Abstract
The adoption of liquid biopsy approaches in clinical practice has triggered a significant paradigm shift in the diagnostic, prognostic, and predictive outcomes for cancer patients. Circulating tumor DNA (ctDNA) is considered a valuable biomarker for monitoring tumor burden and its mutational dynamics. In this context, not all cell-free DNA (cfDNA) molecules are derived from tumor cells. Furthermore, due to tumor heterogeneity, not all ctDNA molecules contain cancer-associated alleles, complicating the direct quantification of the circulating tumor allele fraction (cTF) within the total cfDNA. Cancer arises from the accumulation of multiple genetic and epigenetic changes. Each of these molecular features can be exploited as the basis of methodological strategies used in ctDNA quantification. Different layers of omics data, from genomics, evaluating mutational analysis of somatic single-nucleotide variants and copy number alterations, to epigenomics, primarily consisting of the evaluation of methylation profiles and fragmentation patterns, can be used for this purpose. Some of these approaches can be effective in a multi-modal manner. To date, the quantification approaches for estimating cTF vary enormously, making direct comparisons and an assessment of their translational value challenging. Moreover, the lack of regulatory approval for many of these assays is a critical barrier to their widespread clinical adoption. This review explores the different omics approaches described for ctDNA quantification, outlining strengths and limitations, and highlighting their valuable applications in clinical settings.
Keywords:
genomics; epigenomics; transcriptomics; omics; ctDNA; cTF; cfDNA; liquid biopsy; methylation; NGS 1. Introduction
Liquid biopsy (LBx) involves the comprehensive evaluation of circulating biomolecules as circulating tumor cells (CTCs), cell-free DNA (cfDNA), cell-free RNA (cfRNA), proteins, lipids, and metabolites [1]. LBx has gained an increasingly important role in various clinical contexts, notably in non-invasive prenatal testing (NIPT) and as a significant non-invasive biomarker in oncology [2]. Focusing on cfDNA, an increase in its total content in body fluids can be linked to several physiological or para-physiological conditions, including pregnancy and transplantation, as well as pathological states like diabetes, inflammation, sepsis, and autoimmune processes [3,4,5]. Circulating tumor DNA (ctDNA) constitutes the fraction of the total cfDNA that is derived from tumor cells [6]. The current and potential clinical applications of LBx in oncology range from the early detection and risk stratification of cancer to monitoring treatment response, which allows for the detection of molecular resistances and minimal residual disease (MRD) [2]. Additionally, an ambitious approach involves multicancer early detection (MCED), where LBx serves as a marker for the early identification of a malignant event in seemingly healthy individuals [7]. One of the key advantages of ctDNA analysis is its ability to provide real-time monitoring and to reflect the dynamic genomic landscape of cancer, offering insights into tumoral heterogeneity without the need for solid biopsies of invasive tissue (TBx) [8,9]. Furthermore, total ctDNA levels have been proven to have prognostic value and are strongly correlated to clinical features and outcomes [10,11,12,13,14].
In the last decade, the establishment of the NIPT, with its evaluation of fetal cfDNA in maternal plasma, has supported and promoted the parallel improvement of LBx, particularly from a methodological standpoint [15]. Despite long-standing efforts, the widespread clinical application of LBx has been slow in most contexts. This is primarily due to a few major challenges, such as the complexity of adopting and standardizing established methods and the need for a critical interpretation of the molecular results. In the plethora of clinical applications and studies, the effective discrimination of the ctDNA ratio from the cfDNA background is critical for obtaining reliable and reproducible results, especially when interpreting a true negative genotyping. In LBx, the absence of tumor-specific alterations may genuinely reflect the tumor genotype (i.e., true negative) or may be a result of insufficient ctDNA shedding, low content, or inadequate detection sensitivity (i.e., false negative) [6]. For this reason, the Food and Drug Administration (FDA) label for approved LBx applications recommended a reflex test for TBx for negative LBx results across several tumor types [16].
With these premises, the accurate estimation of the ctDNA rate, and thus the circulating tumor allele fraction (cTF), is a challenging topic when it comes to ensuring the actionability of molecular findings from LBx. To advance this field, an improved understanding of the biological properties of cfDNA has accelerated the development of new diagnostic and methodological tools.
To date, the literature lacks an established and well-accepted consensus on the best methodological practice for ctDNA quantification. This review describes the main approaches and methodologies currently available to estimate the ctDNA fraction, while also providing an overview of the biological characteristics of ctDNA that are relevant to the purpose of this manuscript. The ultimate goal is to provide clinicians and experts with a valuable reference of the current state of the art, underscoring the need for clear guidelines on this topic.
2. Biological Characteristics of cfDNA and ctDNA
cfDNA refers to fragmented DNA molecules that circulate in body fluids like blood plasma and serum. These fragments originate from various physiological processes, including apoptosis, necrosis, and active secretion [17,18,19,20]. Subcomponents of cfDNA primarily consist of nuclear and mitochondrial DNA. Minor biological fractions are also present, such as circular DNA (eccDNA), microDNA, and DNA derived from viruses, bacteria, and food (plants and meat) [21,22,23]. ctDNA is a fraction of cfDNA specifically derived from tumor cells. It ideally carries the same genetic background as the primary tumor and metastatic lesions, making it a non-invasive source of cancer molecular information [2]. Due to the heterozygosity and the tumor heterogeneity, not all ctDNA molecules at a specific locus contain the mutated tumor allele. For this reason, we refer to the term cTF as the fraction of cfDNA that originates from the tumor and contains a tumor-specific allele [7]. Therefore, cTF comprises ctDNA fragments containing tumor-specific somatic single-nucleotide variants (SNVs), copy number alterations (CNAs), or an abnormal epigenomics profile. The cTF is commonly used as a measure of ctDNA signal in many LBx cancer detection tests. Typically, cfDNA fragments range in size from 100 to 800 base pairs, with a prominent peak around 160–180 base pairs, which corresponds to the length of DNA wrapped around a nucleosome [24]. Some individual cfDNA molecules, however, can be much larger, exceeding 20–30 kb [25,26,27]. The ctDNA fragments are often shorter, typically around 130–150 base pairs [24]. The concentration of cfDNA in the blood of healthy subjects ranges between 0 and 100 ng/mL [3,25,26,27]. Studies on tissue-specific methylation patterns in healthy individuals have shown that most cfDNA is of hematopoietic origin, with a fraction also originating from solid tissues as vascular endothelial cells, neurons, and hepatocytes [28,29,30]. In cancer patients, cfDNA levels can rise more than 20-fold, sometimes exceeding 1000 ng/mL, with the ctDNA fraction ranging from 0.1 to 89%. This proportion may increase with disease progression [25,31,32,33,34]. The variability in ctDNA levels depends on several factors, including the tumor type and stage. High ctDNA shedding is observed in some advanced cancers (e.g., non-small cell lung cancer, colorectal cancer, breast cancer, etc.), while lower release occurs in others (e.g., primary brain, renal, and thyroid cancers) [31,35]. Early-stage tumors or benign lesions typically shed lower amounts of cfDNA, which poses a challenge for the clinical use of liquid biopsy in these settings [36,37]. An additional consideration for cfDNA/ctDNA detection is the balance between their release and clearance, which is primarily mediated by the liver and kidneys. In healthy individuals, efficient clearance mechanisms partially account for the low cfDNA levels. The increased content of necrotic/apoptotic cells and the chronic inflammatory context affect these clearance mechanisms, which is a contributing factor to the increased cfDNA/ctDNA concentrations [23].
These biological features and the differences between cfDNA and ctDNA are the conceptual basis for developing and adopting different strategies to estimate the tumor fraction rate of cfDNA.
3. Circulating Tumor Fraction Estimation Technologies
First and foremost, it is important to emphasize that the ability to accurately estimate the cTF from total cfDNA begins with the proper collection of bodily fluid and the efficient isolation of nucleic acids. These pre-analytical steps are crucial for cTF estimation because they can significantly affect the generation of background noise, thereby limiting the probability of analyzing a true tumor-derived signal [1]. A critical analysis of these pre-analytical variables and their impact on LBx performance is beyond the scope of this review, and valuable literature is available [38,39,40].
To the best of our knowledge, there is currently no well-recognized method to discriminate between ctDNA and cfDNA during the nucleic acid isolation step, primarily because both types of nucleic acids share similar characteristics and are extracted under the same conditions. A number of molecular and genomic characteristics can be derived from cfDNA and used to infer the cTF, each with its own performance metrics and limitations. The most common cfDNA post-isolation approaches exploit the evaluation of the following: (1) tumor-specific mutations, (2) quantitative changes, (3) methylation patterns, and (4) fragment profiles.
Most of the technologies described here are based on next-generation sequencing (NGS) coupled with specialized bioinformatics pipelines for data analysis. The success of the NGS approach in estimating cTF relies on its sensitivity and specificity to detect the often low levels of ctDNA. It also allows for the identification of a wide range of rare genomic alterations, including SNVs and CNAs, across the entire genome or within targeted regions. Just as in somatic variants’ identification from a solid biopsy, unique molecular identifiers (UMIs) are incorporated into the common NGS library preparation workflows to reduce background noise and enable the detection of rare variants, especially with low amounts of starting ctDNA. In addition to genetic features, epigenetic modifications have also gained attention as a valuable source of cTF information. Generally speaking, epigenetic signatures have emerged as a superior marker, especially in early-stage settings, when compared with genetic screening [41].
Significant differences in the adoption and optimization of specific approaches for cTF estimation depend on the type of underlying tumor-derived molecular alteration being analyzed (i.e., SNVs, CNAs, or epigenetic features).
3.1. Genomics Analysis of the Circulating Tumor Fraction: Somatic Single-Nucleotide Variants
Early attempts to infer the tumor fraction of cfDNA relied on a personalized approach, where a specific somatic single-nucleotide variant (SSNV) identified in a patient’s TBx was subsequently monitored in their plasma samples over time [42]. Using this targeted approach, the cTF was typically inferred according to the variant allele frequency (VAF) or mutant allele frequency (MAF). These parameters represent the percentage of NGS reads that contain the mutant allele relative to the total number of reads at a given locus [43]. The AF-based approach is a robust and useful method for clinical purposes, allowing for the dynamic monitoring of one or a few given SSNVs. The clinical value of AF as an independent prognostic and predictive biomarker has been well-documented in numerous studies [9,44,45,46,47,48].
Methods for inferring cTF using SSNVs include digital PCR (i.e., droplet digital PCR (ddPCR) and beads, emulsions, amplification, and magnetics (BEAMing)), optimized quantitative real-time PCR (AS-NEPB-PCR, PNA-LNA PCR clamp, COLD-PCR), and targeted deep sequencing (>5000× coverage) with or without error suppression [31,49,50,51]. These approaches are sensitive and fast, capable of quantifying mutant DNA copies down to a 0.01% AF. As a proof-of-concept study, the TRACERx clinical trial used a targeted multiplex PCR approach to monitor ctDNA after whole-exome sequencing genotyping of TBx. The study demonstrated that in 92.9% of early-stage lung cancer relapse cases, ctDNA was detected 70 days before clinical computed tomography (CT) scan confirmation [17].
However, this type of targeted approach requires both TBx and LBx analyses and, in some cases, germline sequencing data. This makes it time-consuming, relatively expensive due to the need for a patient-specific custom design, and not always feasible. In addition, it does not overcome issues related to the unviability of TBx in some clinical contexts (e.g., small biopsy, challenging tissue site location, etc.) and limits the monitoring of tumor evolution and heterogeneity over time [8]. It is also possible to adopt broader, tissue-agnostic NGS multi-gene panels. These approaches use optimized analytical and bioinformatics workflows that overcome critical biases, such as background noise from sources like clonal hematopoiesis of indeterminate potential (CHIP) [52]. In these TBx-free NGS approaches, the evaluation of cTF at multiple loci is combined into a single estimation [7]. To increase assay sensitivity, several optimized strategies have been developed to enrich the proportion of tumor-derived DNA before targeted deep NGS. In 2012, Forshew et al. described the tagged-amplicon deep sequencing technology (TAm-Seq) as a highly sensitive approach able to detect low-frequency mutations (as low as 2%) in cfDNA from patients with high-grade serous ovarian cancer [53]. Since then, several high-sensitivity NGS-based methods have been developed, such as the enhanced TAm-Seq (eTAm-Seq™) technology [54] and the error-suppressed multiplexed deep sequencing method. This method distinguishes true mutations in ctDNA from amplification artifacts and sequencing errors, increasing detection sensitivity to as low as 0.02% [55]. Cancer personalized profiling by deep sequencing (CAPP-Seq) is another approach designed for improved sensitivity and specificity (up to 0.02%) [56]. In the single-molecule amplification and resequencing technology (cSMART) assay, each cfDNA molecule is uniquely barcoded and universally amplified. The amplification products are then circularized and reamplified in a target-specific manner to enrich a pool of target sequences containing hotspot mutations [57,58]. The cSMART analysis has been successfully adopted in the DYNAMIC study, which proved that the clearance of ctDNA in patients with lung cancer accurately reflects real-time tumor burden in response to therapy. In this study, the ctDNA rate was calculated by multiplying the MAF% by the cfDNA concentration [59].
