Integrating CTLA-4 Genetics and Soluble Isoforms for the Stratification of HCV-Related Hepatocellular Carcinoma Risk and Aggressiveness
Abstract
1. Introduction
2. Results
2.1. Genes Correlated with HCC
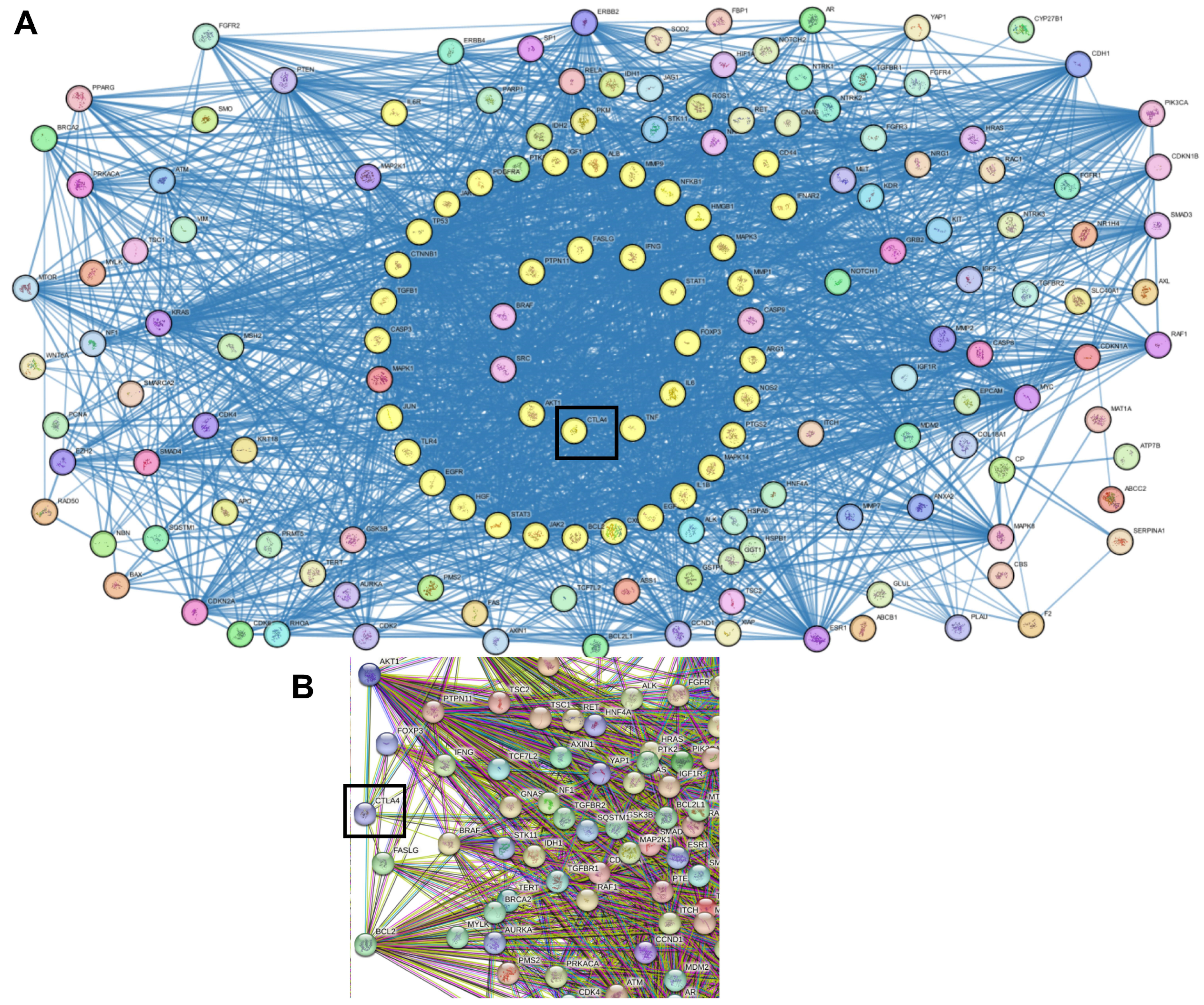
2.2. Protein–Protein Interactions
2.3. Demographic and Clinical Data of the Study Subjects
2.4. Genotyping of CTLA-4 Variants
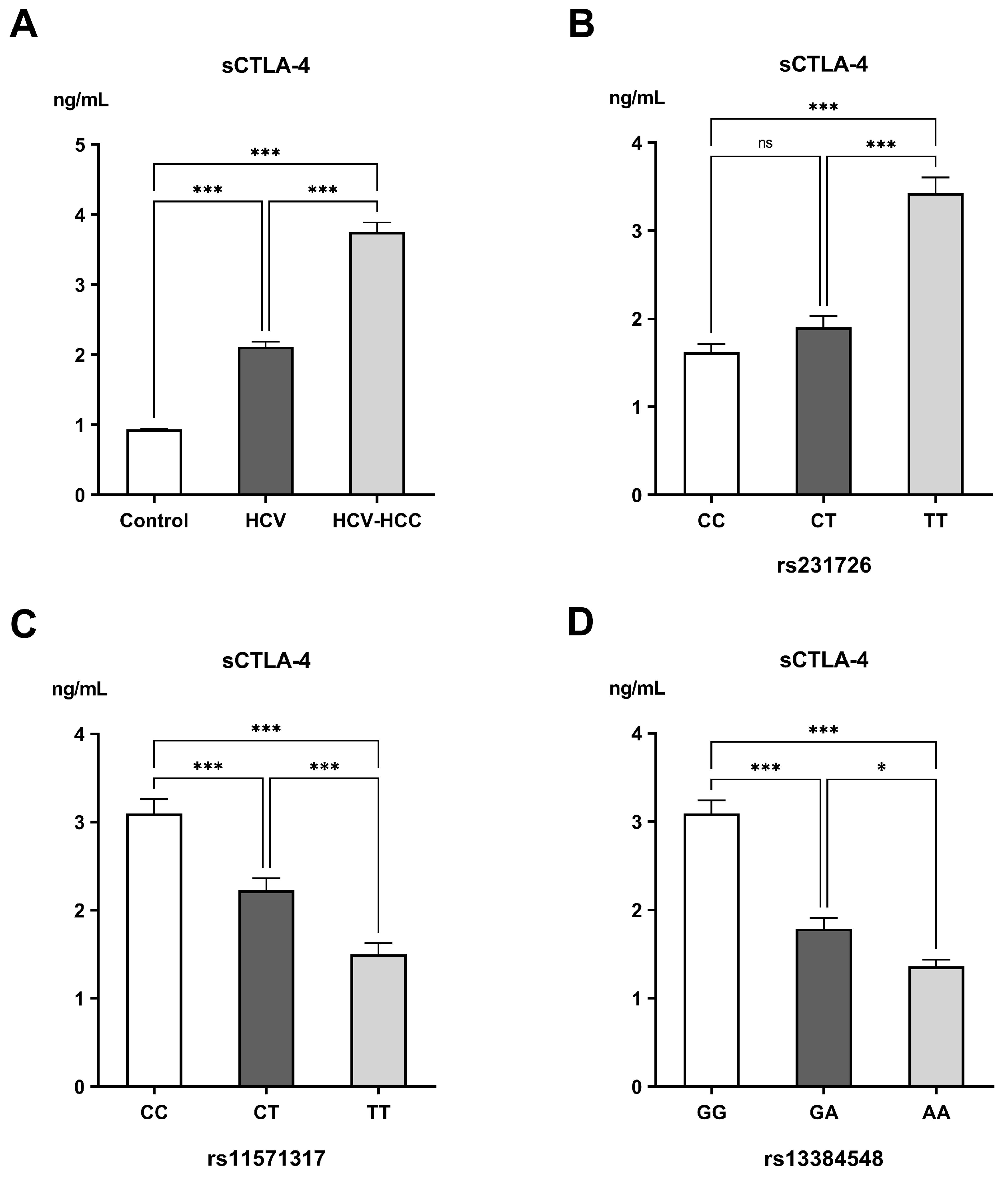
2.5. Measurements of Soluble CTLA-4 (sCTLA-4)
| rs231726 TT n = 43 | p-Value | rs11571317 CC n = 31 | p-Value | rs13384548 GG n = 52 | p-Value | sCTLA-4 | p-Value | |
|---|---|---|---|---|---|---|---|---|
| Child–Pugh score | ||||||||
| A | 8 (18.60%) | 0.006 | 5 (16.10%) | 0.086 | 11 (21.20%) | 0.031 | 3.37 ± 0.29 | 0.031 |
| B | 11 (25.60%) | 11 (35.50%) | 15 (28.80%) | 3.48 ± 0.22 | ||||
| C | 24 (55.80%) | 15 (48.40%) | 26 (50.30%) | 4.15 ± 0.20 | ||||
| MELD/Na score | ||||||||
| ≤9 | 4 (9.30%) | 0.023 | 2 (6.50%) | 0.006 | 4 (7.70%) | 0.001 | 3.41 ± 0.32 | 0.479 |
| 10–19 | 17 (39.50%) | 14 (45.20%) | 21 (40.40%) | 4.02 ± 0.24 | ||||
| 20–29 | 14 (32.60%) | 11 (35.50%) | 19 (36.50%) | 3.66 ± 0.23 | ||||
| ≥30 | 8 (18.60%) | 4 (12.90%) | 8 (15.40%) | 3.72 ± 0.40 | ||||
| BCLC stage | ||||||||
| A | 5 (11.60%) | 0.209 | 3 (9.70%) | 0.110 | 6 (11.50%) | 0.028 | 3.11 ± 0.39 | 0.226 |
| B | 14 (32.60%) | 11 (35.50%) | 21 (40.40%) | 3.95 ± 0.24 | ||||
| C | 13 (30.20%) | 11(35.50%) | 14 (26.90%) | 3.76 ± 0.23 | ||||
| D | 11 (25.60%) | 6 (19.40%) | 11 (21.20%) | 3.91 ± 0.31 | ||||
| Number of lesions | ||||||||
| <3 | 21 (48.80%) | 0.879 | 15 (48.40%) | 0.857 | 28 (53.80%) | 0.579 | 3.69 ± 0.21 | 0.661 |
| ≥3 | 22 (51.20%) | 16 (51.6%) | 24 (46.20%) | 3.82 ± 0.17 | ||||
| Largest lesion | ||||||||
| <3 cm | 10 (23.30%) | <0.001 | 7 (22.6%) | 0.002 | 17 (32.70%) | 0.013 | 3.40 ± 023 | 0.073 |
| ≥3 cm | 33 (76.70%) | 24 (77.4%) | 35 (67.30%) | 3.92 ± 0.16 |
3. Discussion
4. Materials and Methods
4.1. Study Population and Design
4.2. Retrieving Genes Correlated with HCC
4.3. Gene Ontology
4.4. Genotyping of CTLA-4 Variants
4.5. Measurement of Soluble CTLA-4 (sCTLA-4)
4.6. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AFP | Alpha-fetoprotein |
| ALT | Alanine aminotransferase |
