Evolutionary Conservation and Regulatory Diversification of AS1 Homologs in Soybean
Abstract
1. Introduction
2. Results
2.1. Phylogenetic, Gene Structure, and Conserved Motif Analysis of AS1 Genes
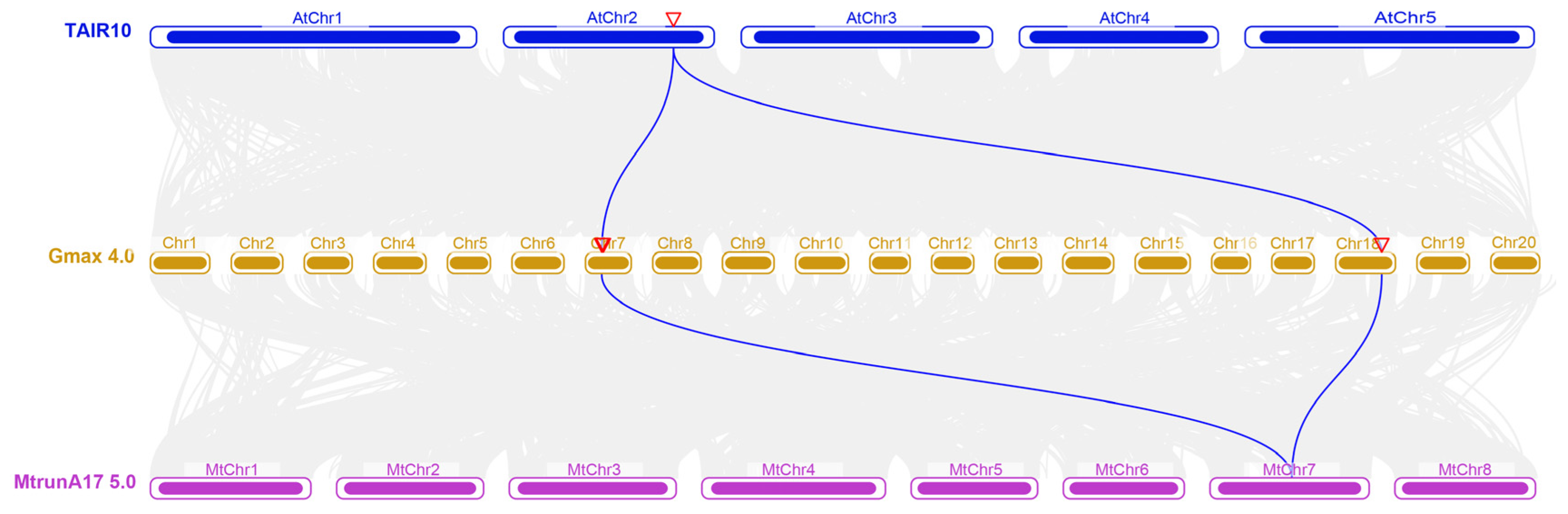
2.2. Collinearity Analysis of the AS1 Genes Among A. thaliana, M. truncatula, and G. max
2.3. Sequence Conservation Among AS1 Proteins in Arabidopsis and Soybean
2.4. Expression Pattern of GmAS1 Genes
2.5. Genetic Complementation Analysis
2.6. Prediction of Cis-Acting Elements
3. Discussion
3.1. Evolutionary Conservation of AS1 Function
3.2. Regulatory Diversification of GmAS1 Paralogs
3.3. Evolutionary and Agronomic Implications
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification of AS1 Genes in Plant Species
4.3. Phylogenetic, Gene Structure, and Conserved Motif Analysis of AS1 Homologs
4.4. Collinearity Analysis of the AS1 Genes
4.5. Mutiple Sequence Alignment of AS1 Homologs
4.6. RNA Extraction and Quantitative RT-PCR (qRT-PCR) Analysis
4.7. RNA in Situ Hybridization
4.8. Genetic Complementation Analysis
4.9. Prediction of Cis-Acting Elements
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hay, A.; Tsiantis, M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 2006, 38, 942–947. [Google Scholar] [CrossRef] [PubMed]
- Nicotra, A.B.; Leigh, A.; Boyce, C.K.; Jones, C.S.; Niklas, K.J.; Royer, D.L.; Tsukaya, H. The evolution and functional significance of leaf shape in the angiosperms. Funct. Plant Biol. 2011, 38, 535. [Google Scholar] [CrossRef]
- Nakayama, H.; Leichty, A.R.; Sinha, N.R. Molecular mechanisms underlying leaf development, morphological diversification, and beyond. Plant Cell 2022, 34, 2534–2548. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.E.; Barley, R.; Curtis, M.; Arroyo, J.M.; Dunham, M.; Hudson, A.; Martienssen, R.A. Asymmetric Leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 2000, 408, 967–971. [Google Scholar] [CrossRef]
- Timmermans, M.C.; Hudson, A.; Becraft, P.W.; Nelson, T. ROUGH SHEATH2: A myb protein that represses Knox homeobox genes in maize lateral organ primordia. Science 1999, 284, 151–153. [Google Scholar] [CrossRef]
- Waites, R.; Selvadurai, H.R.; Oliver, I.R.; Hudson, A. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 1998, 93, 779–789. [Google Scholar] [CrossRef]
- Waites, R.; Hudson, A. Phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 1995, 121, 2143–2154. [Google Scholar] [CrossRef]
- Guo, M.; Thomas, J.; Collins, G.; Timmermans, M.C. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 2008, 20, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Barkoulas, M.; Tsiantis, M. ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 94, 10467–10472. [Google Scholar]