For estimating ctDNA rates using large, high-sensitivity NGS panels, the maximum somatic allele frequency (MSAF) is often used as a surrogate for ctDNA calculation. Most cfDNA molecules are wild-type; therefore, the median MAF% of somatic alterations is generally low (<0.5%). To infer the ctDNA fraction, parameters such as the higher sum VAF of genomic alterations in ctDNA (%ctDNAsum) and the higher maximum VAF of genomic alterations in ctDNA (%ctDNAmax) have been proposed. The %ctDNAsum represents the sum of the VAF% of the individual alterations identified in the ctDNA (excluding the variants of unknown significance). The %ctDNAmax represents the maximum VAF% of the alterations identified in the ctDNA (excluding the variants of unknown significance). Several studies correlated high loads of genomic alterations, represented by high %ctDNAsum and %ctDNAmax values, with clinical outcomes in various oncological contexts [60,61,62,63,64,65,66]. The use of such a parameter for the ctDNA% estimation is part of several commercial kits, such as the ctTSO500HT (Illumina, San Diego, CA, USA). While the use of large, targeted gene panels expands the ability to evaluate tumor dynamics over time with great sensitivity, the limitations related to the choice of the gene panel content still remain [67].
In addition to a targeted deep sequencing approach, an example of optimized low-coverage genome-wide NGS approaches has been described. One such example is the GEMINI (GEnome-wide Mutational Incidence for Non-Invasive detection of cancer) tool, which consists of sequencing individual cfDNA molecules to enrich for ctDNA somatic mutations without requiring a matched blood and tumor sequencing [68]. Broader sequencing approaches for cTF quantification using SSNV include whole-exome sequencing (WES) and whole-genome sequencing (WGS) (with a sensitivity of 5–10%). These methods ideally allow for the investigation of unknown and novel acquired somatic variants, maximizing the study of tumor evolution. However, the need for high-coverage sequencing to detect alterations with low VAF% limits the clinical applicability of such broader approaches being expensive and not always feasible [67].
Generally speaking, the SSNVs-based approach for ctDNA estimation may be affected by structural alterations occurring in the same genomic region. Additionally, it appears to be less than ideal in early-stage cancer settings due to the following: (1) the expected low somatic mutational rate; (2) the generally low shedding rate; and, consequently, (3) the low proportion of cfDNA fragments harboring somatic tumor-specific alteration, which requires continuous methodological optimization [31,68].
3.2. Genomics Analysis of the Circulating Tumor Fraction: Somatic Copy Number Alterations
Another NGS-based approach for calculating the cTF consists of the evaluation of the SCNAs (somatic copy number alterations and large-scale aneuploidies) in cfDNA using WGS. SCNAs are defined as chromosome-level changes, such as deletions or amplifications of large genomic regions, including both sub-chromosomal parts (i.e., focal CNAs) and entire chromosome arms (aneuploidy) [69]. Compared to SSNVs, the analysis of SCNAs may be more broadly applicable, as it is not dependent on a specific sequence variant and most cancers harbor arm-level CNAs [70].
Some WGS approaches for cTF calculation using SCNAs require a high-coverage depth of up to 100× [71,72]. While this deep-coverage WGS data analysis, which also provides allelic information for sequence variants, is considered the most established SCNA-based method for cTF estimation, it is cost-intensive and requires high-quality cfDNA, making its application in routine clinical pipelines challenging. To overcome these limitations, approaches based on low-pass and ultra-low-pass WGS (LP/ULP-WGS) or shallow WGS (sWGS), with a typical median depth of 0.1×–1×, were adopted as a rapid and low-cost method [73,74]. A number of bioinformatics teams have developed algorithms and computational approaches for SCNA analysis from LBx sWGS data, including ABSOLUTE [71], TITAN [72], QDNAseq [75], WisecondorX [76], SAMURAI [77], PRINCe [78], and ACE [79], among others. These tools are often integrated into custom pipelines to infer cTF. Adalsteinsson and colleagues developed ichorCNA, a ULP-WGS (0.1x) software package that predicts SCNA segments and estimates the tumor fraction (https://github.com/broadinstitute/ichorCNA (accessed on 10 November 2025)). With a lower limit of detection of 0.03, ichorCNA achieves a sensitivity of 0.95 for detecting the presence of a tumor and a specificity of 0.91 for classifying a healthy donor. At a tumor fraction threshold of 0.1, the approach reaches 91% sensitivity and 100% specificity [80]. The successful application of sWGS for detecting aneuploidy in NIPT, which identifies fetus-derived DNA in maternal plasma, paved the way for similar methodological approaches for detecting tumor-associated CNAs [81]. Interestingly, several authors have reported the incidental detection of presymptomatic maternal occult malignancies in this setting [82]. In line with this, commercial kits, such as the Ion ReproSeq PGS™ preimplantation genetic testing kit (ThermoFisher, Waltham, MA, USA), coupled with ichorCNA software (https://github.com/broadinstitute/ichorCNA (accessed on 10 November 2025)), have been successfully used for ctDNA rate calculation [83]. Recently, Lakatos and colleagues developed the LiquidCNA tool for cTF estimation from sWGS, which also allows for the longitudinal tracking of SCNAs across samples from the same patient to characterize subclonal tumor evolution [84]. Other sWGS approaches for analyzing tumor-associated copy number changes include Plasma-Seq [73] and the ThruPLEX® Tag-Seq Kit (Takara, Shiga, Japan) [85]. Interestingly, even third-generation sequencing approaches like Nanopore technology have been successfully used for cTF estimation. While long-read sequencing is generally not ideal for analyzing short cfDNA fragments, additional methodological steps (e.g., enrichment using a specific bead/sample ratio) have improved the detection of CNA profiles from plasma cfDNA [86]. ULP-WGS has been optimized for detecting cancer and estimating cTF, even when the expected tumor fraction is low (i.e., <20%). It is noteworthy, however, that researchers have reported a potential underestimation of the ctDNA rate in cases of theoretically high tumor fractions (i.e., metastatic settings). It has been proposed that this is due to the impact of copy-neutral loss of heterozygosity events that are indistinguishable from truly copy-neutral segments using the resolution of ULP-WGS, resulting in lower tumor fraction estimation [87,88]. Several ULP-WGS applications for ctDNA estimation are available for metastatic prostate [12] or breast cancers [11,80,83]. The ichorCNA algorithm has also been applied to hepatocellular carcinoma (HCC) with aggressive clinical behavior, although it appears not to be sensitive enough for early HCC detection [89,90].
It is important to note that cTF measured using SCNAs is based on quantitative alteration occurring on a genome-wide scale. When compared to the SSNVs-based method, the cTF rate may differ from the observed VAF%. This can be explained by the fact that VAF% is affected by small copy number variation events, especially if they do not occur in copy-number-neutral regions, making the two methods not always comparable [91]. Additionally, it has been reported that SCNAs may fail to provide a robust estimation of cTF due to the lack of wide aneuploidy and chromosomal instability in some tumor types [92,93].
A summary of genomics-based approaches adopted for cTF estimation is collected in Table 1.

Table 1.
Summary of genomics approaches applied to circulating tumor fraction estimation.
3.3. Epigenomics Analysis: Methylation Pattern
DNA methylation is an epigenetic mark that plays a key role in gene expression regulation and in genomic stability. It typically involves the formation of 5-methylcytosine (5mC) through the covalent addition of a methyl group to a cytosine base, primarily in cytosine-guanine dinucleotide (CpG)-rich sequences [99]. More recently, analysis based on the 5-hydroxymethylcytosine (5hmC) has also gained attention. The 5hmC is an intermediate in the process of DNA demethylation consisting in a modified cytosine generated through the oxidation of 5mC [100]. DNA methylation occurs early in tumor cells, and its profile is dynamically regulated during tissue development and differentiation, resulting in tissue- and cell-specific gene expression [101]. The role of the methylation process in cancer development and progression is well documented, though many aspects remain unclear. Abnormal hypo- and hyper-methylation lead to novel oncogenic properties, primarily linked to tumor suppressor gene silencing and chromosomal instability [102]. These aberrations can be seen as follows: (1) a global hypomethylation profile, explained by the higher transcriptional activity of cancer cells; and (2) focal hypermethylation spots occurring at specific gene promoters [103]. The DNA methylation patterns that characterize tumor cells are also preserved in shedding ctDNA. The methylation status of the cfDNA has the theoretical advantages of being stable over solid tumor development and indicative of both cancer presence and tissue of origin. This is important in clinical practice, as some DNA methylation alterations are consistent across various cancer types, while others are unique to a specific cancer [104]. In this context, the term tumor methylated fraction (TMeF) can be used to refer to the cTF with cancer-specific methylated aberrations. The rationale for ctDNA quantification using methylation patterns is based on the existence of differentially methylated regions (DMRs) associated with tumors, used to differentiate cfDNA from cancer-derived ctDNA.
Overall, the value of detecting and quantifying TMeF has been described in the contexts of early cancer diagnosis (MCED), the monitoring of disease response, and progression [68,105,106,107]. Generally, the evaluation of epigenetic markers has been described as superior to SSNVs for early-stage cancer diagnosis [42,108,109]. For instance, the Galleri bisulfite methylation test, applied to 4077 patients, reported an overall sensitivity of 90.1% for the detection of cancer Stage IV but only 27.5% for Stage I and II [110], data that were confirmed in the SYMPLIFY study [111,112].
From a methodological perspective, similar to SSNVs, LBx studies adopting methylation analysis can employ either a targeted or a broader, whole-genome approach [113]. Additionally, methods fall into two main categories, conversion-based and conversion-free. Conversion of unmethylated cytosines to uracil using bisulfite treatment or enzymatic methods before PCR and sequencing is the most common technique for cfDNA methylation analysis. However, it requires chemical treatments that can lead to significant molecule loss and low sequencing efficiency [114]. Alternatively, non-conversion-based methods allow for the targeted capture of specific methylated DNA regions. Examples include the cell-free methylated DNA immunoprecipitation (cfMeDIP) and the cell-free methyl-binding domain (cfMBD). These approaches use 5mC antibodies to enrich methylated cfDNA fragments and identify methylation levels and TMeF through fluorescence [115,116]. Some authors have noted that capture-based, conversion-free methods are less sensitive in the analysis of hypomethylated regions, which may be relevant for determining the tissue of origin of ctDNA [28]. Most methylation-based quantification methods in LBx focused on identifying preselected panels of CpG sites, with differentiated methylation status known to be relevant in a specific clinical context. These approaches generally require prior tissue methylation profiling (tumor vs. normal) during the assay design stage [117]. Methylation-specific qPCR has been developed to analyze one or a few tumor-specific loci that show a correlation with clinical outcomes [118,119,120,121,122,123]. Successful applications have been described in HCC, where approaches effectively detected the presence of HCC and distinguished it from benign hepatic lesions [124]. The SEPT9 bisulfite- and PCR-based assay is FDA approved for colorectal cancer (CRC) screening [125]. However, meta-analyses suggest that the SEPT9 methylation test is inferior to existing fecal immunohistochemical assays in asymptomatic cohorts [126]. An example of a targeted digital PCR approach is the TriMeth assay, which targets three CRC-specific methylation markers (C9orf50, CLIP4, and KCNQ5) [127].
In addition to these successful examples, PCR-based or capture-based approaches can be unfeasible for large multiplex analysis of many targets and are not ideal for broad ctDNA quantitation [128]. Recently, an NGS-based assay covering more than 500 DMRs was developed and successfully applied in several studies [124,129]. This assay, named Northstar Response, compares the count of methylated cfDNA molecules at specific genomic locations, known to be hypermethylated in cancer tissue, to those in normal tissue. To accurately count the number of methylated molecules at each location, the assay leverages quantitative counting templates (QCTs) designed to amplify similarly to sample molecules during the PCR step [130]. Methylation-based quantitation is included in approved commercial kits (Galleri®, GRAIL, Epi proColon 2.0, Menlo Park, CA, USA). The Guardant Reveal® assay (Guardant Health, Palo Alto, CA, USA) estimates cTF by evaluating 300 DMR sites using a bisulfite-free NGS approach. The cTF is estimated by normalizing the count of methylated molecules from cancer-specific DMRs with the count of methylated molecules from a matched control region using a proprietary algorithm.
