Spermatogenic dysfunction [95] |
| rs41257066 | 20 | 35956386 | LIFR | A | Embryo implantation [96] |
| rs41944664 | 20 | 41658534 | PDZD2 | D | Risk factor to recurrent miscarriage [97] |
| rs110116785 | 20 | 41999241 | DROSHA | D | Embryo development [98] |
| rs109535019 | 20 | 46964318 | CDH9 | ADintra | Sperm capacitation and egg—sperm binding [99] |
| rs42986448 | 20 | 51571771 | CDH12 | AAinter | Female fertility [100] |
| rs109901314 | 21 | 59873899 | DICER1 | AAintra | Female fertility [101] |
| rs41993852 | 22 | 1986320 | EOMES | A | Embryo development [102] |
| rs109972368 | 22 | 17708467 | OXTR | AAintra | Uterine contraction and pregnancy maintenance [103] |
| rs137590982 | 22 | 17910318 | LMCD1 | A | Male fertility [104] |
| rs136164685 | 22 | 17954298 | LMCD1 | AAintra | Male fertility [104] |
| rs137797676 | 23 | 9112456 | TCP11 | AAintra | Sperm capacitation, motility, and acrosome reaction [105,106] |
| rs110080847 | 24 | 21456850 | INO80C | A | Spermatogenesis [107] |
| rs42052893 | 24 | 61612123 | KDSR | A | Embryo development [108] |
| rs110028007 | 27 | 40780768 | RARB | AAintra | Embryo development [109] |
| rs134769462 | 27 | 41433477 | THRB | A, D | Female fertility [110] |
| rs133507315 | 29 | 11356071 | DLG2 | A, AAintra | Delayed puberty [111] |
| In ‘Effect Type’ column, A = additive effect, D = dominance effect, AAintra = intra-chromosome A × A effect, AAinter = inter-chromosome A × A effect, ADintra = intra-chromosome A × D effect, ADinter = inter-chromosome A × D effect, and DDintra = intra-chromosome D × D effect, DAinter = inter-chromosome D × A effect. | |||||
References
- VanRaden, P.; Sanders, A.; Tooker, M.; Miller, R.; Norman, H.; Kuhn, M.; Wiggans, G. Development of a national genetic evaluation for cow fertility. J. Dairy Sci. 2004, 87, 2285–2292. [Google Scholar] [CrossRef]
- CDCB. Impact on U.S. Dairy. Available online: https://uscdcb.com/impact/ (accessed on 7 September 2025).
- Auldist, M.; Pyman, M.; Grainger, C.; Macmillan, K. Comparative reproductive performance and early lactation productivity of Jersey × Holstein cows in predominantly Holstein herds in a pasture-based dairying system. J. Dairy Sci. 2007, 90, 4856–4862. [Google Scholar] [CrossRef]
- Begley, N.; Pierce, K.; Buckley, F. Milk production, udder health, body condition score and fertility performance of Holstein-Friesian, Norwegian Red and Norwegian Red × Holstein-Friesian cows on Irish dairy farms. In Breeding for Robustness in Cattle; Wageningen Academic: Wageningen, The Netherlands, 2009; pp. 191–198. [Google Scholar]
- Hazel, A.; Heins, B.J.; Seykora, A.J.; Hansen, L.B. Production, fertility, survival, and body measurements of Montbéliarde-sired crossbreds compared with pure Holsteins during their first 5 lactations. J. Dairy Sci. 2014, 97, 2512–2525. [Google Scholar] [CrossRef]
- Heins, B.J.; Hansen, L.B.; Hazel, A.; Seykora, A.J.; Johnson, D.; Linn, J. Jersey × Holstein crossbreds compared with pure Holsteins for body weight, body condition score, fertility, and survival during the first three lactations. J. Dairy Sci. 2012, 95, 4130–4135. [Google Scholar] [CrossRef]
- Shull, G.H. What is” heterosis”? Genetics 1948, 33, 439. [Google Scholar] [CrossRef]
- Birchler, J.A.; Yao, H.; Chudalayandi, S.; Vaiman, D.; Veitia, R.A. Heterosis. Plant Cell 2010, 22, 2105–2112. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yang, S.; Gong, J.; Zhao, Y.; Feng, Q.; Gong, H.; Li, W.; Zhan, Q.; Cheng, B.; Xia, J. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat. Commun. 2015, 6, 6258. [Google Scholar] [CrossRef]
- Akanno, E.C.; Chen, L.; Abo-Ismail, M.K.; Crowley, J.J.; Wang, Z.; Li, C.; Basarab, J.A.; MacNeil, M.D.; Plastow, G.S. Genome-wide association scan for heterotic quantitative trait loci in multi-breed and crossbred beef cattle. Genet. Sel. Evol. 2018, 50, 48. [Google Scholar] [CrossRef]
- Hollick, J.B.; Chandler, V.L. Epigenetic allelic states of a maize transcriptional regulatory locus exhibit overdominant gene action. Genetics 1998, 150, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Krieger, U.; Lippman, Z.B.; Zamir, D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 2010, 42, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, S.; Wake, T.; Nakamura, H.; Minamiyama, M.; Araki-Nakamura, S.; Ohmae-Shinohara, K.; Koketsu, E.; Okamura, S.; Miura, K.; Kawaguchi, H. The dominance model for heterosis explains culm length genetics in a hybrid sorghum variety. Sci. Rep. 2021, 11, 4532. [Google Scholar] [CrossRef]
- Boeven, P.H.; Zhao, Y.; Thorwarth, P.; Liu, F.; Maurer, H.P.; Gils, M.; Schachschneider, R.; Schacht, J.; Ebmeyer, E.; Kazman, E. Negative dominance and dominance-by-dominance epistatic effects reduce grain-yield heterosis in wide crosses in wheat. Sci. Adv. 2020, 6, eaay4897. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ma, L.; Prakapenka, D.; VanRaden, P.M.; Cole, J.B.; Da, Y. A large-scale genome-wide association study in US Holstein cattle. Front. Genet. 2019, 10, 412. [Google Scholar] [CrossRef]
- Liang, Z.; Prakapenka, D.; VanRaden, P.M.; Jiang, J.; Ma, L.; Da, Y. A Million-cow genome-wide association study of three fertility traits in US Holstein cows. Int. J. Mol. Sci. 2023, 24, 10496. [Google Scholar] [CrossRef] [PubMed]
- Prakapenka, D.; Liang, Z.; Jiang, J.; Ma, L.; Da, Y. A Large-scale genome-wide association study of epistasis effects of production traits and daughter pregnancy rate in US Holstein cattle. Genes 2021, 12, 1089. [Google Scholar] [CrossRef] [PubMed]
- Ingham, A.; Ramaswami, M.; Ramangoudr-Bhojappa, R.; Pladevall-Morera, D.; De Santis, F.; Terriente, J.; Muñoz, I.M.; Rouse, J.; Chandrasekharappa, S.C.; Lopez-Contreras, A.J. Loss of SLX4IP leads to Common Fragile Sites instability and compromises DNA interstrand crosslink repair in vivo. J. Biol. Chem. 2025, 301, 110244. [Google Scholar] [CrossRef]
- CDCB. CDCB Genomic Dictionary. Available online: https://redmine.uscdcb.com/projects/cdcb-customer-service/wiki/CDCB_Genomic_Dictionary (accessed on 7 September 2025).