| AST | Aspartate aminotransferase |
| BCLC | Barcelona Clinic Liver Cancer staging system |
| CTLA-4 | Cytotoxic T-lymphocyte-associated antigen-4 |
| ELISA | Enzyme-linked immunosorbent assay |
| HCC | Hepatocellular carcinoma |
| HCV | Hepatitis C virus |
| MELD/Na | Model for End-Stage Liver Disease Sodium score |
| OR | Odds ratio |
| PC | Prothrombin concentration |
| sCTLA-4 | soluble CTLA-4 |
| SNPs | Single-nucleotide polymorphisms |
References
- Singh, S.P.; Madke, T.; Chand, P. Global Epidemiology of Hepatocellular Carcinoma. J. Clin. Exp. Hepatol. 2025, 15, 102446. [Google Scholar] [CrossRef]
- Wang, J.; Qiu, K.; Zhou, S.; Gan, Y.; Jiang, K.; Wang, D.; Wang, H. Risk factors for hepatocellular carcinoma: An umbrella review of systematic review and meta-analysis. Ann. Med. 2025, 57, 2455539. [Google Scholar] [CrossRef]
- Hassan, M.; Nasr, S.M.; Amin, N.A.; El-Ahwany, E.; Zoheiry, M.; Elzallat, M. Circulating liver cancer stem cells and their stemness-associated MicroRNAs as diagnostic and prognostic biomarkers for viral hepatitis-induced liver cirrhosis and hepatocellular carcinoma. Non-Coding RNA Res. 2023, 8, 155–163. [Google Scholar] [CrossRef]
- Xie, C.; Singal, A.K. Beyond the cure: Navigating hepatocellular risk and surveillance after Hepatitis C eradication in the direct-acting antiviral era. J. Clin. Transl. Hepatol. 2025, 13, 418–424. [Google Scholar] [CrossRef]
- Hassan, M.; Elzallat, M.; Aboushousha, T.; Elhusseny, Y.; El-Ahwany, E. MicroRNA-122 mimic/microRNA-221 inhibitor combination as a novel therapeutic tool against hepatocellular carcinoma. Non-Coding RNA Res. 2023, 8, 126–134. [Google Scholar] [CrossRef]
- Cabral, L.K.; Disoma, C.; Tarchi, P.; El-Khobar, K.E.; Agustiningsih, A.; Dituri, F.; Tiribelli, C.; Sukowati, C. Dual inhibition of SRC family Kinases and Sorafenib enhances anti-tumor activity in hepatocellular carcinoma cells. Int. J. Mol. Sci. 2025, 26, 6506. [Google Scholar] [CrossRef]
- Sallam, M.; Khalil, R. Contemporary insights into hepatitis C virus: A comprehensive review. Microorganisms 2024, 12, 1035. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Fernández-Piñeiro, I.; Badiola, I.; Elzallat, M.; Sánchez, A.; Aboushousha, T.; Hafiz, E.; El-Ahwany, E. Therapeutic potential of miRNA-26a-encapsulated nanoparticles against hepatocellular carcinoma in a murine model. Liver Res. 2025, in press. [Google Scholar] [CrossRef]