Genome-wide shotgun massively parallel bisulfite sequencing is successfully used in several tumor types with the theoretical advantage of requiring only low-depth sequencing [131,132]. This is because the methylation changes are pervasive in the genomes and can be evaluated with a lower number of reads compared to other structural or sequence changes. Some authors have demonstrated the feasibility of this approach with a sequencing depth as low as 10 million reads, making it a relatively cost-effective option [131]. NGS applied to methylation analysis requires specific bioinformatics pipelines, such as Methy-Pipe [133], which counts methylated cytosines along the sequencing reads. Methylation-sensitive restriction enzyme digestion followed by sequencing (MRE-Seq) has also proven to be useful for investigating the cancer signal origin (CSO) using a deep neural network (DNN) analysis in CRC and lung cancer [134]. Overall, analytical workflow that allows for the simultaneous analysis of sequence variants (SSNVs) and methylation status of ctDNA are ideal and the most attractive [135]. Examples of long-read sequencing (third-generation sequencing) are also available in the literature. For instance, the Oxford Nanopore technology directly analyses the native cfDNA molecule without the need for PCR or enrichment steps and provides information about both methylation status and genomic alterations simultaneously.
3.4. Epigenomics Analysis: Fragmentomics
The term “cfDNA fragmentome” refers to the genome-wide profile of cfDNA fragments, including size, distribution, breakpoint locations, and end motif profile. This information provides a comprehensive view of the genomic, epigenomic, and transcriptomic state of the cfDNA, being based on nucleosome positioning with respect to nucleosome center, chromatin structure, and nuclease activity during cell death [136]. The fragmentation of cfDNA is a highly conserved process across mammals, influenced by the nucleosome structure that protects the DNA from cleavage. It is known that cfDNA in human plasma is nonrandomly fragmented by caspase enzymes [137]. In fact, the association of DNA molecules with nucleosomes, the basic unit of DNA packaging, avoids the fragmentation process and protects the nucleic acid. The activity of caspase explains the common profile observed in cfDNA. In healthy subjects, cfDNA fragments generally showed a mode of ~167 bp, in which ~147 bp are wrapped and protected around the nucleosome, with a 10 bp periodicity corresponding to a turn of the DNA helix, and 20 bp consisting of a linker region site of cleavage [22,138,139,140,141]. The cfDNA profile of patients with cancer is characterized by a significant increase in shorter fragments (size range of 20 to 150 bp) and a length reduction predominantly at the C-end [131,142,143]. This can be explained by a more open chromatin state in cancer cells due to greater transcriptional activity, which makes more sites accessible for cleavage [144]. Variability in fragment size distribution has been observed across different cancer types. Mouliere and colleagues observed an enrichment of shorter fragments (20–150 bp) in plasma samples obtained from patients affected by melanoma, breast, ovarian, lung, CRC, and cholangiocarcinoma cancers (called “high ctDNA cancers”), but not in renal, glioblastoma, bladder, or pancreatic ones (called “low ctDNA cancers”) [145]. Additionally, a link between fragmentomic patterns and tissue-specific gene expression has been demonstrated [145,146,147]. The abovementioned two main characteristics of ctDNA, the shortening of fragments and the lowered C-end predominance, have been adopted to distinguish and quantify cTF [137]. While methods like capillary electrophoresis and ddPCR can provide general information on fragment length, a broader approach like WGS allows for a more sensitive assessment of the global cfDNA fragmentation pattern [148,149]. Recently, targeted approaches have also been developed, focusing on fragmentation patterns within the coding regions of oncogenes and oncosuppressors [150]. Methods to evaluate fragmentomics and infer cTF include the following: (1) deep sequencing with molecular barcodes and bioinformatics-based error correction; (2) size selection strategies for the enrichment of a subpopulation of fragments; and (3) low-coverage WGS combined with machine learning analysis. Most studies in the literature have focused on the selection of short fragments (90–150 bp) to enrich for the ctDNA fraction, based on differences in fragment size distribution across a genome [144,151]. The size selection approach has also been used for genomic or methylation-based studies, offering the integration of multiple omics in the same experiment, with the aim of improving the sensitivity of a downstream assay. Wang and colleagues, for example, adopted an optimized cfMeDIP-seq approach to gather information on both methylation and fragmentation patterns in breast cancer [152]. Mouliere and co-authors developed a “double step” of cfDNA size selection. At first, they select 90–150 bp fragments of cfDNA from an agarose cassette, which were then used as an input for sWGS with CNV analysis, followed by a second size selection based on in silico NGS read filtering [153]. Similar enrichment methods have been achieved by excising bands of an appropriate size from polyacrylamide gels, resulting in a significant enrichment of the ctDNA fraction, from 2.5-fold to 9.1-fold [154]. Enrichment methods can also be applied to sequencing reads using bioinformatics pipelines, such as with the INVAR, that integrate variant reads and signal enrichment based on biological fragmentation patterns of ctDNA [155]. Other enrichment techniques include the single-strand binding polymerase chain reaction (SSB-PCR), probe detection systems based on IV endonuclease, rolling circle amplification, and hybrid capture [156]. Recently, cfDNA fragmentome analysis obtained using an LP-WGS combined with the machine learning pipeline DNA evaluation of fragments for early interception (DELFI) demonstrated high sensitivity for ctDNA quantification and early detection in several clinical settings, such as ovarian [136], lung [157,158], and liver [159], among others [69,152,160,161,162]. To note, Markus and colleagues reported the utility of the fragmentomic analysis in ovarian cancer samples not only in the evaluation of fragments shorter than mono-nucleosomes (~167 bp), but also shorter than di-nucleosomes (~240–330 bp). They also observed an enrichment of DNA fragments that start and end at the border or within the nucleosome core in the ctDNA rate. This information sheds light on the complexity of the biological characteristics of ctDNA fragmentation, underlining the utility of an integrated approach to detect tumor-derived fragments [94]. Further information related to ctDNA fragments, such as the analysis of ctDNA-preferred ends, has been explored in multiple studies and relies on the identification of tumor-associated cfDNA-preferred end coordinates and abundance using the NGS approach [163,164]. The integration of both fragmentomic information and epigenetic cfDNA features seems to be a good marker of early cancer detection and residual disease monitoring, boosting the diagnostic capabilities [165]. Furthermore, by combining fragmentomic information and SCNAs, Janke and colleagues demonstrated the utility of multi-omics data integration in the prediction of therapy response in advanced lung cancer at different time points [166].
A summary of epigenomics-based approaches adopted to cTF estimation is collected in Table 2.

Table 2.
Summary of epigenomics approaches applied to circulating tumor fraction estimation.
4. Integration of Multi-Omics Data
The successful implementation of a multimodal strategy in LB relies on several sophisticated computational tools capable of integrating two or more different layers of biological information. These methods advance beyond the single-marker analysis using machine learning (ML) and artificial intelligence (AI) approaches to decode the complex biological signatures that distinguish ctDNA from cfDNA. Given the complexity of cancer biology and maximizing the probability of cancer detection, cTF estimations from each biological layer can be combined into a single and more robust score, potentially weighted by the reliability or strength of the signal derived from each modality for a given cancer type. In this context, examples of ML and AI tools include random forests [167], support vector machines [168], and deep neural networks [169]. These classifiers can be trained on several input biological features extracted from genomics (SSNVs, SCNAs), epigenomics (methylation patterns), and fragmentomics (size distribution, end motifs) datasets to improve the sensitivity and specificity of cancer detection and cTF quantification. Such data integration models have proven to be particularly effective for challenging LB scenarios like ctDNA detection in the early-stage setting [170,171]. Overall, two main ML and AI strategies can be identified. An approach consists of building scores or classifiers based on the combination of individual omic layer score outputs to make an overall prediction. Moldovan and co-authors developed an ML classifier pipeline that combines cfDNA genomic (specifically, SCNAs using ichorCNA) and fragmentomic (specifically, using the open pipeline called FrEIA) features to reach a sensitive detection of tumor-derived cfDNA starting from the same sWGS dataset [172]. The authors evaluated four supervised ML approaches (k-neighbors, logistic regression, random forest, and support vector classifier) starting from four genomics and epigenomics cfDNA datasets, identifying that logistic regression provided the highest estimation of classification performance. A multi-class prediction model trained by a support vector classifier was adopted by Siejka-Zielinska and co-authors using methylation data and fragmentomics analyses and was successfully applied in the early HCC and pancreatic ductal adenocarcinoma settings [173]. Another approach for integrating omic-specific features is to use ensemble classifiers [174,175]. For instance, the ensemble THEMIS classifier was applied to the integration of four input features (methylated fragment ratio, fragment size index, chromosomal aneuploidy, and fragment end motif) to improve the ctDNA detection specificity and sensitivity over the individual features alone [176]. An additional example is the ML method named lung cancer likelihood in plasma (Lung-CLiP) that includes targeted sequencing of plasma cfDNA, matches leukocyte DNA, and integrates SSNV calling (leverages specific biological and technical features as cfDNA fragment size, the gene affected, and the likelihood of CHIP) with the genome-wide SCNAs analysis [177]. In addition to AI and ML bioinformatics pipelines, it is relevant to note that cutting-edge long-read sequencing technologies provide the opportunity to obtain multi-modal data from a single sequencing experiment, analyzing fragmentomics, methylomics, as well as genomic alterations [178,179]. Despite the current limitations of such platforms, primarily related to base-calling accuracy and bioinformatics pipeline implementation, it stands to reason that the continued evolution of technology and algorithms will unlock the full potential of nanopore and PacBio technology for cfDNA-based liquid biopsy analyses.
Looking to the ultimate goal of cancer research in terms of prevention, early detection, personalized paths of treatments, and optimization of disease monitoring, AI approaches can also be adopted for the integration of multiomics LB data for additional clinical, pathological (e.g., biochemistry), and radiological information (Figure 1) [180]. The direction of ML adoption consists of the shift from predictive to generative models that can elaborate on new entities, such as digital human avatars, that serve as integrative predictive models for cancer patients to support clinicians in improving decision-making processes [181,182].
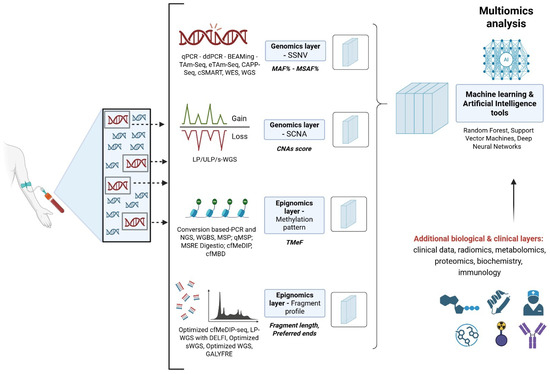
Figure 1.
Integration of multi-omics layers. The Figure shows the schematic multi-modal integration of the genomics and epigenomics strategies for ctDNA estimation. For both genomics and epigenomics layers, we reported the main methodological approaches available (e.g., ddPCR, WGS), the investigated molecular features (e.g., SSNV, SCNA), and the output variable type (e.g., MAF%, TMeF). Examples of machine learning approaches for data integration were also reported. Created in https://BioRender.com.
5. Translational Outlook and Standardization Challenges
The great research and technical improvements of LB approaches hold immense promise for their robust application in clinical oncological settings. However, several hurdles must be overcome to fully move these powerful technologies from the research bench to routine clinical care. Indeed, their translational value is significantly hampered by the lack of standardized protocols and regulatory oversight. Moreover, current approaches face limitations in terms of clinical applicability, and also mainly due to cost, data handling requirements, and turnaround time [183,184]. In both research and clinical contexts, a considerable inter-laboratory variability has clearly emerged in cfDNA processing, sequencing, and proprietary bioinformatic pipelines used for ctDNA estimation and, generally speaking, for molecular data interpretation. Some of the LB technologies and assays described here benefit from regulatory approval status from the FDA and European CE-IVD. These regulatory frameworks differentiate clinically actionable and reimbursed tests from emerging research tools. To date, regulatory approvals mainly cover SSNV-based (both PCR and NGS) and methylation-based assays (e.g., cobas® EGFR Mutation Test v2, FoundationOne® Liquid CDx, Epi pro-Colon®) [17,31,35,48,53,65,97,98,110,125,131]. Other promising assays lack the same rigorous regulatory standing, even if they can be offered as a laboratory-developed test (LDT) in clinical laboratory improvement amendments (CLIA) or the European Union’s in vitro diagnostic medical device regulation (IVDR)-certified labs (Galleri®, Guardant Reveal®). Techniques such as the SCNA-based method and fragmentomics are currently suitable for the research setting. Their current lack of regulatory standardization results in a significant inter-laboratory variability, insufficient clinical validation from large prospective trials, and, consequently, no effective path to a widespread clinical application.
