New Roles of bZIP-Containing Membrane-Bound Transcription Factors in Chromatin Tethering and Karyoptosis
Abstract
1. Introduction
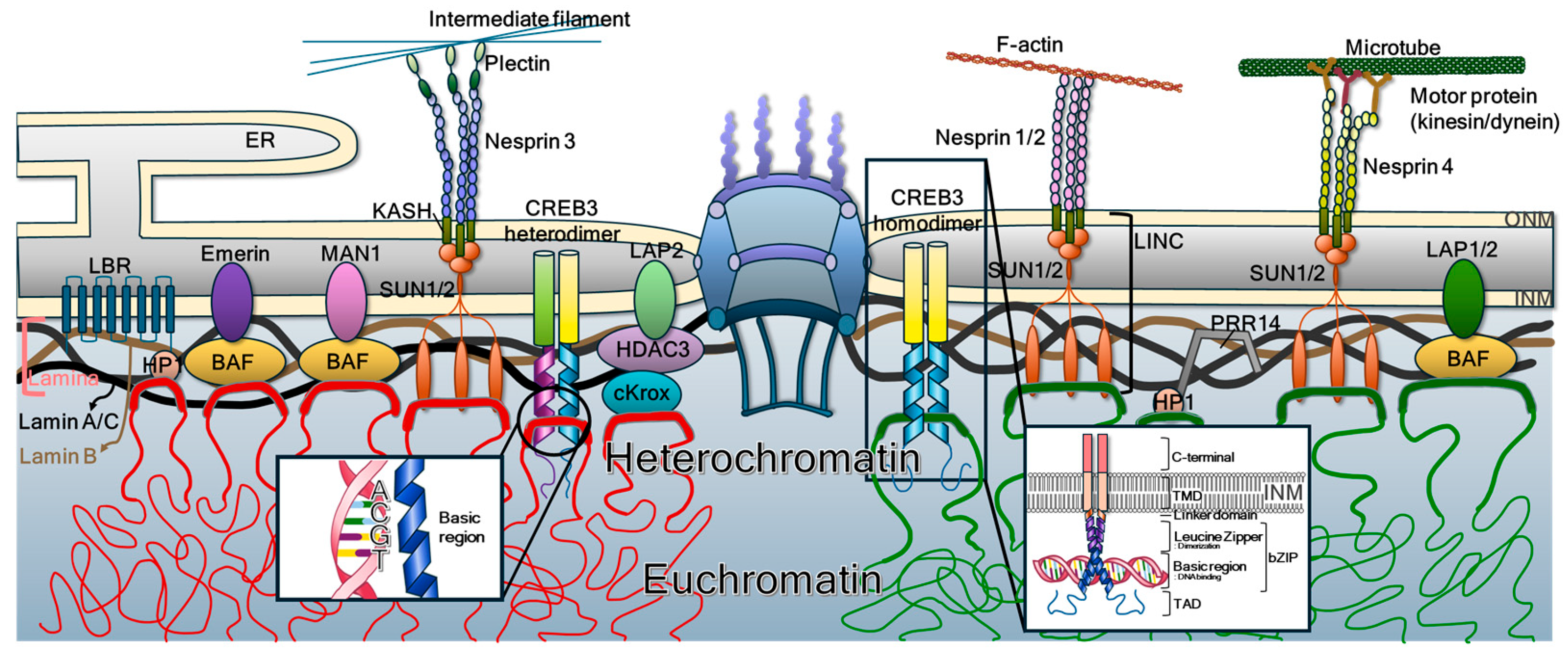
2. Nuclear Membrane Constituents
2.1. Nuclear Pore Complexes
2.2. Integral Proteins in the ONM
2.3. Integral Proteins in the INM
2.4. INM-Localized Integral Proteins Interacting with Chromatin
3. bZIP Family Members
3.1. Background of bZIP Transcription Factors
3.2. bZIP Family Members Lacking Transmembrane Domain
3.3. bZIP Members Containing Transmembrane Domain
4. Nuclear Integrity Regulation
4.1. Chromatin Tethering via Interaction with Chromatin
4.2. Chromatin Tethering via Interaction with Genomic DNA in Chromatin
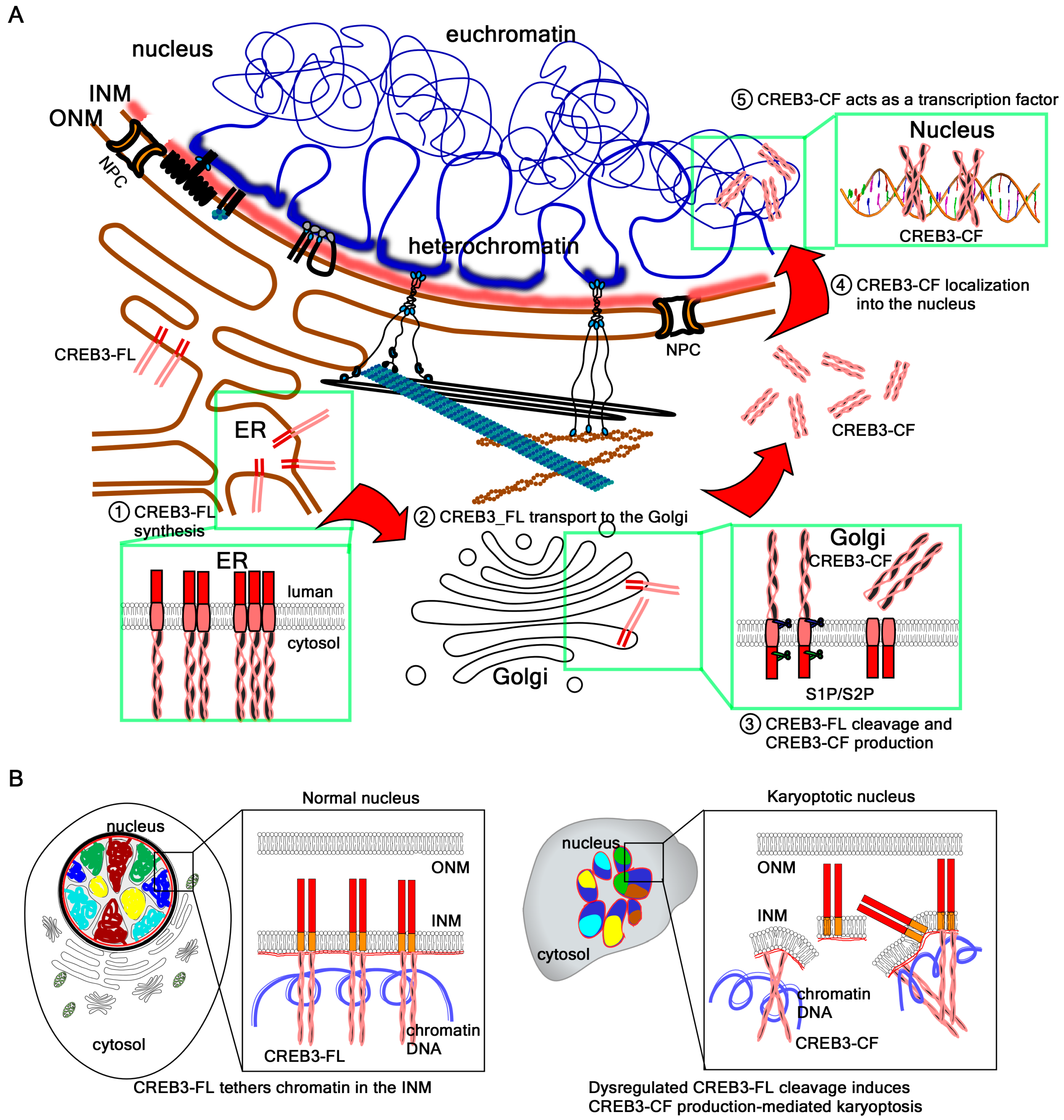
4.3. The Molecular Mechanism of Karyoptosis Involving CREB3-CF
4.4. Potential Roles of Chromatin Acting as a Nucleoskeleton
5. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, W.; Byun, J.; Kang, H.C.; Lee, H.S.; Lee, J.Y.; Kwon, Y.J.; Cho, Y.Y. Karyoptosis as a novel type of UVB-induced regulated cell death. Free Radic. Res. 2024, 58, 796–810. [Google Scholar] [CrossRef] [PubMed]
- Adam, S.A. The Nucleoskeleton. Cold Spring Harb. Perspect. Biol. 2017, 9, a023556. [Google Scholar] [CrossRef] [PubMed]
- Worman, H.J.; Schirmer, E.C. Nuclear membrane diversity: Underlying tissue-specific pathologies in disease? Curr. Opin. Cell Biol. 2015, 34, 101–112. [Google Scholar] [CrossRef]
- Stirling, J.; O’Hare, P. CREB4, a transmembrane bZip transcription factor and potential new substrate for regulation and cleavage by S1P. Mol. Biol. Cell 2006, 17, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Audas, T.E.; Li, Y.; Cockram, G.P.; Dean, J.D.; Martyn, A.C.; Kokame, K.; Lu, R. Luman/CREB3 induces transcription of the endoplasmic reticulum (ER) stress response protein Herp through an ER stress response element. Mol. Cell Biol. 2006, 26, 7999–8010. [Google Scholar] [CrossRef]
- Lee, G.E.; Byun, J.; Lee, C.J.; Cho, Y.Y. Molecular Mechanisms for the Regulation of Nuclear Membrane Integrity. Int. J. Mol. Sci. 2023, 24, 15497. [Google Scholar] [CrossRef]
- Lee, G.E.; Bang, G.; Byun, J.; Lee, C.J.; Chen, W.; Jeung, D.; An, H.J.; Kang, H.C.; Lee, J.Y.; Lee, H.S.; et al. Dysregulated CREB3 cleavage at the nuclear membrane induces karyoptosis-mediated cell death. Exp. Mol. Med. 2024, 56, 686–699. [Google Scholar] [CrossRef]
- Guo, T.; Fang, Y. Functional organization and dynamics of the cell nucleus. Front. Plant Sci. 2014, 5, 378. [Google Scholar] [CrossRef]