- VanRaden, P.M.; Sun, C.; O’Connell, J.R. Fast imputation using medium or low-coverage sequence data. BMC Genet. 2015, 16, 82. [Google Scholar] [CrossRef]
- CDCB Genomic Evaluations. Available online: https://uscdcb.com/genomic-evaluations/ (accessed on 7 September 2025).
- Wiggans, G.; VanRaden, P.; Cooper, T. The genomic evaluation system in the United States: Past, present, future. J. Dairy Sci. 2011, 94, 3202–3211. [Google Scholar] [CrossRef]
- CDCB. QC Metrics for Genotyping Laboratories. Available online: https://redmine.uscdcb.com/projects/cdcb-customer-service/wiki/QC_Metrics_for_Genotyping_Laboratories (accessed on 7 September 2025).
- National Library of Medicine (NCBI). Available online: https://www.ncbi.nlm.nih.gov/genome/82?genome_assembly_id=1850378 (accessed on 7 September 2025).
- Ma, L.; Runesha, H.B.; Dvorkin, D.; Garbe, J.; Da, Y. Parallel and serial computing tools for testing single-locus and epistatic SNP effects of quantitative traits in genome-wide association studies. BMC Bioinform. 2008, 9, 315. [Google Scholar] [CrossRef]
- Weeks, N.T.; Luecke, G.R.; Groth, B.M.; Kraeva, M.; Ma, L.; Kramer, L.M.; Koltes, J.E.; Reecy, J.M. High-performance epistasis detection in quantitative trait GWAS. Int. J. High Perform. Comput. Appl. 2016, 32, 1094342016658110. [Google Scholar] [CrossRef]
- Henderson, C. Applications of Linear Models in Animal Breeding; University of Guelph: Guelph, ON, Canada, 1984. [Google Scholar]
- Mao, Y.; London, N.R.; Ma, L.; Dvorkin, D.; Da, Y. Detection of SNP epistasis effects of quantitative traits using an extended Kempthorne model. Physiol. Genom. 2006, 28, 46–52. [Google Scholar] [CrossRef]
- Kempthorne, O. The correlation between relatives in a random mating population. Proc. R. Soc. London. Ser. B-Biol. Sci. 1954, 143, 103–113. [Google Scholar] [CrossRef]
- Kempthorne, O. An Introduction to Genetic Statistics; Wiley: New York, NY, USA, 1957. [Google Scholar]
- Parnell, E.; Shapiro, L.P.; Voorn, R.A.; Forrest, M.P.; Jalloul, H.A.; Loizzo, D.D.; Penzes, P. KALRN: A central regulator of synaptic function and synaptopathies. Gene 2021, 768, 145306. [Google Scholar] [CrossRef]
- Yang, W.L.; Yu, S.Q.; Peng, J.Z.; Chang, P.H.; Chen, X.Y. FGF12 regulates cell cycle gene expression and promotes follicular granulosa cell proliferation through ERK phosphorylation in geese. Poult. Sci. 2023, 102, 102937. [Google Scholar] [CrossRef]
- Terré, B.; Piergiovanni, G.; Segura-Bayona, S.; Gil-Gómez, G.; Youssef, S.A.; Attolini, C.S.O.; Wilsch-Bräuninger, M.; Jung, C.; Rojas, A.M.; Marjanovic, M.; et al. GEMC1 is a critical regulator of multiciliated cell differentiation. EMBO J. 2016, 35, 942–960. [Google Scholar] [CrossRef]
- Eo, J.; Song, H.; Lim, H.J. Etv5, a transcription factor with versatile functions in male reproduction. Clin. Exp. Reprod. Med. 2012, 39, 41–45. [Google Scholar] [CrossRef]
- Eo, J.; Shin, H.; Kwon, S.; Song, H.; Murphy, K.M.; Lim, J.H. Complex ovarian defects lead to infertility in Etv5-/- female mice. Mol. Hum. Reprod. 2011, 17, 568–576. [Google Scholar] [CrossRef]
- Wang, W.; Liu, H.-C.; Jung, M.; He, Z.; Rosenwaks, Z. Functional uromodulin-like 1 (Umodl1), a glycosylated surface protein is essential to oocyte quality. Fertil. Steril. 2008, 90, S107–S108. [Google Scholar] [CrossRef]
- Baumann, C.; Wang, X.; Yang, L.; Viveiros, M.M. Error-prone meiotic division and subfertility in mice with oocyte-conditional knockdown of pericentrin. J. Cell Sci. 2017, 130, 1251–1262. [Google Scholar] [CrossRef]
- Lin, Y.; Page, D.C. Dazl deficiency leads to embryonic arrest of germ cell development in XY C57BL/6 mice. Dev. Biol. 2005, 288, 309–316. [Google Scholar] [CrossRef]
- Yang, Y.-L.; Sun, L.-F.; Yu, Y.; Xiao, T.-X.; Wang, B.-B.; Ren, P.-G.; Tang, H.-R.; Zhang, J.V. Deficiency of Gpr1 improves steroid hormone abnormality in hyperandrogenized mice. Reprod. Biol. Endocrinol. 2018, 16, 50. [Google Scholar] [CrossRef]
- Naillat, F.; Veikkolainen, V.; Miinalainen, I.; Sipila, P.; Poutanen, M.; Elenius, K.; Vainio, S.J. ErbB4, a receptor tyrosine kinase, coordinates organization of the seminiferous tubules in the developing testis. Mol. Endocrinol. 2014, 28, 1534–1546. [Google Scholar] [CrossRef]
- Wei, M.; Zhang, C.; Tian, Y.; Du, X.; Wang, Q.; Zhao, H. Expression and function of WNT6: From development to disease. Front. Cell Dev. Biol. 2020, 8, 558155. [Google Scholar] [CrossRef]
- Mak, W.; Fang, C.; Holden, T.; Dratver, M.B.; Lin, H. An Important role of pumilio 1 in regulating the development of the mammalian female germline. Biol. Reprod. 2016, 94, 134. [Google Scholar] [CrossRef]