6. Concluding Remarks
The appropriate estimation of cTF is critical for the reliability of the analytical test and for the clinical utility of LBx data. The variability in the methodological approach adopted to estimate the ctDNA rate highlights the capacity to evaluate different features of the ctDNA. The choice of the best methodology depends on the specific cancer biology. The various “omics” approaches (genomics, epigenomics, and fragmentomics) offer a unique way to infer cTF, with their own strengths and limitations. Genomics focuses on the specific SSNVs that are most effective when a target, patient-specific mutation is known from a TBx, but they may be less useful in tumors with low mutation rates. Conversely, SCNAs are broadly applicable, since most cancers have widespread chromosomal aberrations. However, both methods can be challenged by CHIP or insufficient ctDNA shedding. Because methylation patterns can appear early in tumor development, methylation-based methods are particularly promising for early cancer detection. The signal can be stronger and more widespread than a single SSNV, making it a powerful tool for quantification, but it requires specialized assays and analysis pipelines [49]. Fragmentomics can provide insights into tumor biology and is increasingly being combined with machine learning to improve cTF estimation sensitivity. Under the selective pressure of therapy, the biological behavior of tumor cells can lead to evolving genomic and epigenomic profiles over serial sampling. Consequently, the molecular features (e.g., SSNV, CNAs) adopted for cTF calculation in a specific clinical time point could not be equal in another one. Given the complexity and variability of tumor biology, the integration of multiple omics data, often derived from a single sequencing run, is the most promising strategy. The goal is to move towards a multi-modal liquid biopsy that provides a reliable, “all-in-one”, solution for cancer management.
Author Contributions
Conceptualization, E.D.P. and A.M.; methodology, A.C. and M.C.; resources, A.P. and G.A.P.; data curation, E.D.P. and A.P.; writing—original draft preparation, E.D.P.; writing—review and editing, A.M. and A.U.; supervision, A.U.; project administration, E.D.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
We thank the Genomics Core Facility Group at the Gemelli Science and Technology Park (G-STeP) for their valuable contribution.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ctDNA | Circulating tumor DNA |
| cfDNA | Cell-free DNA |
| cTF | Circulating tumor allele fraction |
| LBx | Liquid biopsy |
| CTCs | Circulating tumor cells |
| cfRNA | Cell-free RNA |
| NIPT | Non-invasive prenatal testing |
| MRD | Minimal residual disease |
| MCED | Multicancer early detection |
| TBx | Tissue solid biopsies |
| FDA | Food and Drug Administration |
| eccDNA | Circular DNA |
| SNVs | Single-nucleotide variants |
| CNAs | Copy number alterations |
| NGS | Next-generation sequencing |
| UMIs | Unique molecular identifiers |
| SSNVs | Somatic single-nucleotide variants |
| VAF | Variant allele frequency |
| MAF | Mutant allele frequency |
| ddPCR | Droplet digital PCR |
| BEAMing PCR | Beads, emulsions, amplification, and magnetics PCR |
| AS-NEPB-PCR | Allele-specific non-extendable primer blocker PCR |
| PNA-LNA PCR clamp | Peptide nucleic acid-locked nucleic acid PCR clamp |
| COLD PCR | Co-amplification at lower denaturation temperature PCR |
| PCR | Polymerase chain reaction |
| LOD | Limit of detection |
| LDT | Laboratory-developed test |
| CLIA | Clinical laboratory improvement amendments |
| IVDR | European Union’s in vitro diagnostic medical device regulation |
| RUO | Research use only |
| CT | Computed tomography |
| CHIP | Clonal hematopoiesis of indeterminate potential |
| TAm-Seq | Tagged-amplicon deep sequencing technology |
| eTAm-Seq | Enhanced TAm-Seq |
| CAPP-Seq | Cancer personalized profiling by deep sequencing |
| cSMART | Single-molecule amplification and resequencing technology |
| MSAF | Maximum somatic allele frequency |
| GEMINI | Genome-wide mutational incidence for non-invasive detection of cancer |
| WES | Whole-exome sequencing |
| WGS | Whole-genome sequencing |
| SCNAs | Somatic copy number alterations |
| LP/ULP-WGS | Low-pass/ultra-low-pass WGS |
| sWGS | Shallow WGS |
| HCC | Hepatocellular carcinoma |
| qPCR | Quantitative-PCR |
| ARMS/superARMS PCR | Amplification refractory mutation system PCR |
| TAT | Turnaround time |
| 5mC | 5-methylcytosine |
| CpG | Cytosine-guanine dinucleotide |
| 5hmC | 5-hydroxymethylcytosine |
| TMeF | Tumor methylated fraction |
| DMRs | Differentially methylated regions |
| cfMeDIP | Cell-free methylated DNA immunoprecipitation |
| cfMBD | Cell-free methyl-binding domain |
| CRC | Colorectal cancer |
| QCTs | Quantitative counting templates |
| MRE-Seq | Methylation-sensitive restriction enzyme digestion followed by sequencing |
| DNN | Deep neural network |
| SSB-PCR | Single-strand binding polymerase chain reaction |
| DELFI | DNA evaluation of fragments for early interception |
| MSP | Methylation-specific PCR |
| qMSP | Quantitative methylation-specific PCR |
| MSRE | Methylation-sensitive restriction enzymes digestion |
| WGBS | Whole-genome bisulfite sequencing |
| CE | Capillary electrophoresis |
| GALYFRE | Genome-wide analysis of fragment ends |
| ML | Machine learning |
| AI | Artificial intelligence |
References
- Hasenleithner, S.O.; Speicher, M.R. A clinician’s handbook for using ctDNA throughout the patient journey. Mol. Cancer 2022, 21, 81. [Google Scholar] [CrossRef]
- Sánchez-Herrero, E.; Serna-Blasco, R.; Robado de Lope, L.; González-Rumayor, V.; Romero, A.; Provencio, M. Circulating Tumor DNA as a Cancer Biomarker: An Overview of Biological Features and Factors That may Impact on ctDNA Analysis. Front. Oncol. 2022, 12, 943253. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Sharon, E.; Shi, H.; Kharbanda, S.; Koh, W.; Martin, L.R.; Khush, K.K.; Valantine, H.; Pritchard, J.K.; De Vlaminck, I. Quantification of transplant-derived circulating cell-free DNA in absence of a donorgenotype. PLoS Comput. Biol. 2017, 13, e1005629. [Google Scholar] [CrossRef]
- Burnham, P.; Khush, K.; De Vlaminck, I. Myriad applications of circulating cell-free DNA in precision organ transplant monitoring. Ann. Am. Thorac. Soc. 2017, 14, S237–S241. [Google Scholar] [CrossRef]
- Rolfo, C.D.; Madison, R.W.; Pasquina, L.W.; Brown, D.W.; Huang, Y.; Hughes, J.D.; Graf, R.P.; Oxnard, G.R.; Husain, H. Measurement of ctDNA Tumor Fraction Identifies Informative Negative Liquid Biopsy Results and Informs Value of Tissue Confirmation. Clin. Cancer Res. 2024, 30, 2452–2460. [Google Scholar] [CrossRef] [PubMed]
- Bredno, J.; Venn, O.; Chen, X.; Freese, P.; Ofman, J.J. Circulating Tumor DNA Allele Fraction: A Candidate Biological Signal for Multicancer Early Detection Tests to Assess the Clinical Significance of Cancers. Am. J. Pathol. 2022, 192, 1368–1378. [Google Scholar] [CrossRef]
- Piana, D.; Iavarone, F.; De Paolis, E.; Daniele, G.; Parisella, F.; Minucci, A.; Greco, V.; Urbani, A. Phenotyping Tumor Heterogeneity through Proteogenomics: Study Models and Challenges. Int. J. Mol. Sci. 2024, 25, 8830. [Google Scholar] [CrossRef]
- Nagasaka, M.; Uddin, M.H.; Al-Hallak, M.N.; Rahman, S.; Balasubramanian, S.; Sukari, A.; Azmi, A.S. Liquid biopsy for therapy monitoring in early-stage non-small cell lung cancer. Mol. Cancer 2021, 20, 82. [Google Scholar] [CrossRef]
- O’Leary, B.; Cutts, R.J.; Huang, X.; Hrebien, S.; Liu, Y.; André, F.; Loibl, S.; Loi, S.; Garcia-Murillas, I.; Cristofanilli, M.; et al. Circulating Tumor DNA Markers for Early Progression on Fulvestrant with or Without Palbociclib in ER+ Advanced Breast Cancer. J. Natl. Cancer Inst. 2021, 113, 309–317. [Google Scholar] [CrossRef]
- Stover, D.G.; Parsons, H.A.; Ha, G.; Freeman, S.S.; Barry, W.T.; Guo, H.; Choudhury, A.D.; Gydush, G.; Reed, S.C.; Rhoades, J.; et al. Association of Cell-Free DNA Tumor Fraction and Somatic Copy Number Alterations with Survival in Metastatic Triple-Negative Breast Cancer. J. Clin. Oncol. 2018, 36, 543–553. [Google Scholar] [CrossRef]
- Choudhury, A.D.; Werner, L.; Francini, E.; Wei, X.X.; Ha, G.; Freeman, S.S.; Rhoades, J.; Reed, S.C.; Gydush, G.; Rotem, D.; et al. Tumor fraction in cell-free DNA as a biomarker in prostate cancer. JCI Insight 2018, 3, e122109. [Google Scholar] [CrossRef]
- Reichert, Z.R.; Morgan, T.M.; Li, G.; Castellanos, E.; Snow, T.; Dall’Olio, F.G.; Madison, R.W.; Fine, A.D.; Oxnard, G.R.; Graf, R.P.; et al. Prognostic value of plasma circulating tumor DNA fraction across four common cancer types: A real-world outcomes study. Ann. Oncol. 2023, 34, 111–120. [Google Scholar] [CrossRef]
- Polski, A.; Xu, L.; Prabakar, R.K.; Kim, J.W.; Shah, R.; Jubran, R.; Kuhn, P.; Cobrinik, D.; Hicks, J.; Berry, J.L. Cell-Free DNA Tumor Fraction in the Aqueous Humor Is Associated with Therapeutic Response in Retinoblastoma Patients. Transl. Vis. Sci. Technol. 2020, 9, 30. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.C.; Lo, Y.M.D. Cell-Free DNA Fragmentomics in Liquid Biopsy. Diagnostics 2022, 12, 978. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Solit, D.B. Clinical cancer genomic profiling. Nat. Rev. Genet. 2021, 22, 483–501. [Google Scholar] [CrossRef] [PubMed]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451, Erratum in Nature 2018, 554, 264. [Google Scholar] [CrossRef]
- Cho, M.S.; Park, C.H.; Lee, S.; Park, H.S. Clinicopathological parameters for circulating tumor DNA shedding in surgically resected non-small cell lung cancer with EGFR or KRAS mutation. PLoS ONE 2020, 15, e0230622. [Google Scholar] [CrossRef]
- Heitzer, E.; Auinger, L.; Speicher, M.R. Cell-Free DNA and Apoptosis: How Dead Cells Inform About the Living. Trends Mol. Med. 2020, 26, 519–528. [Google Scholar] [CrossRef]
- Morbelli, S.; Alama, A.; Ferrarazzo, G.; Coco, S.; Genova, C.; Rijavec, E.; Bongioanni, F.; Biello, F.; Dal Bello, M.G.; Barletta, G.; et al. Circulating Tumor DNA Reflects Tumor Metabolism Rather Than Tumor Burden in Chemotherapy-Naive Patients with Advanced Non-Small Cell Lung Cancer: 18F-FDG PET/CT Study. J. Nucl. Med. 2017, 58, 1764–1769. [Google Scholar] [CrossRef]
- Fleischhacker, M.; Schmidt, B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochim. Biophys. Acta 2007, 1775, 181–232. [Google Scholar] [CrossRef]