- Ungricht, R.; Kutay, U. Mechanisms and functions of nuclear envelope remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 229–245. [Google Scholar] [CrossRef]
- Bodoor, K.; Shaikh, S.; Salina, D.; Raharjo, W.H.; Bastos, R.; Lohka, M.; Burke, B. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 1999, 112 Pt 13, 2253–2264. [Google Scholar] [CrossRef]
- Timney, B.L.; Raveh, B.; Mironska, R.; Trivedi, J.M.; Kim, S.J.; Russel, D.; Wente, S.R.; Sali, A.; Rout, M.P. Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 2016, 215, 57–76. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Hurt, E. The nuclear pore complex: Understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2017, 18, 73–89. [Google Scholar] [CrossRef] [PubMed]
- De Jesus-Gonzalez, L.A.; Palacios-Rapalo, S.; Reyes-Ruiz, J.M.; Osuna-Ramos, J.F.; Cordero-Rivera, C.D.; Farfan-Morales, C.N.; Gutierrez-Escolano, A.L.; Del Angel, R.M. The Nuclear Pore Complex Is a Key Target of Viral Proteases to Promote Viral Replication. Viruses 2021, 13, 706. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.; Qu, R.; Ouyang, J.; Dai, J. Role of Nucleoporins and Transport Receptors in Cell Differentiation. Front. Physiol. 2020, 11, 239. [Google Scholar] [CrossRef]
- Capelson, M.; Liang, Y.; Schulte, R.; Mair, W.; Wagner, U.; Hetzer, M.W. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 2010, 140, 372–383. [Google Scholar] [CrossRef]
- Liang, Y.; Hetzer, M.W. Functional interactions between nucleoporins and chromatin. Curr. Opin. Cell Biol. 2011, 23, 65–70. [Google Scholar] [CrossRef]
- Liang, Y.; Franks, T.M.; Marchetto, M.C.; Gage, F.H.; Hetzer, M.W. Dynamic association of NUP98 with the human genome. PLoS Genet. 2013, 9, e1003308. [Google Scholar] [CrossRef]
- Shaulov, L.; Gruber, R.; Cohen, I.; Harel, A. A dominant-negative form of POM121 binds chromatin and disrupts the two separate modes of nuclear pore assembly. J. Cell Sci. 2011, 124, 3822–3834. [Google Scholar] [CrossRef]
- Ungricht, R.; Klann, M.; Horvath, P.; Kutay, U. Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J. Cell Biol. 2015, 209, 687–703. [Google Scholar] [CrossRef]
- Lu, J.; Wu, T.; Zhang, B.; Liu, S.; Song, W.; Qiao, J.; Ruan, H. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun. Signal 2021, 19, 60. [Google Scholar] [CrossRef]