- Ojima, F.; Saito, Y.; Tsuchiya, Y.; Kayo, D.; Taniuchi, S.; Ogoshi, M.; Fukamachi, H.; Takeuchi, S.; Takahashi, S. Runx3 transcription factor regulates ovarian functions and ovulation in female mice. J. Reprod. Dev. 2016, 62, 479–486. [Google Scholar] [CrossRef]
- Lewandowski, J.P.; Dumbović, G.; Watson, A.R.; Hwang, T.; Jacobs-Palmer, E.; Chang, N.; Much, C.; Turner, K.M.; Kirby, C.; Rubinstein, N.D. The Tug1 lncRNA locus is essential for male fertility. Genome Biol. 2020, 21, 237. [Google Scholar] [CrossRef]
- Dissen, G.A.; Romero, C.; Hirshfield, A.N.; Ojeda, S.R. Nerve growth factor is required for early follicular development in the mammalian ovary. Endocrinology 2001, 142, 2078–2086. [Google Scholar] [CrossRef]
- Stabile, A.M.; Pistilli, A.; Moretti, E.; Bartolini, D.; Ruggirello, M.; Rende, M.; Castellini, C.; Mattioli, S.; Ponchia, R.; Tripodi, S.A.; et al. A possible role for nerve growth factor and its receptors in human sperm pathology. Biomedicines 2023, 11, 3345. [Google Scholar] [CrossRef]
- Yuan, S.Q.; Stratton, C.J.; Bao, J.Q.; Zheng, H.L.; Bhetwal, B.P.; Yanagimachi, R.; Yan, W. Spata6 is required for normal assembly of the sperm connecting piece and tight head-tail conjunction. Proc. Natl. Acad. Sci. USA 2015, 112, E430–E439. [Google Scholar] [CrossRef]
- Mi, X.; Chen, C.Y.; Feng, C.; Qin, Y.Y.; Chen, Z.J.; Yang, Y.J.; Zhao, S.D. The functions and application prospects of hepatocyte growth factor in reproduction. Curr. Gene Ther. 2024, 24, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Amato, G.; Izzo, A.; Tucker, A.; Bellastella, A. Insulin-like growth factor binding protein-3 reduction in follicular fluid in spontaneous and stimulated cycles. Fertil. Steril. 1998, 70, 141–144. [Google Scholar] [CrossRef]
- Mense, K.; Heidekorn-Dettmer, J.; Wirthgen, E.; Brockelmann, Y.; Bortfeldt, R.; Peter, S.; Jung, M.; Hoflich, C.; Hoeflich, A.; Schmicke, M. Increased concentrations of insulin-like growth factor binding protein (IGFBP)-2, IGFBP-3, and IGFBP-4 are associated with fetal mortality in pregnant cows. Front. Endocrinol. 2018, 9, 310. [Google Scholar] [CrossRef]
- Fajner, V.; Giavazzi, F.; Sala, S.; Oldani, A.; Martini, E.; Napoletano, F.; Parazzoli, D.; Cesare, G.; Cerbino, R.; Maspero, E. Hecw controls oogenesis and neuronal homeostasis by promoting the liquid state of ribonucleoprotein particles. Nat. Commun. 2021, 12, 5488. [Google Scholar] [CrossRef] [PubMed]
- Soeda, S.; Oyama, M.; Kozuka-Hata, H.; Yamamoto, T. The CCR4-NOT complex suppresses untimely translational activation of maternal mRNAs. Development 2023, 150, dev201773. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Yang, L.; Hinton, B.T. The Role of fibroblast growth factor receptor substrate 2 (FRS2) in the regulation of two activity levels of the components of the extracellular signal-regulated kinase (ERK) pathway in the mouse epididymis. Biol. Reprod. 2013, 89, 48. [Google Scholar] [CrossRef]
- Seaward, A.V.; Burke, S.D.; Croy, B.A. Interferon gamma contributes to preimplantation embryonic development and to implantation site structure in NOD mice. Hum. Reprod. 2010, 25, 2829–2839. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.O.; Li, J.; Davis, B.W.; Upadhyay, S.; Al Muhisen, H.M.; Suva, L.J.; Clement, T.M.; Andersson, L. Hmga2 deficiency is associated with allometric growth retardation, infertility, and behavioral abnormalities in mice. G3 2022, 12, jkab417. [Google Scholar] [CrossRef]
- Grewal, S.; Carver, J.; Ridley, A.J.; Mardon, H.J. Human endometrial stromal cell Rho GTPases have opposing roles in regulating focal adhesion turnover and embryo invasion in vitro. Biol. Reprod. 2010, 83, 75–82. [Google Scholar] [CrossRef]
- Lin, S.; Yang, M.; Zhu, W.; Yang, C.; Chen, Y.; Cong, P.; Liu, X.; He, Z. Heterozygous deletion of exon 17 of the Kit gene impairs mouse spermatogenesis by attenuating MAPK-ERK signaling. Biol. Res. 2025, 58, 28. [Google Scholar] [CrossRef]
- Liu, Z.K.; Wang, R.C.; Han, B.C.; Yang, Y.; Peng, J.P. A novel role of IGFBP7 in mouse uterus: Regulating uterine receptivity through Th1/Th2 lymphocyte balance and decidualization. PLoS ONE 2012, 7, e45224. [Google Scholar] [CrossRef]
- Buensuceso, A.V.; Son, A.I.; Zhou, R.P.; Paquet, M.; Withers, B.M.; Deroo, B.J. Ephrin-A5 is required for optimal fertility and a complete ovulatory response to gonadotropins in the female mouse. Endocrinology 2016, 157, 942–955. [Google Scholar] [CrossRef]
- Horvat, F.; Fulka, H.; Jankele, R.; Malik, R.; Jun, M.; Solcova, K.; Sedlacek, R.; Vlahovicek, K.; Schultz, R.M.; Svoboda, P. Role of in maternal mRNA turnover. Life Sci. Alliance 2018, 1, e201800084. [Google Scholar] [CrossRef]