- Sherafat, N.S.; Keshavarz, A.; Mardi, A.; Mohammadiara, A.; Aghaei, M.; Aghebati-Maleki, L.; Mohammadi, M.H. Rationale of using immune checkpoint inhibitors (ICIs) and anti-angiogenic agents in cancer treatment from a molecular perspective. Clin. Exp. Med. 2025, 25, 238. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Attia, M.S.; Ali-Eldin, Z.; El Attar, G.; Elzallat, M.; Saad, H.H.K.; Isaac, A. Programmed death-ligand 1 (PD-L1) polymorphisms as predictive biomarkers for the development of liver cirrhosis and hepatocellular carcinoma in HCV Egyptian patients. Tumour Virus Res. 2022, 14, 200249. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; El-Maadawy, W.H.; Elhusseny, Y.; Agamy, F.E.; Fahim, S.A.; Balata, M. Genetic variants and soluble isoforms of PD-1/PD-L1 as novel biomarkers for pancreatic ductal adenocarcinoma (PDAC) susceptibility and prognosis. Biomedicines 2025, 13, 2246. [Google Scholar] [CrossRef]
- Mok, S.; Liu, H.; Ağaç Çobanoğlu, D.; Anang, N.-A.A.S.; Mancuso, J.J.; Wherry, E.J.; Allison, J.P. Anti-CTLA-4 generates greater memory response than anti-PD-1 via TCF-1. Proc. Natl. Acad. Sci. USA 2025, 122, e2418985122. [Google Scholar] [CrossRef]
- Hassan, M.; Elzallat, M.; Mohammed, D.M.; Balata, M.; El-Maadawy, W.H. Exploiting regulatory T cells (Tregs): Cutting-edge therapy for autoimmune diseases. Int. Immunopharmacol. 2025, 155, 114624. [Google Scholar] [CrossRef]
- Tufail, M.; Jiang, C.H.; Li, N. Immune evasion in cancer: Mechanisms and cutting-edge therapeutic approaches. Signal Transduct. Target. Ther. 2025, 10, 227. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, P.T.; Saulters, E.L.; Duckworth, A.D.; Lim, Y.J.; Woolley, J.F.; Slupsky, J.R.; Cragg, M.S.; Ward, F.J.; Dahal, L.N. Soluble CTLA-4 attenuates T cell activation and modulates anti-tumor immunity. Mol. Ther. 2024, 32, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Gootjes, C.; Zwaginga, J.J.; Roep, B.O.; Nikolic, T. Functional impact of risk gene variants on the autoimmune responses in type 1 diabetes. Front. Immunol. 2022, 13, 886736. [Google Scholar] [CrossRef] [PubMed]