- Ori, N.; Eshed, Y.; Chuck, G.; Bowman, J.L.; Hake, S. Mechanisms that control Knox gene expression in the Arabidopsis shoot. Development 2000, 127, 5523–5532. [Google Scholar] [CrossRef]
- Phelps-Durr, T.L.; Thomas, J.; Vahab, P.; Timmermans, M.C. Maize Rough Sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain Knox gene silencing and determinacy during organogenesis. Plant Cell 2005, 17, 2886–2898. [Google Scholar] [CrossRef]
- Theodoris, G.; Inada, N.; Freeling, M. Conservation and molecular dissection of ROUGH SHEATH2 and ASYMMETRIC LEAVES1 function in leaf development. Proc. Natl. Acad. Sci. USA 2003, 100, 6837–6842. [Google Scholar] [CrossRef]
- Tsiantis, M.; Schneeberger, R.; Golz, J.F.; Freeling, M.; Langdale, J.A. The maize Rough Sheath2 gene and leaf development programs in monocot and dicot plants. Science 1999, 284, 154–156. [Google Scholar] [CrossRef]
- Xu, L.; Xu, Y.; Dong, A.; Sun, Y.; Pi, L.; Xu, Y.; Huang, H. Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 2003, 130, 4097–4107. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; McCormick, S.; Timmermans, M.; Sinha, N. The expression domain of PHANTASTICA determines leaflet placement in compound leaves. Nature 2003, 424, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Pan, M.; Zhu, Y.; Wang, P.; Jiang, L.; Xu, D.; Wang, X.; Chen, L.; Guo, W.; Yang, H.; et al. CRISPR/Cas9-mediated targeted mutagenesis of GmAS1/2 genes alters leaf shape in soybean. Int. J. Mol. Sci. 2025, 26, 9657. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Cantabrana, H.; Ripoll, J.J.; Ochando, I.; Vera, A.; Ferrándiz, C.; Martínez-Laborda, A. Common regulatory networks in leaf and fruit patterning revealed by mutations in the Arabidopsis ASYMMETRIC LEAVES1 gene. Development 2007, 134, 2663–2671. [Google Scholar] [CrossRef]
- Gonzalez-Reig, S.; Ripoll, J.J.; Vera, A.; Yanofsky, M.F.; Martinez-Laborda, A. Antagonistic gene activities determine the formation of pattern elements along the mediolateral axis of the Arabidopsis fruit. PLoS Genet. 2012, 8, e1003020. [Google Scholar] [CrossRef]
- Gubert, C.M.; Christy, M.E.; Ward, D.L.; Groner, W.D.; Liljegren, S.J. ASYMMETRIC LEAVES1 regulates abscission zone placement in Arabidopsis flowers. BMC Plant Biol. 2014, 14, 195. [Google Scholar] [CrossRef]
- Lee, N.; Hwang, D.Y.; Lee, H.G.; Hwang, H.; Kang, H.W.; Lee, W.; Choi, M.G.; Ahn, Y.J.; Lim, C.; Kim, J.-I. ASYMMETRIC LEAVES1 promotes leaf hyponasty in Arabidopsis by light-mediated auxin signaling. Plant Physiol. 2025, 197, kiae550. [Google Scholar] [CrossRef]