- Liu, J.; Hetzer, M.W. Nuclear pore complex maintenance and implications for age-related diseases. Trends Cell Biol. 2022, 32, 216–227. [Google Scholar] [CrossRef]
- Starr, D.A.; Fridolfsson, H.N. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 2010, 26, 421–444. [Google Scholar] [CrossRef]
- Mellad, J.A.; Warren, D.T.; Shanahan, C.M. Nesprins LINC the nucleus and cytoskeleton. Curr. Opin. Cell Biol. 2011, 23, 47–54. [Google Scholar] [CrossRef]
- Zi-Yi, Z.; Qin, Q.; Fei, Z.; Cun-Yu, C.; Lin, T. Nesprin proteins: Bridging nuclear envelope dynamics to muscular dysfunction. Cell Commun. Signal 2024, 22, 208. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.G.S.; Tinker, A.; Iskratsch, T. The role of the dystrophin glycoprotein complex in muscle cell mechanotransduction. Commun. Biol. 2022, 5, 1022. [Google Scholar] [CrossRef] [PubMed]
- Worman, H.J.; Yuan, J.; Blobel, G.; Georgatos, S.D. A lamin B receptor in the nuclear envelope. Proc. Natl. Acad. Sci. USA 1988, 85, 8531–8534. [Google Scholar] [CrossRef] [PubMed]
- Olins, A.L.; Rhodes, G.; Welch, D.B.; Zwerger, M.; Olins, D.E. Lamin B receptor: Multi-tasking at the nuclear envelope. Nucleus 2010, 1, 53–70. [Google Scholar] [CrossRef]
- Wagner, N.; Krohne, G. LEM-Domain proteins: New insights into lamin-interacting proteins. Int. Rev. Cytol. 2007, 261, 1–46. [Google Scholar] [CrossRef]
- Brachner, A.; Foisner, R. Evolvement of LEM proteins as chromatin tethers at the nuclear periphery. Biochem. Soc. Trans. 2011, 39, 1735–1741. [Google Scholar] [CrossRef]
- Merner, N.D.; Hodgkinson, K.A.; Haywood, A.F.; Connors, S.; French, V.M.; Drenckhahn, J.D.; Kupprion, C.; Ramadanova, K.; Thierfelder, L.; McKenna, W.; et al. Arrhythmogenic right ventricular cardiomyopathy type 5 is a fully penetrant, lethal arrhythmic disorder caused by a missense mutation in the TMEM43 gene. Am. J. Hum. Genet. 2008, 82, 809–821. [Google Scholar] [CrossRef]
- Poleshko, A.; Mansfield, K.M.; Burlingame, C.C.; Andrake, M.D.; Shah, N.R.; Katz, R.A. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep. 2013, 5, 292–301. [Google Scholar] [CrossRef]
- van Steensel, B.; Belmont, A.S. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell 2017, 169, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Wong, X.; Melendez-Perez, A.J.; Reddy, K.L. The Nuclear Lamina. Cold Spring Harb. Perspect. Biol. 2022, 14, a040113. [Google Scholar] [CrossRef] [PubMed]
- Somech, R.; Shaklai, S.; Geller, O.; Amariglio, N.; Simon, A.J.; Rechavi, G.; Gal-Yam, E.N. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J. Cell Sci. 2005, 118, 4017–4025. [Google Scholar] [CrossRef] [PubMed]