- Zhao, X.; Fan, C.; Qie, T.; Fu, X.; Chen, X.; Wang, Y.; Wu, Y.; Fu, X.; Shi, K.; Yan, W.; et al. Diaph1 knockout inhibits mouse primordial germ cell proliferation and affects gonadal development. Reprod. Biol. Endocrinol. 2024, 22, 82. [Google Scholar] [CrossRef]
- Uzumcu, M.; Dirks, K.A.; Skinner, M.K. Inhibition of platelet-derived growth factor actions in the embryonic testis influences normal cord development and morphology. Biol. Reprod. 2002, 66, 745–753. [Google Scholar] [CrossRef]
- Du, R.H.; Chen, H.Y.; Gao, L. Roles of protein tyrosine phosphatases in reproduction and related diseases. Reprod. Dev. Med. 2023, 7, 252–256. [Google Scholar] [CrossRef]
- Li, J.; Zhang, S.; Shen, C.; Niu, Z.; Yang, H.; Zhang, K.; Liu, Z.; Wang, Y.; Lan, X. Indel mutations within the bovine HSD17B3 gene are significantly associated with ovary morphological traits and mature follicle number. J. Steroid Biochem. Mol. Biol. 2021, 209, 105833. [Google Scholar] [CrossRef]
- Nyegaard, M.; Overgaard, M.T.; Su, Y.Q.; Hamilton, A.E.; Kwintkiewicz, J.; Hsieh, M.; Nayak, N.R.; Conti, M.; Conover, C.A.; Giudice, L.C. Lack of functional pregnancy-associated plasma protein-A (PAPPA) compromises mouse ovarian steroidogenesis and female fertility. Biol. Reprod. 2010, 82, 1129–1138. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Ghavi, D.; Mirzaei, Z.; Barati, T.; Mansoori, S. Novel differentially expressed male infertility-associated genes in sperm as prospective diagnostic biomarkers. Hum. Gene 2024, 39, 201255. [Google Scholar] [CrossRef]
- Wang, Q.; Chikina, M.D.; Pincas, H.; Sealfon, S.C. Homer1 alternative splicing is regulated by gonadotropin-releasing hormone and modulates gonadotropin gene expression. Mol. Cell Biol. 2014, 34, 1747–1756. [Google Scholar] [CrossRef]
- Shi, J.X.; Yang, L.; Gan, J.; Gu, W.W.; Gu, Y.; Shi, Y.; Jiang, H.Y.; Xu, H.R.; Yang, S.H.; Zhang, X.; et al. MiR-3074-5p regulates trophoblasts function via EIF2S1/GDF15 pathway in recurrent miscarriage. Reprod. Sci. 2024, 31, 1290–1302. [Google Scholar] [CrossRef]
- Stavros, S.; Potiris, A.; Mavrogianni, D.; Moustakli, E.; Tsiorou, K.; Zikopoulos, A.; Kathopoulis, N.; Theofanakis, C.; Loutradis, D.; Domali, E.; et al. Exploring the potential impact of SERPINE gene expression in cumulus cells during fertility treatments: A single center study. Clin. Pract. 2025, 15, 83. [Google Scholar] [CrossRef]
- Filis, P.; Kind, P.C.; Spears, N. Implantation failure in mice with a disruption in Phospholipase C beta 1 gene: Lack of embryonic attachment, aberrant steroid hormone signalling and defective endocannabinoid metabolism. Mol. Hum. Reprod. 2013, 19, 290–301. [Google Scholar] [CrossRef]
- Svendsen, J.M.; Smogorzewska, A.; Sowa, M.E.; O’Connell, B.C.; Gygi, S.P.; Elledge, S.J.; Harper, J.W. Mammalian BTBD12/SLX4 assembles a holliday junction resolvase and is required for DNA repair. Cell 2009, 138, 63–77. [Google Scholar] [CrossRef]
- Jungnickel, M.K.; Sutton, K.A.; Baker, M.A.; Cohen, M.G.; Sanderson, M.J.; Florman, H.M. The flagellar protein Enkurin is required for mouse sperm motility and for transport through the female reproductive tract. Biol. Reprod. 2018, 99, 789–797. [Google Scholar] [CrossRef]
- Solloway, M.J.; Robertson, E.J. Early embryonic lethality in Bmp5; Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development 1999, 126, 1753–1768. [Google Scholar] [CrossRef]
- Percharde, M.; Lavial, F.; Ng, J.H.; Kumar, V.; Tomaz, R.A.; Martin, N.; Yeo, J.C.; Gil, J.; Prabhakar, S.; Ng, H.H.; et al. Ncoa3 functions as an essential Esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming. Gene Dev. 2012, 26, 2286–2298. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Sucharita, S.; Majhi, R.K.; Tiwari, A.; Ghosh, A.; Pradhan, S.K.; Patra, B.K.; Dash, R.R.; Nayak, R.N.; Giri, S.C.; et al. TRPA1 is selected as a semi-conserved channel during vertebrate evolution due to its involvement in spermatogenesis. Biochem. Biophys. Res. Commun. 2019, 512, 295–302. [Google Scholar] [CrossRef]
- Pohoczky, K.; Kun, J.; Szalontai, B.; Szoke, E.; Saghy, E.; Payrits, M.; Kajtar, B.; Kovacs, K.; Kornyei, J.L.; Garai, J.; et al. Estrogen-dependent up-regulation of TRPA1 and TRPV1 receptor proteins in the rat endometrium. J. Mol. Endocrinol. 2016, 56, 135–149. [Google Scholar] [CrossRef]
- Wang, S.; Yang, H.; Fu, Y.; Teng, X.; Wang, C.; Xu, W. The Key Role of Peroxisomes in Follicular Growth, Oocyte Maturation, Ovulation, and Steroid Biosynthesis. Oxid. Med. Cell Longev. 2022, 2022, 7982344. [Google Scholar] [CrossRef]
- Nakayama, M.; Sato, H.; Okuda, T.; Fujisawa, N.; Kono, N.; Arai, H.; Suzuki, E.; Umeda, M.; Ishikawa, H.O.; Matsuno, K. Drosophila carrying pex3 or pex16 mutations are models of Zellweger syndrome that reflect its symptoms associated with the absence of peroxisomes. PLoS ONE 2011, 6, e22984. [Google Scholar] [CrossRef]