- Nurmberg, P.L.; Knox, K.A.; Yun, B.-W.; Morris, P.C.; Shafiei, R.; Hudson, A.; Loake, G.J. The developmental selector AS1 is an evolutionarily conserved regulator of the plant immune response. Proc. Natl. Acad. Sci. USA 2007, 104, 18795–18800. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Lee, I.; Lee, S.Y.; Imaizumi, T.; Hong, J.C. CONSTANS and ASYMMETRIC LEAVES 1 complex is involved in the induction of FLOWERING LOCUS T in photoperiodic flowering in Arabidopsis. Plant J. 2012, 69, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-Y.; Iwasaki, M.; Machida, C.; Machida, Y.; Zhou, X.; Chua, N.-H. βC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 2008, 22, 2564–2577. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.-X.; Xu, Y.-B.; Wang, T.-T.; Ma, X.-F.; Zhao, J.-Q.; Li, Y.; Fan, J.; Wang, W.-M. Proper expression of AS1 is required for RPW8.1-mediated defense against powdery mildew in Arabidopsis. Physiol. Mol. Plant Pathol. 2015, 92, 101–111. [Google Scholar] [CrossRef]
- Ge, L.; Chen, R. PHANTASTICA regulates leaf polarity and petiole identity in Medicago truncatula. Plant Signaling Behav. 2014, 9, e28121. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, D.; Mao, Y.; Zhou, Y.; Zhao, L.; Zhang, C.; Liu, Y.; Chen, J. The organ size and morphological change during the domestication process of soybean. Front. Plant Sci. 2022, 13, 913238. [Google Scholar] [CrossRef]
- Wilson, R.F. Soybean: Market driven research needs. In Genetics and Genomics of Soybean; Springer: New York, NY, USA, 2008; pp. 3–15. [Google Scholar]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar] [CrossRef]
- Hughes, A.L. The evolution of functionally novel proteins after gene duplication. Proc. R. Soc. Lond. B Biol. Sci. 1994, 256, 119–124. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Xu, C.; Nadon, B.D.; Kim, K.D.; Jackson, S.A. Genetic and epigenetic divergence of duplicate genes in two legume species. Plant Cell Environ. 2018, 41, 2033–2044. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, R.C.; Polzin, K.; Labate, J.; Specht, J.; Brummer, E.C.; Olson, T.; Young, N.; Concibido, V.; Wilcox, J.; Tamulonis, J.P. Genome duplication in soybean (Glycine subgenus soja). Genetics 1996, 144, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhou, H.; Zhang, Y.; Zhao, Y.; Zhang, Y.; Feng, X.; Lin, H. Diverse roles of MYB transcription factors in plants. J. Integr. Plant Biol. 2025, 67, 539–562. [Google Scholar] [CrossRef]
- Schlueter, J.A.; Lin, J.-Y.; Schlueter, S.D.; Vasylenko-Sanders, I.F.; Deshpande, S.; Yi, J.; O’Bleness, M.; Roe, B.A.; Nelson, R.T.; Scheffler, B.E.; et al. Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genom. 2007, 8, 330. [Google Scholar] [CrossRef]
- Libault, M.; Joshi, T.; Takahashi, K.; Hurley-Sommer, A.; Puricelli, K.; Blake, S.; Finger, R.E.; Taylor, C.G.; Xu, D.; Nguyen, H.T. Large-scale analysis of putative soybean regulatory gene expression identifies a Myb gene involved in soybean nodule development. Plant Physiol. 2009, 151, 1207–1220. [Google Scholar] [CrossRef]