- Demmerle, J.; Koch, A.J.; Holaska, J.M. The nuclear envelope protein emerin binds directly to histone deacetylase 3 (HDAC3) and activates HDAC3 activity. J. Biol. Chem. 2012, 287, 22080–22088. [Google Scholar] [CrossRef]
- Lin, F.; Morrison, J.M.; Wu, W.; Worman, H.J. MAN1, an integral protein of the inner nuclear membrane, binds Smad2 and Smad3 and antagonizes transforming growth factor-beta signaling. Hum. Mol. Genet. 2005, 14, 437–445. [Google Scholar] [CrossRef]
- Lee, H.S.; Cho, S.J.; Kang, H.C.; Lee, J.Y.; Kwon, Y.J.; Cho, Y.Y. RSK2 and its binding partners: An emerging signaling node in cancers. Arch. Pharm. Res. 2025, 48, 365–383. [Google Scholar] [CrossRef]
- Amoutzias, G.D.; Veron, A.S.; Weiner, J., III; Robinson-Rechavi, M.; Bornberg-Bauer, E.; Oliver, S.G.; Robertson, D.L. One billion years of bZIP transcription factor evolution: Conservation and change in dimerization and DNA-binding site specificity. Mol. Biol. Evol. 2007, 24, 827–835. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Droge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Verri, A.; Mazzarello, P.; Biamonti, G.; Spadari, S.; Focher, F. The specific binding of nuclear protein(s) to the cAMP responsive element (CRE) sequence (TGACGTCA) is reduced by the misincorporation of U and increased by the deamination of C. Nucleic Acids Res. 1990, 18, 5775–5780. [Google Scholar] [CrossRef]
- Kheradpour, P.; Kellis, M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014, 42, 2976–2987. [Google Scholar] [CrossRef]
- Llorca, C.M.; Potschin, M.; Zentgraf, U. bZIPs and WRKYs: Two large transcription factor families executing two different functional strategies. Front. Plant Sci. 2014, 5, 169. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Shimizu, T.; Toda, T.; Yanagida, M.; Hakoshima, T. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat. Struct. Biol. 2000, 7, 889–893. [Google Scholar] [CrossRef] [PubMed]
- Seldeen, K.L.; McDonald, C.B.; Deegan, B.J.; Bhat, V.; Farooq, A. Dissecting the role of leucine zippers in the binding of bZIP domains of Jun transcription factor to DNA. Biochem. Biophys. Res. Commun. 2010, 394, 1030–1035. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vinson, C.; Myakishev, M.; Acharya, A.; Mir, A.A.; Moll, J.R.; Bonovich, M. Classification of human B-ZIP proteins based on dimerization properties. Mol. Cell Biol. 2002, 22, 6321–6335. [Google Scholar] [CrossRef]