- Jiang, P.; Chan, C.W.; Chan, K.C.; Cheng, S.H.; Wong, J.; Wong, V.W.; Wong, G.L.; Chan, S.L.; Mok, T.S.; Chan, H.L.; et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl. Acad. Sci. USA 2015, 112, E1317–E1325. [Google Scholar] [CrossRef] [PubMed]
- Kustanovich, A.; Schwartz, R.; Peretz, T.; Grinshpun, A. Life and death of circulating cell-free DNA. Cancer Biol. Ther. 2019, 20, 1057–1067. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef]
- Muhanna, N.; Di Grappa, M.A.; Chan, H.H.L.; Khan, T.; Jin, C.S.; Zheng, Y.; Irish, J.C.; Bratman, S.V. Cell-free DNA kinetics in a pre-clinical model of head and neck cancer. Sci. Rep. 2017, 7, 16723. [Google Scholar] [CrossRef]
- Tug, S.; Helmig, S.; Deichmann, E.R.; Schmeier-Jürchott, A.; Wagner, E.; Zimmermann, T.; Radsak, M.; Giacca, M.; Simon, P. Exercise-induced increases in cell free DNA in human plasma originate predominantly from cells of the haematopoietic lineage. Exerc. Immunol. Rev. 2015, 21, 164–173. [Google Scholar]
- Moss, J.; Magenheim, J.; Neiman, D.; Zemmour, H.; Loyfer, N.; Korach, A.; Samet, Y.; Maoz, M.; Druid, H.; Arner, P.; et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 2018, 9, 5068. [Google Scholar] [CrossRef]
- Wong, F.C.; Sun, K.; Jiang, P.; Cheng, Y.K.; Chan, K.C.; Leung, T.Y.; Chiu, R.W.; Lo, Y.M. Cell-free DNA in maternal plasma and serum: A comparison of quantity, quality and tissue origin using genomic and epigenomic approaches. Clin. Biochem. 2016, 49, 1379–1386. [Google Scholar] [CrossRef]
- Lam, W.K.J.; Gai, W.; Sun, K.; Wong, R.S.M.; Chan, R.W.Y.; Jiang, P.; Chan, N.P.H.; Hui, W.W.I.; Chan, A.W.H.; Szeto, C.C.; et al. DNA of Erythroid Origin Is Present in Human Plasma and Informs the Types of Anemia. Clin. Chem. 2017, 63, 1614–1623. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Wong, S.Q.; Raleigh, J.M.; Callahan, J.; Vergara, I.A.; Ftouni, S.; Hatzimihalis, A.; Colebatch, A.J.; Li, J.; Semple, T.; Doig, K.; et al. Circulating tumor DNA analysis and functional imaging provide complementary approaches for comprehensive disease monitoring in metastatic melanoma. JCO Precis. Oncol. 2017, 1, 1–14. [Google Scholar] [CrossRef]
- Murtaza, M.; Dawson, S.J.; Pogrebniak, K.; Rueda, O.M.; Provenzano, E.; Grant, J.; Chin, S.F.; Tsui, D.W.Y.; Marass, F.; Gale, D.; et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat. Commun. 2015, 6, 8760. [Google Scholar] [CrossRef]
- Neumann, M.H.D.; Bender, S.; Krahn, T.; Schlange, T. ctDNA and CTCs in liquid biopsy–current status and where we need to progress. Comput. Struct. Biotechnol. J. 2018, 16, 190–195. [Google Scholar] [CrossRef]
- Zill, O.A.; Banks, K.C.; Fairclough, S.R.; Mortimer, S.A.; Vowles, J.V.; Mokhtari, R.; Gandara, D.R.; Mack, P.C.; Odegaard, J.I.; Nagy, R.J.; et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin. Cancer Res. 2018, 24, 3528–3538. [Google Scholar] [CrossRef]
- Myint, N.N.M.; Verma, A.M.; Fernandez-Garcia, D.; Sarmah, P.; Tarpey, P.S.; Al-Aqbi, S.S.; Cai, H.; Trigg, R.; West, K.; Howells, L.M.; et al. Circulating tumor DNA in patients with colorectal adenomas: Assessment of detectability and genetic heterogeneity. Cell Death Dis. 2018, 9, 894. [Google Scholar] [CrossRef]
- Sassorossi, C.; Evangelista, J.; Stefani, A.; Chiappetta, M.; Martino, A.; Campanella, A.; De Paolis, E.; Nachira, D.; Del Re, M.; Guerrera, F.; et al. The Role of ctDNA for Diagnosis and Histological Prediction in Early Stage Non-Small-Cell Lung Cancer: A Narrative Review. Diagnostics 2025, 15, 904. [Google Scholar] [CrossRef]
- Ntzifa, A.; Lianidou, E. Pre-analytical conditions and implementation of quality control steps in liquid biopsy analysis. Crit. Rev. Clin. Lab. Sci. 2023, 60, 573–594. [Google Scholar] [CrossRef] [PubMed]
- Lucien, F.; Gustafson, D. Standardized reporting of pre-analytical variables and quality control of plasma and serum to enhance rigor and reproducibility in liquid biopsy research. J. Liq. Biopsy 2024, 6, 100163. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, C.M.; Borsu, L.; Cankovic, M.; Earle, J.S.L.; Gocke, C.D.; Hameed, M.; Jordan, D.; Lopategui, J.R.; Pullambhatla, M.; Reuther, J.; et al. Recommendations for Cell-Free DNA Assay Validations: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2023, 25, 876–897. [Google Scholar] [CrossRef]
- Leygo, C.; Williams, M.; Jin, H.C.; Chan, M.W.Y.; Chu, W.K.; Grusch, M.; Cheng, Y.Y. DNA Methylation as a Noninvasive Epigenetic Biomarker for the Detection of Cancer. Dis. Markers 2017, 2017, 3726595. [Google Scholar] [CrossRef]
- McDonald, B.R.; Contente-Cuomo, T.; Sammut, S.J.; Odenheimer-Bergman, A.; Ernst, B.; Perdigones, N.; Chin, S.F.; Farooq, M.; Mejia, R.; Cronin, P.A.; et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci. Transl. Med. 2019, 11, eaax7392. [Google Scholar] [CrossRef]
- Malapelle, U.; Buono, M.; Pisapia, P.; Russo, G.; Tufano, R.; Pepe, F.; Rolfo, C.; Troncone, G. Circulating tumor DNA in cancer: Predictive molecular pathology meets mathematics. Crit. Rev. Oncol. Hematol. 2021, 163, 103394. [Google Scholar] [CrossRef] [PubMed]
- Pairawan, S.; Hess, K.; Janku, F.; Sanchez, N.; Shaw, K.; Eng, C.; Damodaran, S.; Javle, M.; Kaseb, A.; Hong, D.; et al. Cell-Free Circulating Tumor DNA Variant Allele Frequency Associates with Survival in Metastatic Cancer. Clin. Cancer Res. 2020, 26, 1924–1931. [Google Scholar] [CrossRef] [PubMed]
- Manca, P.; Corallo, S.; Lonardi, S.; Fucà, G.; Busico, A.; Leone, A.G.; Corti, F.; Antoniotti, C.; Procaccio, L.; Smiroldo, V.; et al. Variant Allele Frequency in Baseline Circulating Tumour DNA to Measure Tumour Burden and to Stratify Outcomes in Patients with RAS Wild-Type Metastatic Colorectal Cancer: A Translational Objective of the Valentino Study. Br. J. Cancer 2022, 126, 449–455. [Google Scholar] [CrossRef]
- Berchuck, J.E.; Facchinetti, F.; DiToro, D.F.; Baiev, I.; Majeed, U.; Reyes, S.; Chen, C.; Zhang, K.; Sharman, R.; Uson, J.P.L.S.; et al. The Clinical Landscape of Cell-Free DNA Alterations in 1671 Patients with Advanced Biliary Tract Cancer. Ann. Oncol. 2022, 33, 1269–1283. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, C.A.; Gale, D.; Piskorz, A.M.; Biggs, H.; Hodgkin, C.; Addley, H.; Freeman, S.; Moyle, P.; Sala, E.; Sayal, K.; et al. Exploratory Analysis of TP53 Mutations in Circulating Tumour DNA as Biomarkers of Treatment Response for Patients with Relapsed High-Grade Serous Ovarian Carcinoma: A Retrospective Study. PLoS Med. 2016, 13, e1002198. [Google Scholar] [CrossRef]
- Galant, N.; Nicoś, M.; Kuźnar-Kamińska, B.; Krawczyk, P. Variant Allele Frequency Analysis of Circulating Tumor DNA as a Promising Tool in Assessing the Effectiveness of Treatment in Non-Small Cell Lung Carcinoma Patients. Cancers 2024, 16, 782. [Google Scholar] [CrossRef]
- Newman, A.M.; Lovejoy, A.F.; Klass, D.M.; Kurtz, D.M.; Chabon, J.J.; Scherer, F.; Stehr, H.; Liu, C.L.; Bratman, S.V.; Say, C.; et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016, 34, 547–555. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Frazzi, R.; Bizzarri, V.; Albertazzi, L.; Cusenza, V.Y.; Coppolecchia, L.; Luminari, S.; Ilariucci, F. Droplet digital PCR is a sensitive tool for the detection of TP53 deletions and point mutations in chronic lymphocytic leukaemia. Br. J. Haematol. 2020, 189, e49–e52. [Google Scholar] [CrossRef]
- Fairchild, L.; Whalen, J.; D’Aco, K.; Wu, J.; Gustafson, C.B.; Solovieff, N.; Su, F.; Leary, R.J.; Campbell, C.D.; Balbin, O.A. Clonal hematopoiesis detection in patients with cancer using cell-free DNA sequencing. Sci. Transl. Med. 2023, 15, eabm8729. [Google Scholar] [CrossRef] [PubMed]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.; Kaper, F.; Dawson, S.J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012, 4, 136ra68. [Google Scholar] [CrossRef]
- Gale, D.; Lawson, A.R.J.; Howarth, K.; Madi, M.; Durham, B.; Smalley, S.; Calaway, J.; Blais, S.; Jones, G.; Clark, J.; et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS ONE 2018, 13, e0194630. [Google Scholar] [CrossRef]
- Narayan, A.; Carriero, N.J.; Gettinger, S.N.; Kluytenaar, J.; Kozak, K.R.; Yock, T.I.; Muscato, N.E.; Ugarelli, P.; Decker, R.H.; Patel, A.A. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 2012, 72, 3492–3498. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Zhou, Z.; Xie, Y.; Huang, Z.; Lu, S.; Liu, C.; Wang, J.; Li, X. Methodology established for the detection of circulating tumor DNA by hybridization capture. Biotechniques 2022, 73, 151–158. [Google Scholar] [CrossRef]
- Lv, W.; Wei, X.; Guo, R.; Liu, Q.; Zheng, Y.; Chang, J.; Bai, T.; Li, H.; Zhang, J.; Song, Z.; et al. Noninvasive prenatal testing for Wilson disease by use of circulating single-molecule amplification and resequencing technology (cSMART). Clin. Chem. 2015, 61, 172–181. [Google Scholar] [CrossRef]
- Wang, Z.; Cheng, G.; Han, X.; Mu, X.; Zhang, Y.; Cui, D.; Liu, C.; Zhang, L.; Fan, Z.; Ma, L.; et al. Application of Single-Molecule Amplification and Resequencing Technology for Broad Surveillance of Plasma Mutations in Patients with Advanced Lung Adenocarcinoma. J. Mol. Diagn. 2017, 19, 169–181. [Google Scholar] [CrossRef]
- Chen, K.; Zhao, H.; Shi, Y.; Yang, F.; Wang, L.T.; Kang, G.; Nie, Y.; Wang, J. Perioperative Dynamic Changes in Circulating Tumor DNA in Patients with Lung Cancer (DYNAMIC). Clin. Cancer Res. 2019, 25, 7058–7067. [Google Scholar] [CrossRef] [PubMed]
- Haghighat Jahromi, A.; Zabel, M.; Okamura, R.; Hoh, C.K.; Kurzrock, R. Variant allele fraction of genomic alterations in circulating tumor DNA (%ctDNA) correlates with SUVmax in PET scan. Am. J. Nucl. Med. Mol. Imaging 2021, 11, 307–312. [Google Scholar]
- Silvoniemi, A.; Laine, J.; Aro, K.; Nissi, L.; Bäck, L.; Schildt, J.; Hirvonen, J.; Hagström, J.; Irjala, H.; Aaltonen, L.M.; et al. Circulating Tumor DNA in Head and Neck Squamous Cell Carcinoma: Association with Metabolic Tumor Burden Determined with FDG-PET/CT. Cancers 2023, 15, 3970. [Google Scholar] [CrossRef]
- Ottestad, A.L.; Johansen, H.; Halvorsen, T.O.; Dai, H.Y.; Wahl, S.G.F.; Emdal, E.F.; Grønberg, B.H. Associations between detectable circulating tumor DNA and tumor glucose uptake measured by 18F-FDG PET/CT in early-stage non-small cell lung cancer. BMC Cancer 2023, 23, 646, Erratum in BMC Cancer 2023, 23, 685. [Google Scholar] [CrossRef]