- Zhao, N.; Ding, X.; Lian, T.; Wang, M.; Tong, Y.; Liang, D.; An, Q.; Sun, S.; Jackson, S.A.; Liu, B. The effects of gene duplication modes on the evolution of regulatory divergence in wild and cultivated soybean. Front. Genet. 2020, 11, 601003. [Google Scholar] [CrossRef]
- Harrison, C.J.; Corley, S.B.; Moylan, E.C.; Alexander, D.L.; Scotland, R.W.; Langdale, J.A. Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature 2005, 434, 509–514. [Google Scholar] [CrossRef]
- Altschmied, J.; Delfgaauw, J.; Wilde, B.; Duschl, J.; Bouneau, L.; Volff, J.-N.; Schartl, M. Subfunctionalization of duplicate mitf genes associated with differential degeneration of alternative exons in fish. Genetics 2002, 161, 259–267. [Google Scholar] [CrossRef]
- Huminiecki, L.; Wolfe, K.H. Divergence of spatial gene expression profiles following species-specific gene duplications in human and mouse. Genome Res. 2004, 14, 1870–1879. [Google Scholar] [CrossRef]
- Zhou, X.; Fan, H.; Feng, X.; Ruan, Z.; Yuan, J.; Han, Q.; He, Z.; You, Y.; Chao, H.; Chen, M.; et al. PGCP: A comprehensive database of plant genomes for comparative phylogenomics. Plant Biotechnol. J. 2025, 23, 2928–2930. [Google Scholar] [CrossRef] [PubMed]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Yim, A.K.Y.; Wong, J.W.H.; Ku, Y.S.; Qin, H.; Chan, T.F.; Lam, H.M. Using RNA-Seq data to evaluate reference genes suitable for gene expression studies in soybean. PLoS ONE 2015, 10, e0136343. [Google Scholar] [CrossRef] [PubMed]
- Coen, E.S.; Romero, J.; Doyle, S.; Elliott, R.; Murphy, G.; Carpenter, R. Floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell 1990, 63, 1311–1322. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Zhou, X.; Wang, D.; Yang, X.; He, Y.; Xia, Z.; Chen, J.; Zhao, W. Evolutionary Conservation and Regulatory Diversification of AS1 Homologs in Soybean. Int. J. Mol. Sci. 2025, 26, 11089. https://doi.org/10.3390/ijms262211089
Wang D, Zhou X, Wang D, Yang X, He Y, Xia Z, Chen J, Zhao W. Evolutionary Conservation and Regulatory Diversification of AS1 Homologs in Soybean. International Journal of Molecular Sciences. 2025; 26(22):11089. https://doi.org/10.3390/ijms262211089
Chicago/Turabian StyleWang, Dan, Xuan Zhou, Dongfa Wang, Xiangtao Yang, Yexin He, Zhengjun Xia, Jianghua Chen, and Weiyue Zhao. 2025. "Evolutionary Conservation and Regulatory Diversification of AS1 Homologs in Soybean" International Journal of Molecular Sciences 26, no. 22: 11089. https://doi.org/10.3390/ijms262211089
APA StyleWang, D., Zhou, X., Wang, D., Yang, X., He, Y., Xia, Z., Chen, J., & Zhao, W. (2025). Evolutionary Conservation and Regulatory Diversification of AS1 Homologs in Soybean. International Journal of Molecular Sciences, 26(22), 11089. https://doi.org/10.3390/ijms262211089