- Ryu, T.; Jung, J.; Lee, S.; Nam, H.J.; Hong, S.W.; Yoo, J.W.; Lee, D.K.; Lee, D. bZIPDB: A database of regulatory information for human bZIP transcription factors. BMC Genom. 2007, 8, 136. [Google Scholar] [CrossRef][Green Version]
- Rodriguez-Martinez, J.A.; Reinke, A.W.; Bhimsaria, D.; Keating, A.E.; Ansari, A.Z. Combinatorial bZIP dimers display complex DNA-binding specificity landscapes. Elife 2017, 6, e19272. [Google Scholar] [CrossRef]
- Glembotski, C.C.; Arrieta, A.; Blackwood, E.A.; Stauffer, W.T. ATF6 as a Nodal Regulator of Proteostasis in the Heart. Front. Physiol. 2020, 11, 267. [Google Scholar] [CrossRef]
- Yuxiong, W.; Faping, L.; Bin, L.; Yanghe, Z.; Yao, L.; Yunkuo, L.; Yishu, W.; Honglan, Z. Regulatory mechanisms of the cAMP-responsive element binding protein 3 (CREB3) family in cancers. Biomed. Pharmacother. 2023, 166, 115335. [Google Scholar] [CrossRef]
- Lei, Y.; Yu, H.; Ding, S.; Liu, H.; Liu, C.; Fu, R. Molecular mechanism of ATF6 in unfolded protein response and its role in disease. Heliyon 2024, 10, e25937. [Google Scholar] [CrossRef]
- Eferl, R.; Wagner, E.F. AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer 2003, 3, 859–868. [Google Scholar] [CrossRef]
- Hess, J.; Angel, P.; Schorpp-Kistner, M. AP-1 subunits: Quarrel and harmony among siblings. J. Cell Sci. 2004, 117, 5965–5973. [Google Scholar] [CrossRef] [PubMed]
- Masquilier, D.; Sassone-Corsi, P. Transcriptional cross-talk: Nuclear factors CREM and CREB bind to AP-1 sites and inhibit activation by Jun. J. Biol. Chem. 1992, 267, 22460–22466. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Ding, Y.; Wild, C.; Shen, Q.; Zhou, J. Small molecule inhibitors targeting activator protein 1 (AP-1). J. Med. Chem. 2014, 57, 6930–6948. [Google Scholar] [CrossRef]
- Foletta, V.C.; Segal, D.H.; Cohen, D.R. Transcriptional regulation in the immune system: All roads lead to AP-1. J. Leukoc. Biol. 1998, 63, 139–152. [Google Scholar] [CrossRef]
- Gachon, F. Physiological function of PARbZip circadian clock-controlled transcription factors. Ann. Med. 2007, 39, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Lavery, D.J.; Lopez-Molina, L.; Margueron, R.; Fleury-Olela, F.; Conquet, F.; Schibler, U.; Bonfils, C. Circadian expression of the steroid 15 alpha-hydroxylase (Cyp2a4) and coumarin 7-hydroxylase (Cyp2a5) genes in mouse liver is regulated by the PAR leucine zipper transcription factor DBP. Mol. Cell Biol. 1999, 19, 6488–6499. [Google Scholar] [CrossRef]