- Asadollahpour Nanaei, H.; Ayatollahi Mehrgardi, A.; Esmailizadeh, A. Whole-genome sequence analysis reveals candidate genomic footprints and genes associated with reproductive traits in Thoroughbred horse. Reprod. Domest. Anim. 2020, 55, 200–208. [Google Scholar] [CrossRef]
- Lavagi, I.; Krebs, S.; Simmet, K.; Beck, A.; Zakhartchenko, V.; Wolf, E.; Blum, H. Single-cell RNA sequencing reveals developmental heterogeneity of blastomeres during major genome activation in bovine embryos. Sci. Rep-UK 2018, 8, 4071. [Google Scholar] [CrossRef]
- Nykamp, K.; Lee, M.H.; Kimble, J. C. elegans La-related protein, LARP-1, localizes to germline P bodies and attenuates Ras-MAPK signaling during oogenesis. RNA 2008, 14, 1378–1389. [Google Scholar] [CrossRef]
- Borghei, A.; Ouyang, Y.-B.; Westmuckett, A.D.; Marcello, M.R.; Landel, C.P.; Evans, J.P.; Moore, K.L. Targeted disruption of tyrosylprotein sulfotransferase-2, an enzyme that catalyzes post-translational protein tyrosine O-sulfation, causes male infertility. J. Biol. Chem. 2006, 281, 9423–9431. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Yu, W.; Menden, H.; Younger, S.T.; Sampath, V. FOXC2 autoregulates its expression in the pulmonary endothelium after endotoxin stimulation in a histone acetylation-dependent manner. Front. Cell Dev. Biol. 2021, 9, 657662. [Google Scholar] [CrossRef] [PubMed]
- Kinsler, V.A.; Abu-Amero, S.; Budd, P.; Jackson, I.J.; Ring, S.M.; Northstone, K.; Atherton, D.J.; Bulstrode, N.W.; Stanier, P.; Hennekam, R.C.; et al. Germline melanocortin-1-receptor genotype is associated with severity of cutaneous phenotype in congenital melanocytic nevi: A role for MC1R in human fetal development. J. Investig. Dermatol. 2012, 132, 2026–2032. [Google Scholar] [CrossRef]
- Chambers, I.G.; Kumar, P.; Lichtenberg, J.; Wang, P.; Yu, J.; Phillips, J.D.; Kane, M.A.; Bodine, D.; Hamza, I. MRP5 and MRP9 play a concerted role in male reproduction and mitochondrial function. Proc. Natl. Acad. Sci. USA 2022, 119, e2111617119. [Google Scholar] [CrossRef]
- Olbrich, H.; Schmidts, M.; Werner, C.; Onoufriadis, A.; Loges, N.T.; Raidt, J.; Banki, N.F.; Shoemark, A.; Burgoyne, T.; Al Turki, S. Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am. J. Hum. Genet. 2012, 91, 672–684. [Google Scholar] [CrossRef]
- Liang, Z.; Prakapenka, D.; Zaabza, H.B.; VanRaden, P.M.; Van Tassell, C.P.; Da, Y. A million-cow genome-wide association study of productive life in US Holstein cows. Genet. Sel. Evol. 2024, 56, 67. [Google Scholar] [CrossRef]
- Azhar, M.; Xu, C.; Jiang, X.; Li, W.; Cao, Y.; Zhu, X.; Xing, X.; Wu, L.; Zou, J.; Meng, L.; et al. The arginine methyltransferase Prmt1 coordinates the germline arginine methylome essential for spermatogonial homeostasis and male fertility. Nucleic Acids Res. 2023, 51, 10428–10450. [Google Scholar] [CrossRef]
- Hashimoto, M.; Fukamizu, A.; Nakagawa, T.; Kizuka, Y. Roles of protein arginine methyltransferase 1 (PRMT1) in brain development and disease. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129776. [Google Scholar] [CrossRef]
- Stewart, M.K.; Mattiske, D.M.; Pask, A.J. Estrogen suppresses SOX9 and activates markers of female development in a human testis-derived cell line. BMC Mol. Cell Biol. 2020, 21, 66. [Google Scholar] [CrossRef]
- Warr, N.; Siggers, P.; Carre, G.A.; Wells, S.; Greenfield, A. Genetic analyses reveal functions for MAP2K3 and MAP2K6 in mouse testis determination. Biol. Reprod. 2016, 94, 103. [Google Scholar] [CrossRef]
- Wang, M.; Huang, Y.P.; Wu, H.; Song, K.; Wan, C.; Chi, A.N.; Xiao, Y.M.; Zhao, X.Y. Mitochondrial complex I deficiency leads to the retardation of early embryonic development in knockout mice. PeerJ 2017, 5, e3339. [Google Scholar] [CrossRef]
- Yeh, J.; Kim, B.S.; Gaines, L.; Peresie, J.; Page, C.; Arroyo, A. The expression of hyperpolarization activated cyclic nucleotide gated (HCN) channels in the rat ovary are dependent on the type of cell and the reproductive age of the animal: A laboratory investigation. Reprod. Biol. Endocrinol. 2008, 6, 35. [Google Scholar] [CrossRef]
- Bachelot, A.; Monget, P.; Imbert-Bolloré, P.; Coshigano, K.; Kopchick, J.J.; Kelly, P.A.; Binart, N. Growth hormone is required for ovarian follicular growth. Endocrinology 2002, 143, 4104–4112. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zheng, Z.; Ruan, J.; Li, Z.; Zhuang, X.; Tzeng, C.M. Integrated analysis miRNA and mRNA profiling in patients with severe oligozoospermia reveals miR-34c-3p downregulates PLCXD3 expression. Oncotarget 2016, 7, 52781–52796. [Google Scholar] [CrossRef] [PubMed]