- Ighid, N.; El Akil, S.; Elouilamine, E.; Izaabel, E.H. A negative association of CTLA-4 genetic variant (rs11571317) with breast cancer risk in Moroccan population. Gene Rep. 2022, 26, 101513. [Google Scholar] [CrossRef]
- Zhou, B.; Chen, M.; Shang, S.; Zhao, J. Association of CTLA-4 gene polymorphisms and alopecia areata: A systematic review and meta-analysis. Biomarkers 2022, 27, 338–348. [Google Scholar] [CrossRef]
- Abdelaziz, A.I.; Abdelsameea, E.; Wahdan, S.A.; Elsherbiny, D.; Zakaria, Z.; Azab, S.S. Unveiling the nexus between direct-acting antivirals in hepatitis C virus elimination and immune response. Clin. Exp. Med. 2025, 25, 269. [Google Scholar] [CrossRef]
- Elkady, A.; Hassan, M.; Hagag, M.F.; El-Ahwany, E.; Helal, O.M.; Zoheiry, M.; Abdalla, M.A.; Elzallat, M. Innovative model of surface-enhanced Raman spectroscopy for exosomes identification: An approach for the diagnosis of hepatocellular carcinoma. Clin. Chim. Acta 2023, 540, 117228. [Google Scholar] [CrossRef]
- Elzallat, M.; Hassan, M.; Elkramani, N.; Aboushousha, T.; AbdelLatif, A.; Helal, N.; Abu-Taleb, H.; El-Ahwany, E. Nanoconjugated long non-coding RNA MEG3 as a new therapeutic approach for hepatocellular carcinoma. Heliyon 2023, 9, e15288. [Google Scholar] [CrossRef]
- Hassan, M.; Nasr, S.M.; Elzallat, M. Effect of CD133 polymorphisms on the risk of developing liver cirrhosis and hepatocellular carcinoma induced by viral hepatitis. Virus Res. 2022, 312, 198714. [Google Scholar] [CrossRef]
- Okasha, H.; Hassan, M.; Aboushousha, T.; Samir, S. Effect of Interferon-Beta (IFN-β) on tumor suppressor and apoptotic markers in hepatocellular carcinoma cell line. Int. J. Res. Pharm. Sci. 2019, 10, 2936–2943. [Google Scholar] [CrossRef]
- Ward, F.J.; Kennedy, P.T.; Al-Fatyan, F.; Dahal, L.N.; Abu-Eid, R. CTLA-4-two pathways to anti-Tumour immunity? Immunother. Adv. 2025, 5, ltaf008. [Google Scholar] [CrossRef]
- Ali, N.A.; Hamdy, N.M.; Gibriel, A.A.; EL Mesallamy, H.O. Investigation of the relationship between CTLA4 and the tumor suppressor RASSF1A and the possible mediating role of STAT4 in a cohort of Egyptian patients infected with hepatitis C virus with and without hepatocellular carcinoma. Arch. Virol. 2021, 166, 1643–1651. [Google Scholar] [CrossRef]