- Jacquemin, P.; Martial, J.A.; Davidson, I. Human TEF-5 is preferentially expressed in placenta and binds to multiple functional elements of the human chorionic somatomammotropin-B gene enhancer. J. Biol. Chem. 1997, 272, 12928–12937. [Google Scholar] [CrossRef]
- Je, E.M.; Choi, Y.J.; Chung, Y.J.; Yoo, N.J.; Lee, S.H. TEAD2, a Hippo pathway gene, is somatically mutated in gastric and colorectal cancers with high microsatellite instability. APMIS 2015, 123, 359–360. [Google Scholar] [CrossRef]
- Hunger, S.P.; Li, S.; Fall, M.Z.; Naumovski, L.; Cleary, M.L. The proto-oncogene HLF and the related basic leucine zipper protein TEF display highly similar DNA-binding and transcriptional regulatory properties. Blood 1996, 87, 4607–4617. [Google Scholar] [CrossRef]
- Lehnertz, B.; Chagraoui, J.; MacRae, T.; Tomellini, E.; Corneau, S.; Mayotte, N.; Boivin, I.; Durand, A.; Gracias, D.; Sauvageau, G. HLF expression defines the human hematopoietic stem cell state. Blood 2021, 138, 2642–2654, Erratum in Blood 2022, 140, 2761. [Google Scholar] [CrossRef] [PubMed]
- Li, M.J.; Yu, C.H.; Chou, S.W.; Su, Y.H.; Liao, K.W.; Chang, H.H.; Yang, Y.L. TCF3-HLF-Positive Acute Lymphoblastic Leukemia Resembling Burkitt Leukemia: Cell Morphologic and Immunophenotypic Findings. JCO Precis. Oncol. 2022, 6, e2200236. [Google Scholar] [CrossRef] [PubMed]
- Mohler, J.; Vani, K.; Leung, S.; Epstein, A. Segmentally restricted, cephalic expression of a leucine zipper gene during Drosophila embryogenesis. Mech. Dev. 1991, 34, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Katsuoka, F.; Yamamoto, M. Small Maf proteins (MafF, MafG, MafK): History, structure and function. Gene 2016, 586, 197–205. [Google Scholar] [CrossRef]
- Hirotsu, Y.; Katsuoka, F.; Funayama, R.; Nagashima, T.; Nishida, Y.; Nakayama, K.; Engel, J.D.; Yamamoto, M. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012, 40, 10228–10239. [Google Scholar] [CrossRef]
- Chowdhury, A.; Katoh, H.; Hatanaka, A.; Iwanari, H.; Nakamura, N.; Hamakubo, T.; Natsume, T.; Waku, T.; Kobayashi, A. Multiple regulatory mechanisms of the biological function of NRF3 (NFE2L3) control cancer cell proliferation. Sci. Rep. 2017, 7, 12494. [Google Scholar] [CrossRef]
- Kobayashi, A. Roles of NRF3 in the Hallmarks of Cancer: Proteasomal Inactivation of Tumor Suppressors. Cancers 2020, 12, 2681. [Google Scholar] [CrossRef]