- Terakawa, J.; Nakamura, S.; Ohtomo, M.; Uehara, S.; Kawata, Y.; Takarabe, S.; Sugita, H.; Namiki, T.; Kageyama, A.; Noguchi, M. LIFR-mediated ERBB2 signaling is essential for successful embryo implantation in mice. Biomolecules 2025, 15, 698. [Google Scholar] [CrossRef]
- Nagirnaja, L.; Palta, P.; Kasak, L.; Rull, K.; Christiansen, O.B.; Nielsen, H.S.; Steffensen, R.; Esko, T.; Remm, M.; Laan, M. Structural genomic variation as risk factor for idiopathic recurrent miscarriage. Hum. Mutat. 2014, 35, 972–982. [Google Scholar] [CrossRef]
- Zhang, C.; Long, X.; Ding, Y.; Chen, X.; He, J.; Liu, S.; Geng, Y.; Wang, Y.; Liu, X. Expression of DROSHA in the uterus of mice in early pregnancy and its potential significance during embryo implantation. Reprod. Sci. 2016, 23, 154–162. [Google Scholar] [CrossRef]
- Matsunaga, E.; Nambu, S.; Oka, M.; Iriki, A. Complex and dynamic expression of cadherins in the embryonic marmoset cerebral cortex. Dev. Growth Differ. 2015, 57, 474–483. [Google Scholar] [CrossRef]
- Golawski, K.; Soczewica, R.; Kacperczyk-Bartnik, J.; Manka, G.; Kiecka, M.; Lipa, M.; Warzecha, D.; Spaczynski, R.; Piekarski, P.; Banaszewska, B.; et al. The role of cadherin 12 (CDH12) in the peritoneal fluid among patients with endometriosis and endometriosis-related infertility. Int. J. Environ. Res. Public Health 2022, 19, 11586. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.M.; Luense, L.J.; McGinnis, L.K.; Nothnick, W.B.; Christenson, L.K. Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology 2008, 149, 6207–6212. [Google Scholar] [CrossRef]
- Talbot, C.D.; Walsh, M.D.; Cutty, S.J.; Elsayed, R.; Vlachaki, E.; Bruce, A.E.E.; Wardle, F.C.; Nelson, A.C. Eomes function is conserved between zebrafish and mouse and controls left-right organiser progenitor gene expression via interlocking feedforward loops. Front. Cell Dev. Biol. 2022, 10, 982477. [Google Scholar] [CrossRef]
- Akdemir, N.; Cinemre, F.B.; Cinemre, H.; Sevinc, L.; Aydemir, B.; Coban, B.; Cevrioglu, A.S.; Ozden, S. Polymorphism of the Oxytocin Receptor (OXTR) gene affects the circulating oxytocin receptor levels in late-term pregnancy in a Turkish population. Gynecol. Obstet. Investig. 2020, 85, 343–351. [Google Scholar] [CrossRef]
- Jin, X.H.; Zhang, S.; Ding, T.B.; Zhao, P.T.; Zhang, C.L.; Zhang, Y.X.; Li, W. Testicular Lmcd1 regulates phagocytosis by Sertoli cells through modulation of NFAT1/Txlna signaling pathway. Aging Cell 2020, 19, e13217. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Jiang, M.; Li, C.; Yang, P.; Sun, H.Q.; Tao, D.C.; Zhang, S.H.; Ma, Y.X. Human t-complex protein 11 (TCP11), a testis-specific gene Product, is a potential determinant of the sperm morphology. Tohoku J. Exp. Med. 2011, 224, 111–117. [Google Scholar] [CrossRef]
- Fraser, L.R.; Hosseini, R.; Hanyalogou, A.; Talmor, A.; Dudley, R.K. TCP-11, the product of a mouse t-complex gene, plays a role in stimulation of capacitation and inhibition of the spontaneous acrosome reaction. Mol. Reprod. Dev. Inc. Gamete Res. 1997, 48, 375–382. [Google Scholar] [CrossRef]
- Serber, D.W.; Runge, J.S.; Menon, D.U.; Magnuson, T. The Mouse INO80 chromatin-remodeling complex is an essential meiotic factor for spermatogenesis. Biol. Reprod. 2016, 94, 8. [Google Scholar] [CrossRef]
- Park, K.H.; Ye, Z.W.; Zhang, J.; Hammad, S.M.; Townsend, D.M.; Rockey, D.C.; Kim, S.H. 3-ketodihydrosphingosine reductase mutation induces steatosis and hepatic injury in zebrafish. Sci. Rep. 2019, 9, 1138. [Google Scholar] [CrossRef]
- Janesick, A.; Tang, W.Y.; Nguyen, T.T.L.; Blumberg, B. RARβ2 is required for vertebrate somitogenesis. Development 2017, 144, 1997–2008. [Google Scholar] [CrossRef] [PubMed]
- Ai, N.; Han, C.R.; Zhao, H.; Cheng, S.Y.; Ge, W. Disruption of thyroid hormone receptor thrab leads to female infertility in Zebrafish. Endocrinology 2024, 165, bqae037. [Google Scholar] [CrossRef] [PubMed]
- Jee, Y.H.; Won, S.; Lui, J.C.; Jennings, M.; Whalen, P.; Yue, S.; Temnycky, A.G.; Barnes, K.M.; Cheetham, T.; Boden, M.G.; et al. DLG2 variants in patients with pubertal disorders. Genet. Med. 2020, 22, 1329–1337. [Google Scholar] [CrossRef] [PubMed]





| SNP | Chr | Position | Candidate Gene | α | al+ | ae+ | f_al+ | al- | ae- | f_al- | log10(1/p) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs108997231 | 13 | 3906114 | SLX4IP | 2.116 | 1 | 0.966 | 0.543 | 2 | −1.150 | 0.457 | 30.49 |
| rs110789800 | 1 | 76375060 | LOC614619; GMNC | −2.005 | 2 | 0.655 | 0.673 | 1 | −1.350 | 0.327 | 27.00 |
| rs3423093496 | 26 | 29435606 | MIR2285K-4(d) | −1.897 | 2 | 0.847 | 0.553 | 1 | −1.050 | 0.447 | 26.69 |
| rs109778958 | 17 | 29595964 | LARP1B | 1.928 | 1 | 0.698 | 0.638 | 2 | −1.230 | 0.362 | 26.49 |
| rs41638404 | 20 | 25229625 | ARL15; NDUFS4 | 1.870 | 1 | 0.750 | 0.600 | 2 | −1.120 | 0.400 | 26.30 |