- Graham, D.S.C.; Wong, A.K.; McHugh, N.J.; Whittaker, J.; Vyse, T.J. Evidence for unique association signals in SLE at the CD28-CTLA4-ICOS locus in a family-based study. Hum. Mol. Genet. 2006, 15, 3195–3205. [Google Scholar] [CrossRef] [PubMed]
- Palacios, R.; Comas, D.; Elorza, J.; Villoslada, P. Genomic regulation of CTLA4 and multiple sclerosis. J. Neuroimmunol. 2008, 203, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Ueda, H.; Howson, J.M.M.; Esposito, L.; Heward, J.; Snook, H.; Chamberlain, G.; Rainbow, D.B.; Hunter, K.M.D.; Smith, A.N.; Di Genova, G.; et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003, 423, 506–511. [Google Scholar] [CrossRef]
- Goske, M.; Ramachander, V.R.V.; Komaravalli, P.L.; Rahman, P.F.; Rao, C.; Jahan, P. CTLA-4 genetic variants (rs11571317 and rs3087243): Role in susceptibility and progression of breast cancer. World J. Oncol. 2017, 8, 162–170. [Google Scholar] [CrossRef]
- Hassan, M.; Elhusseny, Y.; Agamy, F.E.; Azmy, M.M.; Balata, M. Soluble CTLA-4 and high-risk genetic variants: A new frontier in pancreatic ductal adenocarcinoma (PDAC) biomarkers. Biochem. Biophys. Rep. 2025, 44, 102304. [Google Scholar] [CrossRef]
- Darmadi, D.; Lindarto, D.; Siregar, J.; Widyawati, T.; Rusda, M.; Amin, M.; Yusuf, F.; Eyanoer, P.; Lubis, M.; Rey, I. Association between serum Cytotoxic T Lymphocyte Antigen (CTLA)-4 level and disease progression in patients With chronic hepatitis B. Med. Arch. 2023, 77, 142–145. [Google Scholar] [CrossRef]
- Liu, J.; Tian, X.; Wang, Y.; Kang, X.; Song, W. Soluble cytotoxic T-lymphocyte–associated antigen 4 (sCTLA-4) as a potential biomarker for diagnosis and evaluation of the prognosis in Glioma. BMC Immunol. 2021, 22, 33. [Google Scholar] [CrossRef]
- Osaki, M.; Sakaguchi, S. Soluble CTLA-4 regulates immune homeostasis and promotes resolution of inflammation by suppressing type 1 but allowing type 2 immunity. Immunity 2025, 58, 889–908.e13. [Google Scholar] [CrossRef] [PubMed]