- Sun, J.; Brand, M.; Zenke, Y.; Tashiro, S.; Groudine, M.; Igarashi, K. Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc. Natl. Acad. Sci. USA 2004, 101, 1461–1466. [Google Scholar] [CrossRef]
- Muto, A.; Ochiai, K.; Kimura, Y.; Itoh-Nakadai, A.; Calame, K.L.; Ikebe, D.; Tashiro, S.; Igarashi, K. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 2010, 29, 4048–4061. [Google Scholar] [CrossRef]
- Takahashi, S. Functional analysis of large MAF transcription factors and elucidation of their relationships with human diseases. Exp. Anim. 2021, 70, 264–271. [Google Scholar] [CrossRef]
- Fujino, M.; Ojima, M.; Takahashi, S. Exploring Large MAF Transcription Factors: Functions, Pathology, and Mouse Models with Point Mutations. Genes 2023, 14, 1883. [Google Scholar] [CrossRef]
- Kim, J.I.; Ho, I.C.; Grusby, M.J.; Glimcher, L.H. The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity 1999, 10, 745–751. [Google Scholar] [CrossRef]
- Lee, K.; Gudapati, P.; Dragovic, S.; Spencer, C.; Joyce, S.; Killeen, N.; Magnuson, M.A.; Boothby, M. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity 2010, 32, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yang, Y.; Qiu, G.; Lal, G.; Wu, Z.; Levy, D.E.; Ochando, J.C.; Bromberg, J.S.; Ding, Y. c-Maf regulates IL-10 expression during Th17 polarization. J. Immunol. 2009, 182, 6226–6236. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, Y.; Suto, A.; Kashiwakuma, D.; Kanari, H.; Kagami, S.; Ikeda, K.; Hirose, K.; Watanabe, N.; Grusby, M.J.; Iwamoto, I.; et al. c-Maf activates the promoter and enhancer of the IL-21 gene, and TGF-beta inhibits c-Maf-induced IL-21 production in CD4+ T cells. J. Leukoc. Biol. 2010, 87, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Moriguchi, T.; Kajihara, M.; Esaki, R.; Harada, A.; Shimohata, H.; Oishi, H.; Hamada, M.; Morito, N.; Hasegawa, K.; et al. MafA is a key regulator of glucose-stimulated insulin secretion. Mol. Cell Biol. 2005, 25, 4969–4976. [Google Scholar] [CrossRef]
- Vanderford, N.L.; Andrali, S.S.; Ozcan, S. Glucose induces MafA expression in pancreatic beta cell lines via the hexosamine biosynthetic pathway. J. Biol. Chem. 2007, 282, 1577–1584. [Google Scholar] [CrossRef]
- Kim, J.W.; Jang, S.M.; Kim, C.H.; An, J.H.; Choi, K.H. Transcriptional activity of neural retina leucine zipper (Nrl) is regulated by c-Jun N-terminal kinase and Tip60 during retina development. Mol. Cell Biol. 2012, 32, 1720–1732. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, L.; Naik, I.; Braunstein, Z.; Zhong, J.; Ren, B. Transcription Factor C/EBP Homologous Protein in Health and Diseases. Front. Immunol. 2017, 8, 1612. [Google Scholar] [CrossRef]