| rs110560119 | 26 | 22679137 | ARMH3 | 1.835 | 1 | 0.735 | 0.599 | 2 | −1.100 | 0.401 | 25.19 |
| rs43292973 | 2 | 22416125 | CIR1 | 1.870 | 1 | 0.790 | 0.577 | 2 | −1.080 | 0.423 | 25.10 |
| rs111018678 | 14 | 3308936 | TRAPPC9 | −1.913 | 2 | 0.553 | 0.711 | 1 | −1.360 | 0.289 | 24.43 |
| rs109798047 | 10 | 5830788 | LOC100138889; LOC781028 | −1.845 | 2 | 0.958 | 0.481 | 1 | −0.887 | 0.519 | 24.41 |
| rs110511554 | 10 | 36374183 | PPP1R14D | 1.856 | 1 | 0.991 | 0.466 | 2 | −0.865 | 0.534 | 24.33 |
| rs110600324 | 15 | 75950150 | PEX16 | 1.869 | 1 | 1.070 | 0.427 | 2 | −0.799 | 0.573 | 23.80 |
| rs43438147 | 5 | 43898744 | FRS2 | −1.781 | 2 | 0.681 | 0.617 | 1 | −1.100 | 0.383 | 23.65 |
| rs3423141818 | 2 | 103153421 | ABCA12 | −1.819 | 2 | 0.845 | 0.535 | 1 | −0.974 | 0.465 | 23.64 |
| rs3423165011 | 3 | 118672880 | LOC101907483; LOC112446044 | 1.811 | 1 | 0.651 | 0.640 | 2 | −1.160 | 0.360 | 23.63 |
| rs43708865 | 9 | 45754622 | LOC785087; LOC112448181 | −1.969 | 2 | 0.589 | 0.701 | 1 | −1.380 | 0.299 | 23.59 |
| rs29019531 | 1 | 97643741 | LOC112448275; LOC107132196 | −1.718 | 2 | 0.809 | 0.529 | 1 | −0.909 | 0.471 | 23.45 |
| rs110080847 | 24 | 21456850 | GALNT1; INO80C | −1.944 | 2 | 0.674 | 0.653 | 1 | −1.270 | 0.347 | 23.40 |
| rs43464313 | 6 | 48853315 | LOC100298058; LOC112447186 | 1.790 | 1 | 0.710 | 0.603 | 2 | −1.080 | 0.397 | 23.37 |
| rs3423092932 | 14 | 1854457 | ADGRB1 | 1.757 | 1 | 0.597 | 0.660 | 2 | −1.160 | 0.340 | 23.09 |
| rs42438948 | 17 | 7548169 | LRBA | −1.883 | 2 | 0.633 | 0.663 | 1 | −1.250 | 0.337 | 22.87 |
| SNP | Chr | Position | Candidate Gene | δ | d_12 | f_12 | d_11 | f_11 | d_22 | f_22 | log10(1/p) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs109688013 | 18 | 14705671 | MC1R | −2.555 | −1.280 | 0.421 | 0.629 | 0.445 | 1.920 | 0.134 | 18.19 |
| rs109588756 | X | 56705838 | VSIG1 | −2.136 | −0.846 | 0.529 | 1.850 | 0.092 | 0.730 | 0.379 | 11.83 |
| rs134257923 | 10 | 79265033 | EIF2S1 | −1.973 | −0.854 | 0.511 | 0.667 | 0.368 | 1.570 | 0.121 | 11.17 |
| rs135634336 | 20 | 27116615 | - | −1.826 | −0.784 | 0.563 | 0.953 | 0.292 | 1.130 | 0.145 | 10.87 |
| rs109658603 | 20 | 26870070 | - | −1.873 | −0.775 | 0.560 | 1.340 | 0.121 | 0.855 | 0.319 | 10.60 |
| rs29018884 | 20 | 27087053 | - | −1.848 | −0.767 | 0.559 | 0.841 | 0.320 | 1.320 | 0.121 | 10.54 |
| rs110147881 | 12 | 20815304 | SERPINE3 | −1.751 | −0.783 | 0.553 | 0.973 | 0.183 | 0.962 | 0.264 | 10.37 |
| rs135312762 | 20 | 28614378 | EMB | −1.988 | −0.788 | 0.511 | 1.790 | 0.088 | 0.610 | 0.402 | 10.35 |
| rs111004899 | 20 | 35745330 | LOC104975274; LOC101906686 | −1.742 | −0.750 | 0.567 | 1.020 | 0.151 | 0.964 | 0.282 | 10.03 |
| rs109513293 | 29 | 47636529 | SHANK2 | −1.714 | −0.740 | 0.570 | 0.857 | 0.201 | 1.090 | 0.230 | 9.92 |
| rs41608172 | 13 | 27232535 | TRNAE-UUC_63 (d) | −1.694 | −0.752 | 0.554 | 0.971 | 0.168 | 0.912 | 0.278 | 9.83 |
| rs109628824 | 20 | 27104916 | - | −1.791 | −0.745 | 0.559 | 0.821 | 0.320 | 1.270 | 0.121 | 9.78 |
| rs109680456 | 12 | 42526721 | LOC107132995 (d) | −1.693 | −0.713 | 0.571 | 0.660 | 0.235 | 1.300 | 0.193 | 9.74 |
| rs41653703 | 4 | 76449707 | RAMP3 | −1.677 | −0.729 | 0.567 | 0.924 | 0.165 | 0.972 | 0.268 | 9.63 |
| rs109588503 | 4 | 76953836 | NUDCD3 | −1.675 | −0.727 | 0.568 | 0.922 | 0.165 | 0.973 | 0.268 | 9.59 |
| rs133706373 | 18 | 56085584 | PRMT1 | −1.723 | −0.761 | 0.543 | 0.803 | 0.308 | 1.120 | 0.148 | 9.58 |
| rs41632642 | 15 | 80951917 | SELENOH | −1.651 | −0.754 | 0.544 | 0.950 | 0.245 | 0.844 | 0.211 | 9.53 |
| rs41889435 | 18 | 56040467 | PRR12; RRAS | −1.659 | −0.747 | 0.549 | 0.905 | 0.261 | 0.918 | 0.189 | 9.46 |
| rs41654346 | 6 | 72581115 | IGFBP7; TRNAK-UUU_12 | −1.681 | −0.754 | 0.536 | 0.773 | 0.314 | 1.080 | 0.150 | 9.40 |
| rs109489228 | 10 | 78362097 | LOC112448513 | −1.930 | −0.786 | 0.512 | 1.670 | 0.097 | 0.617 | 0.391 | 9.39 |
| SNP-1 | Chr | Position-1 | Candidate Gene-1 | SNP-2 | Position-2 | Candidate Gene-2 | αα | log10(1/p) |
|---|---|---|---|---|---|---|---|---|
| rs136164685 | 22 | 17954298 | LMCD1 | rs109133545 | 17997196 | LOC112443504 | 2.880 | 24.42 |
| rs3423093320 | 14 | 35595531 | LOC101903752; TRPA1 | rs3423093613 | 35751572 | TRPA1; LOC112449577 | 3.290 | 21.27 |
| rs41579785 | 19 | 14094861 | ENSBTAG00000053331 | rs3423464242 | 14110279 | LOC104969035; HEATR6 | 3.490 | 21.09 |
| rs3423363450 | 14 | 35558190 | LOC101903752; TRPA1 | rs3423093613 | 35751572 | TRPA1; LOC112449577 | 3.290 | 21.07 |
| rs133491240 | 17 | 23169353 | LOC783956 (u) | rs110348666 | 25067804 | PCDH10 | 2.600 | 20.97 |
| rs111002920 | 3 | 27938935 | LOC782058; NGF | rs110020978 | 28078792 | NGF; LOC112445967 | 3.410 | 20.52 |