- Dong, Y.; Zhu, Y.; Zhuo, M.; Chen, X.; Xie, Y.; Duan, J.; Bai, H.; Hao, S.; Yu, Z.; Yi, Y.; et al. Maximum Somatic Allele Frequency-Adjusted Blood-Based Tumor Mutational Burden Predicts the Efficacy of Immune Checkpoint Inhibitors in Advanced Non-Small Cell Lung Cancer. Cancers 2022, 14, 5649. [Google Scholar] [CrossRef]
- Nie, W.; Wang, Z.J.; Zhang, K.; Li, B.; Cai, Y.R.; Wen, F.C.; Zhang, D.; Bai, Y.Z.; Zhang, X.Y.; Wang, S.Y.; et al. ctDNA-adjusted bTMB as a predictive biomarker for patients with NSCLC treated with PD-(L)1 inhibitors. BMC Med. 2022, 20, 170. [Google Scholar] [CrossRef]
- Chen, Y.T.; Seeruttun, S.R.; Wu, X.Y.; Wang, Z.X. Maximum Somatic Allele Frequency in Combination with Blood-Based Tumor Mutational Burden to Predict the Efficacy of Atezolizumab in Advanced Non-small Cell Lung Cancer: A Pooled Analysis of the Randomized POPLAR and OAK Studies. Front. Oncol. 2019, 9, 1432. [Google Scholar] [CrossRef]
- Finkle, J.D.; Boulos, H.; Driessen, T.M.; Lo, C.; Blidner, R.A.; Hafez, A.; Khan, A.A.; Lozac’hmeur, A.; McKinnon, K.E.; Perera, J.; et al. Validation of a liquid biopsy assay with molecular and clinical profiling of circulating tumor DNA. NPJ Precis. Oncol. 2021, 5, 63. [Google Scholar] [CrossRef]
- Tivey, A.; Lee, R.J.; Clipson, A.; Hill, S.M.; Lorigan, P.; Rothwell, D.G.; Dive, C.; Mouliere, F. Mining nucleic acid “omics” to boost liquid biopsy in cancer. Cell Rep. Med. 2024, 5, 101736. [Google Scholar] [CrossRef] [PubMed]
- Bruhm, D.C.; Mathios, D.; Foda, Z.H.; Annapragada, A.V.; Medina, J.E.; Adleff, V.; Chiao, E.J.; Ferreira, L.; Cristiano, S.; White, J.R.; et al. Single-molecule genome-wide mutation profiles of cell-free DNA for non-invasive detection of cancer. Nat. Genet. 2023, 55, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- Shahrouzi, P.; Forouz, F.; Mathelier, A.; Kristensen, V.N.; Duijf, P.H.G. Copy number alterations: A catastrophic orchestration of the breast cancer genome. Trends Mol. Med. 2024, 30, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef]
- Carter, S.L.; Cibulskis, C.; Helman, E.; McKenna, A.; Shen, H.; Zack, T.; Laird, P.W.; Onofrio, R.C.; Winckler, W.; Weir, B.A.; et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012, 30, 413–421. [Google Scholar] [CrossRef]
- Ha, G.; Roth, A.; Khattra, J.; Ho, J.; Yap, D.; Prentice, L.M.; Melnyk, N.; McPherson, A.; Bashashati, A.; Laks, E.; et al. TITAN: Inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res. 2014, 24, 1881–1893. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Ulz, P.; Belic, J.; Gutschi, S.; Quehenberger, F.; Fischereder, K.; Benezeder, T.; Auer, M.; Pischler, C.; Mannweiler, S.; et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Leary, R.J.; Sausen, M.; Kinde, I.; Papadopoulos, N.; Carpten, J.D.; Craig, D.; O’Shaughnessy, J.; Kinzler, K.W.; Parmigiani, G.; Vogelstein, B.; et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci. Transl. Med. 2012, 4, 162ra154. [Google Scholar] [CrossRef] [PubMed]
- Scheinin, I.; Sie, D.; Bengtsson, H.; van de Wiel, M.A.; Olshen, A.B.; van Thuijl, H.F.; van Essen, H.F.; Eijk, P.P.; Rustenburg, F.; Meijer, G.A.; et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014, 24, 2022–2032. [Google Scholar] [CrossRef]
- Raman, L.; Dheedene, A.; De Smet, M.; Van Dorpe, J.; Menten, B. WisecondorX: Improved copy number detection for routine shallow whole-genome sequencing. Nucleic Acids Res. 2019, 47, 1605–1614. [Google Scholar] [CrossRef]
- Potente, S.; Boscarino, D.; Paladin, D.; Marchini, S.; Beltrame, L.; Romualdi, C. SAMURAI: Shallow analysis of copy number alterations using a reproducible and integrated bioinformatics pipeline. Brief Bioinform. 2024, 26, bbaf035. [Google Scholar] [CrossRef]
- Hovelson, D.H.; Liu, C.J.; Wang, Y.; Kang, Q.; Henderson, J.; Gursky, A.; Brockman, S.; Ramnath, N.; Krauss, J.C.; Talpaz, M.; et al. Rapid, ultra low coverage copy number profiling of cell-free DNA as a precision oncology screening strategy. Oncotarget 2017, 8, 89848–89866. [Google Scholar] [CrossRef]
- Poell, J.B.; Mendeville, M.; Sie, D.; Brink, A.; Brakenhoff, R.H.; Ylstra, B. ACE: Absolute copy number estimation from low-coverage whole-genome sequencing data. Bioinformatics 2019, 35, 2847–2849. [Google Scholar] [CrossRef]
- Adalsteinsson, V.A.; Ha, G.; Freeman, S.S.; Choudhury, A.D.; Stover, D.G.; Parsons, H.A.; Gydush, G.; Reed, S.C.; Rotem, D.; Rhoades, J.; et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 2017, 8, 1324. [Google Scholar] [CrossRef]
- Benn, P.; Cuckle, H.; Pergament, E. Non-invasive prenatal testing for aneuploidy: Current status and future prospects. Ultrasound Obstet. Gynecol. 2013, 42, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, D.W.; Chudova, D.; Sehnert, A.J.; Bhatt, S.; Murray, K.; Prosen, T.L.; Garber, J.E.; Wilkins-Haug, L.; Vora, N.L.; Warsof, S.; et al. Noninvasive Prenatal Testing and Incidental Detection of Occult Maternal Malignancies. JAMA 2015, 314, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Allsopp, R.C.; Page, K.; Ambasager, B.; Wadsley, M.K.; Acheampong, E.; Ntereke, T.P.; Guo, Q.; Lall, G.M.; Gleason, K.L.T.; Wren, E.; et al. A Rapid, Shallow Whole Genome Sequencing Workflow Applicable to Limiting Amounts of Cell-Free DNA. Clin. Chem. 2023, 69, 510–518. [Google Scholar] [CrossRef]
- Lakatos, E.; Hockings, H.; Mossner, M.; Huang, W.; Lockley, M.; Graham, T.A. LiquidCNA: Tracking subclonal evolution from longitudinal liquid biopsies using somatic copy number alterations. iScience 2021, 24, 102889. [Google Scholar] [CrossRef]
- Bouzidi, A.; Labreche, K.; Baron, M.; Veyri, M.; Denis, J.A.; Touat, M.; Sanson, M.; Davi, F.; Guillerm, E.; Jouannet, S.; et al. Low-Coverage Whole Genome Sequencing of Cell-Free DNA from Immunosuppressed Cancer Patients Enables Tumor Fraction Determination and Reveals Relevant Copy Number Alterations. Front. Cell Dev. Biol. 2021, 9, 661272. [Google Scholar] [CrossRef]
- Martignano, F.; Munagala, U.; Crucitta, S.; Mingrino, A.; Semeraro, R.; Del Re, M.; Petrini, I.; Magi, A.; Conticello, S.G. Nanopore sequencing from liquid biopsy: Analysis of copy number variations from cell-free DNA of lung cancer patients. Mol. Cancer 2021, 20, 32. [Google Scholar] [CrossRef]
- Rickles-Young, M.; Tinoco, G.; Tsuji, J.; Pollock, S.; Haynam, M.; Lefebvre, H.; Glover, K.; Owen, D.H.; Collier, K.A.; Ha, G.; et al. Assay Validation of Cell-Free DNA Shallow Whole-Genome Sequencing to Determine Tumor Fraction in Advanced Cancers. J. Mol. Diagn. 2024, 26, 413–422. [Google Scholar] [CrossRef]
- Yeung, C.C.S.; McElhone, S.; Chen, X.Y.; Ng, D.; Storer, B.E.; Deeg, H.J.; Fang, M. Impact of copy neutral loss of heterozygosity and total genome aberrations on survival in myelodysplastic syndrome. Mod. Pathol. 2018, 31, 569–580. [Google Scholar] [CrossRef]
- Pinto, E.; Lazzarini, E.; Pelizzaro, F.; Gambato, M.; Santarelli, L.; Potente, S.; Zanaga, P.; Zappitelli, T.; Cardin, R.; Burra, P.; et al. Somatic Copy Number Alterations in Circulating Cell-Free DNA as a Prognostic Biomarker for Hepatocellular Carcinoma: Insights from a Proof-of-Concept Study. Cancers 2025, 17, 1115. [Google Scholar] [CrossRef]
- Sogbe, M.; Bilbao, I.; Marchese, F.P.; Zazpe, J.; De Vito, A.; Pozuelo, M.; D’Avola, D.; Iñarrairaegui, M.; Berasain, C.; Arechederra, M.; et al. Prognostic value of ultra-low-pass whole-genome sequencing of circulating tumor DNA in hepatocellular carcinoma under systemic treatment. Clin. Mol. Hepatol. 2024, 30, 177–190. [Google Scholar] [CrossRef]
- Markus, H.; Chandrananda, D.; Moore, E.; Mouliere, F.; Morris, J.; Brenton, J.D.; Smith, C.G.; Rosenfeld, N. Refined characterization of circulating tumor DNA through biological feature integration. Sci. Rep. 2022, 12, 1928. [Google Scholar] [CrossRef]
- Albertson, D.G.; Collins, C.; McCormick, F.; Gray, J.W. Chromosome aberrations in solid tumors. Nat. Genet. 2003, 34, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.M.; Shih, J.; Ha, G.; Gao, G.F.; Zhang, X.; Berger, A.C.; Schumacher, S.E.; Wang, C.; Hu, H.; Liu, J.; et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018, 33, 676–689.e3. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Barrios, C.; Nieto-Alcolado, I.; Torrente, M.; Jiménez-Sánchez, C.; Calvo, V.; Gutierrez-Sanz, L.; Palka, M.; Donoso-Navarro, E.; Provencio, M.; Romero, A. Comparison of methods for circulating cell-free DNA isolation using blood from cancer patients: Impact on biomarker testing. Transl. Lung Cancer Res. 2016, 5, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.; Mouliere, F.; El Messaoudi, S.; Mollevi, C.; Lopez-Crapez, E.; Rolet, F.; Gillet, B.; Gongora, C.; Dechelotte, P.; Robert, B.; et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat. Med. 2014, 20, 430–435. [Google Scholar] [CrossRef]
- Veldore, V.H.; Choughule, A.; Routhu, T.; Mandloi, N.; Noronha, V.; Joshi, A.; Dutt, A.; Gupta, R.; Vedam, R.; Prabhash, K. Validation of liquid biopsy: Plasma cell-free DNA testing in clinical management of advanced non-small cell lung cancer. Lung Cancer Targets Ther. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Woodhouse, R.; Li, M.; Hughes, J.; Delfosse, D.; Skoletsky, J.; Ma, P.; Meng, W.; Dewal, N.; Milbury, C.; Clark, T.; et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS ONE 2020, 15, e0237802. [Google Scholar] [CrossRef]
- Caputo, V.; Ciardiello, F.; Corte, C.M.D.; Martini, G.; Troiani, T.; Napolitano, S. Diagnostic value of liquid biopsy in the era of precision medicine: 10 years of clinical evidence in cancer. Explor. Target. Antitumor Ther. 2023, 4, 102–138. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.P.; Rand, K.N.; Molloy, P.L. Hypomethylation of repeated DNA sequences in cancer. Epigenomics 2010, 2, 245–269. [Google Scholar] [CrossRef] [PubMed]
- Sonar, S.; Nyahatkar, S.; Kalele, K.; Adhikari, M.D. Role of DNA methylation in cancer development and its clinical applications. Clin. Transl. Disc. 2024, 4, e279. [Google Scholar] [CrossRef]
- Li, L.; Sun, Y. Circulating tumor DNA methylation detection as biomarker and its application in tumor liquid biopsy: Advances and challenges. MedComm 2024, 5, e766. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Dietrich, D.; Weider, S.; de Vos, L.; Vogt, T.J.; Färber, M.; Zarbl, R.; Hunecke, A.; Glosch, A.K.; Gabrielpillai, J.; Bootz, F.; et al. Circulating Cell-Free SEPT9 DNA Methylation in Blood Is a Biomarker for Minimal Residual Disease Detection in Head and Neck Squamous Cell Carcinoma Patients. Clin. Chem. 2023, 69, 1050–1061. [Google Scholar] [CrossRef]