- Duffy, A.G.; Ulahannan, S.V.; Makorova-Rusher, O.; Rahma, O.; Wedemeyer, H.; Pratt, D.; Davis, J.L.; Hughes, M.S.; Heller, T.; ElGindi, M.; et al. Tremelimumab in combination with ablation in patients with advanced hepatocellular carcinoma. J. Hepatol. 2017, 66, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Stein, T.I.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H.; et al. GeneCards Version 3: The human gene integrator. Database 2010, 2010, baq020. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, Y.H.; Wang, S.P.; Zhang, Y.; Huang, T.; Cai, Y.-D. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS ONE 2017, 12, e0184129. [Google Scholar] [CrossRef]
| Groups | p-Value | |||
|---|---|---|---|---|
| Control (n = 75) | HCV (n = 75) | HCV-HCC (n = 75) | ||
| Age (year) | 59.93 ± 0.93 | 61.36 ± 0.82 | 60.67 ± 0.802 | 0.500 |
| Gender | ||||
| Males | 36 (48.00%) | 35 (46.67%) | 40 (53.33%) | 0.688 |
| Females | 39 (52.00%) | 40 (53.33%) | 35 (46.67%) | |
| Smoking | ||||
| No | 40 (53.33%) | 42 (56%) | 34 (45.33%) | 0.396 |
| Yes | 35 (46.67%) | 33 (44%) | 41 (54.67%) | |
| Laboratory data | ||||
| AST (IU/L) | 23.43 ± 0.874 | 54.87 ± 3.719 | 195.17 ± 33.326 # | <0.001 |
| ALT (IU/L) | 24.68 ± 1.517 | 47.37 ± 2.78 ** | 86.63 ± 9.97 # | <0.001 |
| Total bilirubin (mg/dL) | 0.59 ± 0.03 | 2.55 ± 0.43 * | 6.45 ± 1.00 # | <0.001 |
| Serum albumin (g/dL) | 4.07 ± 0.05 | 3.40 ± 0.09 *** | 2.48 ± 0.06 # | <0.001 |
| PC (%) | 95.86 ± 0.78 | 76.18 ± 2.29 *** | 57.65 ± 2.41 # | <0.001 |
| AFP (ng/mL) | 4.42 ± 0.21 | 8.57 ± 0.53 | 16,051.87 ± 3463.00 # | <0.001 |
| Control (n = 75) | HCV (n = 75) | HCV-HCC (n = 75) | p-Value | Odds Ratio (95% CI) of HCC vs. Control | p-Value | Odds Ratio (95% CI) of HCC vs. HCV | p-Value | |
|---|---|---|---|---|---|---|---|---|
| Genotype | ||||||||
| CC | 36 (48.00%) | 33 (44.00%) | 14 (18.70%) # | <0.001 | ||||
| CT | 27 (36.00%) | 28 (37.30%) | 18 (24.00%) | |||||
| TT | 12 (16.00%) | 14 (18.70%) | 43 (57.30%) # | |||||
| Genotype | ||||||||
| CC | 36 (48.00%) | 33 (44.00%) | 14 (18.70%) # | <0.001 | 0.25 | <0.001 | 0.29 | 0.001 |
| CT + TT | 39 (52.00%) | 42 (56.00%) | 61 (81.30%) # | (0.12–0.52) | (0.14–0.61) | |||
| Genotype | ||||||||
| CT | 27 (36.00%) | 28 (37.30%) | 18 (24.00%) | 0.158 | 0.56 | 0.109 | 0.53 | 0.077 |