| rs41656697 | 10 | 10474235 | HOMER1 | rs29019953 | 10520078 | HOMER1 | −2.350 | 20.16 |
| rs42982833 | 20 | 45269875 | LOC112443070 (u) | rs137619389 | 45663409 | LOC112443070; LOC112442955 | 3.330 | 20.04 |
| rs109874240 | 1 | 142636491 | TFF1; TMPRSS3 | rs42141252 | 142787751 | SLC37A1 | −2.810 | 19.93 |
| rs109520109 | 5 | 42304422 | CPNE8 | rs43436966 | 43506809 | LOC104972423; MYRFL | −2.700 | 19.76 |
| rs134052004 | 14 | 40390594 | PEX2; LOC101904449 | rs41625934 | 40600797 | LOC101904449; LOC782385 | −3.830 | 19.75 |
| rs42445061 | 3 | 60310725 | TTLL7(d) | rs134212595 | 60964021 | - | 2.470 | 19.70 |
| rs111002920 | 3 | 27938935 | LOC782058; NGF | rs109896720 | 28047336 | NGF; LOC112445967 | 3.430 | 19.61 |
| rs109621977 | 3 | 48432235 | ALG14 | rs109849660 | 49515221 | ABCA4 | −2.390 | 19.21 |
| rs109619154 | 29 | 8688612 | LOC101905908; PRSS23 | rs110088807 | 9023536 | ME3 | 2.920 | 19.09 |
| rs135072252 | 18 | 40169055 | HYDIN | rs110785717 | 40219814 | HYDIN; LOC112442400 | 2.730 | 19.09 |
| rs109367522 | 5 | 42871428 | PTPRR | rs42791351 | 43371061 | LOC104972423; MYRFL | −2.700 | 19.09 |
| rs110991306 | 2 | 94592808 | GPR1; ZDBF2 | rs110975979 | 95586874 | LOC112442377; LOC100847666 | 2.360 | 19.06 |
| rs43183410 | 20 | 45022442 | LOC112443070 (u) | rs137619389 | 45663409 | LOC112443070; LOC112442955 | −3.230 | 19.02 |
| rs3423093320 | 14 | 35595531 | LOC101903752; TRPA1 | rs3423093914 | 35815088 | LOC112449577; LOC112449516 | 2.340 | 18.83 |
| SNP-1 | Chr-1 | Position-1 | Candidate Gene-1 | SNP-2 | Chr-2 | Position-2 | Candidate Gene-2 | δδ | log10(1/p) |
|---|---|---|---|---|---|---|---|---|---|
| rs42670220 | 20 | 39445232 | RAI14 | rs41942366 | 20 | 41041011 | LOC104975283; SUB1 | 5.010 | 15.33 |
| rs110573919 | 25 | 3828387 | DNAAF8 | rs134957967 | 25 | 3920360 | UBN1 | 4.350 | 14.32 |
| rs109289243 | 17 | 66235451 | TPST2 | rs109255241 | 17 | 66309374 | LOC100847159; LOC614881 | 4.710 | 14.28 |
| rs132697751 | 13 | 25821584 | ENKUR | rs109352819 | 13 | 25911339 | GPR158 | 4.750 | 14.25 |
| rs41942122 | 20 | 34603769 | LOC782462; LOC112443045 | rs109633897 | 20 | 34759872 | LOC782462; LOC112443045 | 5.230 | 13.94 |
| rs41874708 | 18 | 32330449 | LOC112442399; CDH11 | rs41637282 | 18 | 32771279 | CDH11 | 4.950 | 13.90 |
| rs132912044 | 4 | 6268973 | LOC101904266 (d) | rs3423164105 | 4 | 6379484 | LOC101904266 (d) | 4.600 | 13.87 |
| rs110405028 | 18 | 14014113 | APRT | rs110180580 | 18 | 14050260 | CBFA2T3 | 4.500 | 13.48 |
| rs41594638 | 7 | 75473623 | LOC100296952; LOC112447598 | rs41667214 | 7 | 76297401 | LOC112447528 (u) | 4.250 | 13.29 |
| rs42640157 | 20 | 58364680 | ANKH | rs110840725 | 20 | 58566498 | OTULINL | 3.930 | 12.93 |
| rs109014148 | 20 | 33024411 | PLCXD3 | rs43001858 | 20 | 33323766 | C6 | 5.010 | 12.77 |
| rs43278453 | 1 | 142383844 | UMODL1 | rs136138690 | 1 | 143444786 | CRYAA; LOC112448267 | 4.130 | 12.72 |
| rs41951335 | 20 | 58357512 | ANKH | rs110840725 | 20 | 58566498 | OTULINL | 3.900 | 12.68 |
| rs134010835 | 22 | 35456434 | MAGI1 | rs110776523 | 22 | 35535752 | MAGI1 | 4.950 | 12.53 |
| rs41894282 | 18 | 54627066 | BICRA; EHD2 | rs3423454787 | 18 | 54680124 | SELENOW; LOC101902766 | 5.650 | 12.52 |
| rs135176671 | 1 | 144551556 | TSPEAR | rs109840008 | 1 | 146320854 | PRMT2 | 4.030 | 12.37 |
| rs137324140 | 13 | 2145874 | PLCB4 | rs134403374 | 13 | 2232129 | ENSBTAG00000054174 | 4.470 | 12.14 |
| rs3423449147 | 20 | 67492982 | LOC104969150; ICE1 | rs41964197 | 20 | 68697504 | LOC112443018 (d) | 4.880 | 12.03 |
| Crossbred | Jersey | Holstein | |
|---|---|---|---|
| Mean | 54.9703 ± 33.178 | 53.434 ± 33.199 | 49.06 ± 32.01 |
| n | 31,338 | 722,949 | 4,956,093 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, R.; Liang, Z.; Prakapenka, D.; Ma, L.; Da, Y. Genome-Wide Association Study of Daughter Pregnancy Rate in Crossbred Dairy Cows. Int. J. Mol. Sci. 2025, 26, 11149. https://doi.org/10.3390/ijms262211149
Yang R, Liang Z, Prakapenka D, Ma L, Da Y. Genome-Wide Association Study of Daughter Pregnancy Rate in Crossbred Dairy Cows. International Journal of Molecular Sciences. 2025; 26(22):11149. https://doi.org/10.3390/ijms262211149
Chicago/Turabian StyleYang, Ruifei, Zuoxiang Liang, Dzianis Prakapenka, Li Ma, and Yang Da. 2025. "Genome-Wide Association Study of Daughter Pregnancy Rate in Crossbred Dairy Cows" International Journal of Molecular Sciences 26, no. 22: 11149. https://doi.org/10.3390/ijms262211149
APA StyleYang, R., Liang, Z., Prakapenka, D., Ma, L., & Da, Y. (2025). Genome-Wide Association Study of Daughter Pregnancy Rate in Crossbred Dairy Cows. International Journal of Molecular Sciences, 26(22), 11149. https://doi.org/10.3390/ijms262211149