- Li, W.; Li, Q.; Kang, S.; Same, M.; Zhou, Y.; Sun, C.; Liu, C.C.; Matsuoka, L.; Sher, L.; Wong, W.H.; et al. CancerDetector: Ultrasensitive and non-invasive cancer detection at the resolution of individual reads using cell-free DNA methylation sequencing data. Nucleic Acids Res. 2018, 46, e89. [Google Scholar] [CrossRef]
- Yizhak, K.; Aguet, F.; Kim, J.; Hess, J.M.; Kübler, K.; Grimsby, J.; Frazer, R.; Zhang, H.; Haradhvala, N.J.; Rosebrock, D.; et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 2019, 364, eaaw0726. [Google Scholar] [CrossRef]
- Guo, S.; Diep, D.; Plongthongkum, N.; Fung, H.L.; Zhang, K.; Zhang, K. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat. Genet. 2017, 49, 635–642. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Klein, E.A.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef]
- Chung, D.C.; Gray, D.M.; Singh, H.; Issaka, R.B.; Raymond, V.M.; Eagle, C.; Hu, S.; Chudova, D.I.; Talasaz, A.; Greenson, J.K.; et al. A Cell-free DNA Blood-Based Test for Colorectal Cancer Screening. N. Engl. J. Med. 2024, 390, 973–983. [Google Scholar] [CrossRef]
- Nicholson, B.D.; Oke, J.; Virdee, P.S.; Harris, D.A.; O’Doherty, C.; Park, J.E.; Hamady, Z.; Sehgal, V.; Millar, A.; Medley, L.; et al. Multi-cancer early detection test in symptomatic patients referred for cancer investigation in England and Wales (SYMPLIFY): A large-scale, observational cohort study. Lancet Oncol. 2023, 24, 733–743, Erratum in Lancet Oncol. 2024, 25, e401. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, T.; Xu, D.; Shi, H.; Zhang, C.; Mo, Y.Y.; Wang, Z. Predicting DNA Methylation State of CpG Dinucleotide Using Genome Topological Features and Deep Networks. Sci. Rep. 2016, 6, 19598. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Genereux, D.P.; Johnson, W.C.; Burden, A.F.; Stöger, R.; Laird, C.D. Errors in the bisulfite conversion of DNA: Modulating inappropriate- and failed-conversion frequenci.es. Nucleic Acids Res. 2008, 36, e150, Erratum in Nucleic Acids Res. 2009, 37, 5235. [Google Scholar] [CrossRef] [PubMed]
- Geertsen, L.; Koldby, K.M.; Thomassen, M.; Kruse, T.; Lund, L. Circulating Tumor DNA in Patients with Renal Cell Carcinoma. A Systematic Review of the Literature. Eur. Urol. Open Sci. 2022, 37, 27–35. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tzanikou, E.; Lianidou, E. The potential of ctDNA analysis in breast cancer. Crit. Rev. Clin. Lab. Sci. 2020, 57, 54–72. [Google Scholar] [CrossRef] [PubMed]
- Rendek, T.; Pos, O.; Duranova, T.; Saade, R.; Budis, J.; Repiska, V.; Szemes, T. Current Challenges of Methylation-Based Liquid Biopsies in Cancer Diagnostics. Cancers 2024, 16, 2001. [Google Scholar] [CrossRef] [PubMed]
- Yasui, K.; Toshima, T.; Inada, R.; Umeda, Y.; Yano, S.; Tanioka, H.; Nyuya, A.; Fujiwara, T.; Yamada, T.; Naomoto, Y.; et al. Circulating cell-free DNA methylation patterns as non-invasive biomarkers to monitor colorectal cancer treatment efficacy without referencing primary site mutation profiles. Mol. Cancer 2024, 23, 1. [Google Scholar] [CrossRef] [PubMed]
- Manoochehri, M.; Borhani, N.; Gerhäuser, C.; Assenov, Y.; Schönung, M.; Hielscher, T.; Christensen, B.C.; Lee, M.K.; Gröne, H.J.; Lipka, D.B.; et al. DNA methylation biomarkers for noninvasive detection of triple-negative breast cancer using liquid biopsy. Int. J. Cancer 2023, 152, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- de Vos, L.; Jung, M.; Koerber, R.M.; Bawden, E.G.; Holderried, T.A.W.; Dietrich, J.; Bootz, F.; Brossart, P.; Kristiansen, G.; Dietrich, D. Treatment Response Monitoring in Patients with Advanced Malignancies Using Cell-Free SHOX2 and SEPT9 DNA Methylation in Blood: An Observational Prospective Study. J. Mol. Diagn. 2020, 22, 920–933. [Google Scholar] [CrossRef]
- Fackler, M.J.; Lopez Bujanda, Z.; Umbricht, C.; Teo, W.W.; Cho, S.; Zhang, Z.; Visvanathan, K.; Jeter, S.; Argani, P.; Wang, C.; et al. Novel Methylated Biomarkers and a Robust Assay to Detect Circulating Tumor DNA in Metastatic Breast Cancer. Cancer Res. 2014, 74, 2160–2170, Erratum in Cancer Res. 2014, 74, 3196. [Google Scholar] [CrossRef]
- Symonds, E.L.; Pedersen, S.K.; Yeo, B.; Al Naji, H.; Byrne, S.E.; Roy, A.; Young, G.P. Assessment of tumor burden and response to therapy in patients with colorectal cancer using a quantitative ctDNA test for methylated BCAT1/IKZF1. Mol. Oncol. 2022, 16, 2031–2041. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bjerre, M.T.; Nørgaard, M.; Larsen, O.H.; Jensen, S.Ø.; Strand, S.H.; Østergren, P.; Fode, M.; Fredsøe, J.; Ulhøi, B.P.; Mortensen, M.M.; et al. Epigenetic Analysis of Circulating Tumor DNA in Localized and Metastatic Prostate Cancer: Evaluation of Clinical Biomarker Potential. Cells 2020, 9, 1362. [Google Scholar] [CrossRef]
- Angeli-Pahim, I.; Chambers, A.; Duarte, S.; Soma, D.; Beduschi, T.; Sahin, I.; Hughes, S.; Zarrinpar, A. Methylated ctDNA Quantification: Noninvasive Approach to Monitoring Hepatocellular Carcinoma Burden. J. Am. Coll. Surg. 2024, 238, 770–778. [Google Scholar] [CrossRef]
- Church, T.R.; Wandell, M.; Lofton-Day, C.; Mongin, S.J.; Burger, M.; Payne, S.R.; Castaños-Vélez, E.; Blumenstein, B.A.; Rösch, T.; Osborn, N.; et al. PRESEPT Clinical Study Steering Committee, Investigators and Study Team. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014, 63, 317–325. [Google Scholar] [CrossRef]
- Song, L.; Jia, J.; Peng, X.; Xiao, W.; Li, Y. The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: A meta-analysis. Sci. Rep. 2017, 7, 3032. [Google Scholar] [CrossRef] [PubMed]
- Ogaard, N.; Reinert, T.; Henriksen, T.V.; Frydendahl, A.; Aagaard, E.; Ørntoft, M.W.; Larsen, M.Ø.; Knudsen, A.R.; Mortensen, F.V.; Andersen, C.L. Tumour-agnostic circulating tumour DNA analysis for improved recurrence surveillance after resection of colorectal liver metastases: A prospective cohort study. Eur. J. Cancer. 2022, 163, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Kim, J.H.; Kim, Y.J.; Jeon, H.; Kim, B.C.; Jeon, Y.; Kim, Y.; Bak, H.; Kang, Y.; Kim, C.; et al. Quantification method of ctDNA using cell-free DNA methylation profile for noninvasive screening and monitoring of colon cancer. Clin. Epigenet. 2024, 16, 95. [Google Scholar] [CrossRef]
- Hsiao, A.; Woodward, B.; Ye, P.; Varga, M.G.; Altaie, G.; Lu, K.; Searle, N.; Viens, R.; Langpap, S.; Li, Z.; et al. Brief Report: Methylation-Based ctDNA Serial Monitoring Correlates with Immunotherapy Response in NSCLC. Clin. Lung Cancer 2025, 26, 72–77. [Google Scholar] [CrossRef]
- Ye, P.P.; Viens, R.; Shelburne, K.E.; Langpap, S.S.; Bower, X.S.; Shi, J.J.; Zhou, W.; Wignall, J.C.; Zhu, J.J.; Woodward, B.D.; et al. Molecular counting enables accurate and precise quantification of methylated ctDNA for tumor-naive cancer therapy response monitoring. Sci. Rep. 2025, 15, 5869. [Google Scholar] [CrossRef]
- Chan, K.C.; Jiang, P.; Chan, C.W.; Sun, K.; Wong, J.; Hui, E.P.; Chan, S.L.; Chan, W.C.; Hui, D.S.; Ng, S.S.; et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 18761–18768. [Google Scholar] [CrossRef]
- Gao, Y.; Zhao, H.; An, K.; Liu, Z.; Hai, L.; Li, R.; Zhou, Y.; Zhao, W.; Jia, Y.; Wu, N.; et al. Whole-genome bisulfite sequencing analysis of circulating tumour DNA for the detection and molecular classification of cancer. Clin. Transl. Med. 2022, 12, e1014. [Google Scholar] [CrossRef] [PubMed]
- Lun, F.M.; Chiu, R.W.; Sun, K.; Leung, T.Y.; Jiang, P.; Chan, K.C.; Sun, H.; Lo, Y.M. Noninvasive prenatal methylomic analysis by genomewide bisulfite sequencing of maternal plasma DNA. Clin. Chem. 2013, 59, 1583–1594. [Google Scholar] [CrossRef]
- Kwon, H.J.; Shin, S.H.; Kim, H.H.; Min, N.Y.; Lim, Y.; Joo, T.-W.; Lee, K.J.; Jeong, M.-S.; Kim, H.; Yun, S.-Y.; et al. Advances in methylation analysis of liquid biopsy in early cancer detection of colorectal and lung cancer. Sci. Rep. 2023, 13, 13502. [Google Scholar] [CrossRef] [PubMed]
- Füllgrabe, J.; Gosal, W.S.; Creed, P.; Liu, S.; Lumby, C.K.; Morley, D.J.; Ost, T.W.B.; Vilella, A.J.; Yu, S.; Bignell, H.; et al. Simultaneous sequencing of genetic and epigenetic bases in DNA. Nat. Biotechnol. 2023, 41, 1457–1464. [Google Scholar] [CrossRef]
- Medina, J.E.; Annapragada, A.V.; Lof, P.; Short, S.; Bartolomucci, A.L.; Mathios, D.; Koul, S.; Niknafs, N.; Noë, M.; Foda, Z.H.; et al. Early Detection of Ovarian Cancer Using Cell-Free DNA Fragmentomes and Protein Biomarkers. Cancer Discov. 2025, 15, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Wang, H.; Wu, W.; Geng, S.; Zhong, G.; Li, Y.; Guo, H.; Long, G.; Ren, Q.; Luan, Y.; et al. Circulating cell-free DNA fragmentation is a stepwise and conserved process linked to apoptosis. BMC Biol. 2023, 21, 253. [Google Scholar] [CrossRef]
- Fan, H.C.; Blumenfeld, Y.J.; Chitkara, U.; Hudgins, L.; Quake, S.R. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. USA 2008, 105, 16266–16271. [Google Scholar] [CrossRef]
- Lo, Y.M.; Chan, K.C.; Sun, H.; Chen, E.Z.; Jiang, P.; Lun, F.M.; Zheng, Y.W.; Leung, T.Y.; Lau, T.K.; Cantor, C.R.; et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2010, 2, 61ra91. [Google Scholar] [CrossRef]
- Zheng, Y.W.L.; Chan, K.C.A.; Sun, H.; Jiang, P.Y.; Su, X.X.; Chen, E.Z.; Lun, F.M.F.; Hung, E.C.W.; Lee, V.; Wong, J.; et al. Nonhematopoietically Derived DNA Is Shorter than Hematopoietically Derived DNA in Plasma: A Transplantation Model. Clin. Chem. 2012, 58, 549–558. [Google Scholar] [CrossRef]
- Heitzer, E.; Speicher, M.R. One size does not fit all: Size-based plasma DNA diagnostics. Sci. Transl. Med. 2018, 10, eaav3873. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.C.; Lee, S.W.; Jiang, P.; Leung, T.Y.; Chan, K.C.; Chiu, R.W.; Lo, Y.M. High-resolution profiling of fetal DNA clearance from maternal plasma by massively parallel sequencing. Clin. Chem. 2013, 59, 1228–1237. [Google Scholar] [CrossRef]
- De Maio, N.; Shaw, L.P.; Hubbard, A.; George, S.; Sanderson, N.D.; Swann, J.; Wick, R.; AbuOun, M.; Stubberfield, E.; Hoosdally, S.J.; et al. Comparison of long-read sequencing technologies in the hybrid assembly of complex bacterial genomes. Microb. Genom. 2019, 5, e000294. [Google Scholar] [CrossRef]
- Mouliere, F.; Robert, B.; Peyrotte, E.A.; Del Rio, M.; Ychou, M.; Molina, F.; Gongora, C.; Thierry, A.R. High fragmentation characterizes tumour-derived circulating DNA. PLoS ONE. 2011, 6, e23418. [Google Scholar] [CrossRef]