| CC + TT | 48 (64.00%) | 47 (62.70%) | 57 (76.00%) | (0.28–1.14) | (0.26–1.08) | |||
| Genotype | ||||||||
| TT | 12 (16.00%) | 14 (18.70%) | 43 (57.30%) # | <0.001 | 7.06 | <0.001 | 5.86 | <0.001 |
| CC + CT | 63 (84.00%) | 61 (81.30%) | 32 (42.70%) # | (3.27–15.21) | (2.80–12.26) | |||
| Allele | ||||||||
| C | 99 (66.00%) | 94 (62.70%) | 46 (30.70%) # | <0.001 | 0.23 | <0.001 | 0.26 | <0.001 |
| T | 51 (34.00%) | 56 (36.30%) | 104 (69.30%) # | (0.14–0.37) | (0.16–0.43) |
| Control (n = 75) | HCV (n = 75) | HCV-HCC (n = 75) | p-Value | Odds Ratio (95% CI) of HCV vs. Control | p-Value | Odds Ratio (95% CI) of HCC vs. Control | p-Value | |
|---|---|---|---|---|---|---|---|---|
| Genotype | ||||||||
| CC | 5 (6.70%) | 37 (49.30%) ** | 31 (41.33%) ** | <0.001 | ||||
| CT | 19 (25.30%) | 30 (40.00%) | 28 (37.33%) | |||||
| TT | 51 (68.00%) | 8 (10.70%) ** | 16 (21.33%) ** | |||||
| Genotype | ||||||||
| CC | 5 (6.70%) | 37 (49.30%) ** | 31 (41.30%) ** | <0.001 | 13.63 | <0.001 | 9.86 | <0.001 |
| CT + TT | 70 (93.30%) | 38 (50.70%) ** | 44 (58.70%) ** | (4.95–37.58) | (3.57–27.27) | |||
| Genotype | ||||||||
| CT | 19 (25.30%) | 30 (40.00%) | 28 (37.30%) | 0.131 | 1.97 | 0.055 | 1.76 | 0.113 |
| CC + TT | 56 (74.70%) | 45 (60.00%) | 47 (62.70%) | (0.98–3.94) | (0.87–3.54) | |||
| Genotype | ||||||||
| TT | 51 (68.00%) | 8 (10.70%) ** | 16 (21.30%) ** | <0.001 | 0.06 | <0.001 | 0.128 | <0.001 |
| CC + CT | 24 (32.00%) | 67 (89.30%) ** | 59 (78.70%) ** | (0.02–0.14) | (0.06–0.27) | |||
| Allele | ||||||||
| C | 29 (19.30%) | 104 (69.30%) ** | 90 (60.00%) ** | <0.001 | 9.43 | <0.001 | 6.26 | <0.001 |
| T | 121 (80.70%) | 46 (30.70%) ** | 60 (40.00%) ** | (5.53–16.08) | (3.72–10.53) |
| Control (n = 75) | HCV (n = 75) | HCV-HCC (n = 75) | Odds Ratio (95% CI) of HCV vs. Control | p-Value | Odds Ratio (95% CI) of HCC vs. Control | p-Value | Odds Ratio (95% CI) of HCC vs. HCV | p-Value | |
|---|---|---|---|---|---|---|---|---|---|
| Genotype | |||||||||
| GG | 14 (18.70%) | 34 (45.30%) | 52 (69.30%) | ||||||
| GA | 32 (42.70%) | 23 (30.70%) | 16 (21.30%) | ||||||
| AA | 29 (38.70%) | 18 (24.00%) | 7 (9.30%) | ||||||
| Genotype | |||||||||