- Esfahani, M.S.; Hamilton, E.G.; Mehrmohamadi, M.; Nabet, B.Y.; Alig, S.K.; King, D.A.; Steen, C.B.; Macaulay, C.W.; Schultz, A.; Nesselbush, M.C.; et al. Inferring gene expression from cell-free DNA fragmentation profiles. Nat. Biotechnol. 2022, 40, 585–597. [Google Scholar] [CrossRef]
- Stanley, K.E.; Jatsenko, T.; Tuveri, S.; Sudhakaran, D.; Lannoo, L.; Van Calsteren, K.; de Borre, M.; Van Parijs, I.; Van Coillie, L.; Bogaert, K.V.D.; et al. Cell type signatures in cell-free DNA fragmentation profiles reveal disease biology. Nat. Commun. 2024, 15, 2220. [Google Scholar] [CrossRef]
- Maansson, C.T.; Thomsen, L.S.; Meldgaard, P.; Nielsen, A.L.; Sorensen, B.S. Integration of cell-free DNA end motifs and fragment lengths can identify active genes in liquid biopsies. Int. J. Mol. Sci. 2024, 25, 1243. [Google Scholar] [CrossRef] [PubMed]
- Lapin, M.; Oltedal, S.; Tjensvoll, K.; Buhl, T.; Smaaland, R.; Garresori, H.; Javle, M.; Glenjen, N.I.; Abelseth, B.K.; Gilje, B.; et al. Fragment size and level of cell-free DNA provide prognostic information in patients with advanced pancreatic cancer. J. Transl. Med. 2018, 16, 300. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Koval, A.P.; Khromova, A.S.; Blagodatskikh, K.A.; Zhitnyuk, Y.V.; Shtykova, Y.A.; Alferov, A.A.; Kushlinskii, N.E.; Shcherbo, D.S. Application of PCR-based approaches for evaluation of cell-free DNA fragmentation in colorectal cancer. Front. Mol. Biosci. 2023, 10, 1101179. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Helzer, K.; Sharifi, M.; Sperger, J.; Shi, Y.; Annala, M.; Bootsma, M.; Reese, S.; Taylor, A.; Kaufmann, K.; Krause, H.; et al. Fragmentomic analysis of circulating tumor DNA-targeted cancer panels. Ann. Oncol. 2023, 34, 813–825. [Google Scholar] [CrossRef]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.Ø.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef]
- Wang, J.; Niu, Y.; Yang, M.; Shu, L.; Wang, H.; Wu, X.; He, Y.; Chen, P.; Zhong, G.; Tang, Z.; et al. Altered cfDNA fragmentation profile in hypomethylated regions as diagnostic markers in breast cancer. Epigenet. Chromatin 2023, 16, 33. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; Heider, K.; et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018, 10, eaat4921. [Google Scholar] [CrossRef]
- Underhill, H.R.; Kitzman, J.O.; Hellwig, S.; Welker, N.C.; Daza, R.; Baker, D.N.; Gligorich, K.M.; Rostomily, R.C.; Bronner, M.P.; Shendure, J. Fragment length of circulating tumor DNA. PLOS Genet. 2016, 12, e1006162. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Heider, K.; Gale, D.; Murphy, S.; Fisher, E.; Mouliere, F.; Ruiz-Valdepenas, A.; Santonja, A.; Morris, J.; Chandrananda, D.; et al. ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci. Transl. Med. 2020, 12, eaaz8084. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lin, N.; Niu, M.; Yin, B. Circulating tumor DNA mutation analysis: Advances in its application for early diagnosis of hepatocellular carcinoma and therapeutic efficacy monitoring. Aging 2024, 16, 11460–11474. [Google Scholar] [CrossRef] [PubMed]
- Mathios, D.; Johansen, J.S.; Cristiano, S.; Medina, J.E.; Phallen, J.; Larsen, K.R.; Bruhm, D.C.; Niknafs, N.; Ferreira, L.; Adleff, V.; et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat. Commun. 2021, 12, 5060. [Google Scholar] [CrossRef] [PubMed]
- Cotton, L.B.; Bach, P.B.; Cisar, C.; Schonewolf, C.A.; Tennefoss, D.; Vachani, A.; Carter-Bawa, L.; Zaidi, A.H. Innovations in Early Lung Cancer Detection: Tracing the Evolution and Advancements in Screening. J. Clin. Med. 2024, 13, 4911. [Google Scholar] [CrossRef]
- Foda, Z.H.; Annapragada, A.V.; Boyapati, K.; Bruhm, D.C.; Vulpescu, N.A.; Medina, J.E.; Mathios, D.; Cristiano, S.; Niknafs, N.; Luu, H.T.; et al. Detecting liver cancer using cell-free DNA fragmentomes. Cancer Discov. 2023, 13, 616–631. [Google Scholar] [CrossRef]
- Annapragada, A.V.; Niknafs, N.; White, J.R.; Bruhm, D.C.; Cherry, C.; Medina, J.E.; Adleff, V.; Hruban, C.; Mathios, D.; Foda, Z.H.; et al. Genome-wide repeat landscapes in cancer and cell-free DNA. Sci. Transl. Med. 2024, 16, eadj9283. [Google Scholar] [CrossRef] [PubMed]
- Noë, M.; Mathios, D.; Annapragada, A.V.; Koul, S.; Foda, Z.H.; Medina, J.E.; Cristiano, S.; Cherry, C.; Bruhm, D.C.; Niknafs, N.; et al. DNA methylation and gene expression as determinants of genomewide cell-free DNA fragmentation. Nat. Commun. 2024, 15, 6690. [Google Scholar] [CrossRef]
- Dong, W.; Hu, W.; Lu, Y.; Zheng, Q. Cell-free DNA fragmentomics: A universal framework for early cancer detection and monitoring. Am. J. Clin. Exp. Immunol. 2025, 14, 237–240. [Google Scholar] [CrossRef]
- Jiang, P.; Sun, K.; Tong, Y.K.; Cheng, S.H.; Cheng, T.H.T.; Heung, M.M.S.; Wong, J.; Wong, V.W.S.; Chan, H.L.Y.; Chan, K.C.A.; et al. Chiu RWK. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 2018, 115, E10925–E10933. [Google Scholar] [CrossRef]
- Budhraja, K.K.; McDonald, B.R.; Stephens, M.D.; Contente-Cuomo, T.; Markus, H.; Farooq, M.; Favaro, P.F.; Connor, S.; Byron, S.A.; Egan, J.B.; et al. Genome-wide analysis of aberrant position and sequence of plasma DNA fragment ends in patients with cancer. Sci. Transl. Med. 2023, 15, eabm6863. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, Q.; Huang, Z.; Song, L.; Zhao, F.; Gu, T.; Feng, Z.; Wang, H.; Li, B.; Wang, D.; et al. Cell-free epigenomes enhanced fragmentomics-based model for early detection of lung cancer. Clin. Transl. Med. 2025, 15, e70225. [Google Scholar] [CrossRef] [PubMed]
- Janke, F.; Gasser, M.; Angeles, A.K.; Riediger, A.L.; Görtz, M.; Appenheimer, L.; Laut, A.K.; Ogrodnik, S.; Gerhardt, S.; Stenzinger, A.; et al. Low-coverage whole genome sequencing of cell-free DNA to predict and track immunotherapy response in advanced non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2025, 44, 87. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Rabia Saleem, B.-Y.; Fatih Kurugollu, A.A.; Lu, L. Explaining deep neural networks: A survey on the global interpretation methods. Neurocomputing 2022, 513, 165–180. [Google Scholar] [CrossRef]
- Stutheit-Zhao, E.Y.; Sanz-Garcia, E.; Liu, Z.A.; Wong, D.; Marsh, K.; Abdul Razak, A.R.; Spreafico, A.; Bedard, P.L.; Hansen, A.R.; Lheureux, S.; et al. Early changes in tumor-naive cell-free methylomes and fragmentomes predict outcomes in pembrolizumab-treated solid tumors. Cancer Discov. 2024, 14, 1048–1063. [Google Scholar] [CrossRef]
- Sahoo, K.; Lingasamy, P.; Khatun, M.; Sudhakaran, S.L.; Salumets, A.; Sundararajan, V.; Modhukur, V. Artificial Intelligence in cancer epigenomics: A review on advances in pan-cancer detection and precision medicine. Epigenet. Chromatin 2025, 18, 35, Erratum in Epigenet. Chromatin 2025, 18, 42. https://doi.org/10.1186/s13072-025-00604-7. [Google Scholar] [CrossRef]
- Moldovan, N.; Pol, Y.; van der Ende, T.; van den Boers, D.; Verkuijlen, S.; Creemers, A.; Ramaker, J.; Vu, T.; Bootsma, S.; Lenos, K.J.; et al. Multi-modal cell-free DNA genomic and fragmentomic patterns enhance cancer survival and recurrence analysis. Cell Rep. 2024, 5, 101349. [Google Scholar] [CrossRef] [PubMed]
- Siejka-Zielińska, P.; Cheng, J.; Jackson, F.; Liu, Y.; Soonawalla, Z.; Reddy, S.; Silva, M.; Puta, L.; McCain, M.V.; Culver, E.L.; et al. Cell-free DNA TAPS provides multimodal information for early cancer detection. Sci. Adv. 2021, 7, eabh0534. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Jeong, S.; Lee, W.; Jeon, Y.; Kim, Y.-J.; Park, S.; Lee, D.; Go, D.; Song, S.-H.; Lee, S.; et al. Cancer signature ensemble integrating cfDNA methylation, copy number, and fragmentation facilitates multi-cancer early detection. Exp. Mol. Med. 2023, 55, 2445–2460. [Google Scholar] [CrossRef]
- Nguyen, V.T.C.; Nguyen, T.H.; Doan, N.N.T.; Pham, T.M.Q.; Nguyen, G.T.H.; Nguyen, T.D.; Tran, T.T.T.; Vo, D.L.; Phan, T.H.; Jasmine, T.X.; et al. Multimodal analysis of methylomics and fragmentomics in plasma cell-free DNA for multi-cancer early detection and localization. eLife 2023, 12, RP89083. [Google Scholar] [CrossRef]
- Bie, F.; Wang, Z.; Li, Y.; Guo, W.; Hong, Y.; Han, T.; Lv, F.; Yang, S.; Li, S.; Li, X.; et al. Multimodal analysis of cell-free DNA whole-methylome sequencing for cancer detection and localization. Nat. Commun. 2023, 14, 6042. [Google Scholar] [CrossRef]
- Chabon, J.J.; Hamilton, E.G.; Kurtz, D.M.; Esfahani, M.S.; Moding, E.J.; Stehr, H.; Schroers-Martin, J.; Nabet, B.Y.; Chen, B.; Chaudhuri, A.A.; et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 2020, 580, 245–251. [Google Scholar] [CrossRef]
- Si, H.Q.; Wang, P.; Long, F.; Zhong, W.; Meng, Y.D.; Rong, Y.; Meng, X.Y.; Wang, F.B. Cancer liquid biopsies by Oxford Nanopore Technologies sequencing of cell-free DNA: From basic research to clinical applications. Mol. Cancer 2024, 23, 265. [Google Scholar] [CrossRef]
- Pando-Caciano, A.; Trivedi, R.; Pauwels, J.; Nowakowska, J.; Cavina, B.; Falkman, L.; Debattista, J.; Belényesi, S.K.; Radhakrishnan, P.; Molina, M.A. Unlocking the promise of liquid biopsies in precision oncology. J. Liq. Biopsy 2024, 3, 100151. [Google Scholar] [CrossRef]
- Cherukuri, S.P.; Kaur, A.; Goyal, B.; Kukunoor, H.R.; Sahito, A.F.; Sachdeva, P.; Yerrapragada, G.; Elangovan, P.; Shariff, M.N.; Natarajan, T.; et al. Artificial Intelligence-Enhanced Liquid Biopsy and Radiomics in Early-Stage Lung Cancer Detection: A Precision Oncology Paradigm. Cancers 2025, 17, 3165. [Google Scholar] [CrossRef]
- Greener, J.G.; Kandathil, S.M.; Moffat, L.; Jones, D.T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022, 23, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Lococo, F.; Boldrini, L.; Diepriye, C.D.; Evangelista, J.; Nero, C.; Flamini, S.; Minucci, A.; De Paolis, E.; Vita, E.; Cesario, A.; et al. Lung cancer multi-omics digital human avatars for integrating precision medicine into clinical practice: The LANTERN study. BMC Cancer 2023, 23, 540, Erratum in BMC Cancer 2023, 23, 1082. https://doi.org/10.1186/s12885-023-11606-7. [Google Scholar] [CrossRef] [PubMed]
- Kerachian, M.A.; Azghandi, M.; Mozaffari-Jovin, S.; Thierry, A.R. Guidelines for pre-analytical conditions for assessing the methylation of circulating cell-free DNA. Clin. Epigenet. 2021, 13, 193. [Google Scholar] [CrossRef] [PubMed]
- Meddeb, R.; Pisareva, E.; Thierry, A.R. Guidelines for the Preanalytical Conditions for Analyzing Circulating Cell-Free DNA. Clin. Chem. 2019, 65, 623–633. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).