| GG | 14 (18.70%) | 34 (45.30%) ** | 52 (69.30%) **, @ | 3.61 | <0.001 | 9.85 | <0.001 | 2.73 | 0.003 |
| GA + AA | 61 (81.30%) | 41 (54.70%) ** | 23 (30.70%) **, @ | (1.73–7.56) | (4.61–21.07) | (1.40–5.32) | |||
| Genotype | |||||||||
| GA | 32 (42.70%) | 23 (30.70%) | 16 (21.30%) * | 0.59 | 0.127 | 0.36 | 0.005 | 0.61 | 0.193 |
| GG + AA | 43 (57.30%) | 52 (69.30%) | 59 (78.70%) * | (0.30–1.16) | (0.18–0.75) | (0.29–1.28) | |||
| Genotype | |||||||||
| AA | 29 (38.70%) | 18 (24.00%) | 7 (9.30%) **, $ | 0.50 | 0.053 | 0.16 | <0.001 | 0.33 | 0.016 |
| GG + GA | 46 (61.30%) | 57 (76.00%) | 68 (90.70%) **, $ | (0.25–1.01) | (0.07–0.40) | (0.13–0.84) | |||
| Allele | |||||||||
| G | 60 (40.00%) | 91 (60.70%) ** | 120 (80.00%) # | 2.31 | <0.001 | 6 | <0.001 | 2.59 | <0.001 |
| A | 90 (60.00%) | 59 (39.30%) ** | 30 (20.00%) # | (1.46–3.68) | (3.58–10.06) | (1.55–4.35) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, M.; El-Maadawy, W.H.; Fahim, S.A.; Youssef, S.M.; Badran, O.M.; Balata, M. Integrating CTLA-4 Genetics and Soluble Isoforms for the Stratification of HCV-Related Hepatocellular Carcinoma Risk and Aggressiveness. Int. J. Mol. Sci. 2025, 26, 11067. https://doi.org/10.3390/ijms262211067
Hassan M, El-Maadawy WH, Fahim SA, Youssef SM, Badran OM, Balata M. Integrating CTLA-4 Genetics and Soluble Isoforms for the Stratification of HCV-Related Hepatocellular Carcinoma Risk and Aggressiveness. International Journal of Molecular Sciences. 2025; 26(22):11067. https://doi.org/10.3390/ijms262211067
Chicago/Turabian StyleHassan, Marwa, Walaa H. El-Maadawy, Sally A. Fahim, Sherihan M. Youssef, Omaima Mostafa Badran, and Mahmoud Balata. 2025. "Integrating CTLA-4 Genetics and Soluble Isoforms for the Stratification of HCV-Related Hepatocellular Carcinoma Risk and Aggressiveness" International Journal of Molecular Sciences 26, no. 22: 11067. https://doi.org/10.3390/ijms262211067
APA StyleHassan, M., El-Maadawy, W. H., Fahim, S. A., Youssef, S. M., Badran, O. M., & Balata, M. (2025). Integrating CTLA-4 Genetics and Soluble Isoforms for the Stratification of HCV-Related Hepatocellular Carcinoma Risk and Aggressiveness. International Journal of Molecular Sciences, 26(22), 11067. https://doi.org/10.3390/ijms262211067