- Tolomeo, M.; Grimaudo, S. The “Janus” Role of C/EBPs Family Members in Cancer Progression. Int. J. Mol. Sci. 2020, 21, 4308. [Google Scholar] [CrossRef]
- Koschmieder, S.; Halmos, B.; Levantini, E.; Tenen, D.G. Dysregulation of the C/EBPalpha differentiation pathway in human cancer. J. Clin. Oncol. 2009, 27, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C.; Calkhoven, C.F. Emerging Role of C/EBPbeta and Epigenetic DNA Methylation in Ageing. Trends Genet. 2020, 36, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Liu, Z.; Wu, L.; Yin, G.; Xie, X.; Kong, W.; Zhou, J.; Liu, S. C/EBPbeta: The structure, regulation, and its roles in inflammation-related diseases. Biomed. Pharmacother. 2023, 169, 115938. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Liang, J.; Lei, X.; Sun, F.; Gong, M.; Liu, B.; Zhou, Z. C/EBPbeta in Alzheimer’s disease: An integrative regulator of pathological mechanisms. Brain Res. Bull. 2025, 221, 111198. [Google Scholar] [CrossRef]
- Parkin, S.E.; Baer, M.; Copeland, T.D.; Schwartz, R.C.; Johnson, P.F. Regulation of CCAAT/enhancer-binding protein (C/EBP) activator proteins by heterodimerization with C/EBPgamma (Ig/EBP). J. Biol. Chem. 2002, 277, 23563–23572. [Google Scholar] [CrossRef]
- Song, B.; Scheuner, D.; Ron, D.; Pennathur, S.; Kaufman, R.J. Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Invest. 2008, 118, 3378–3389. [Google Scholar] [CrossRef]
- Li, Y.; Guo, Y.; Tang, J.; Jiang, J.; Chen, Z. New insights into the roles of CHOP-induced apoptosis in ER stress. Acta Biochim. Biophys. Sin. 2014, 46, 629–640. [Google Scholar] [CrossRef]
- Hu, H.; Tian, M.; Ding, C.; Yu, S. The C/EBP Homologous Protein (CHOP) Transcription Factor Functions in Endoplasmic Reticulum Stress-Induced Apoptosis and Microbial Infection. Front. Immunol. 2018, 9, 3083. [Google Scholar] [CrossRef]
- Mayr, B.; Montminy, M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2001, 2, 599–609. [Google Scholar] [CrossRef]
- Steven, A.; Friedrich, M.; Jank, P.; Heimer, N.; Budczies, J.; Denkert, C.; Seliger, B. What turns CREB on? And off? And why does it matter? Cell Mol. Life Sci. 2020, 77, 4049–4067. [Google Scholar] [CrossRef]
- Gao, Y.P.; Hu, C.; Hu, M.; Dong, W.S.; Li, K.; Ye, Y.J.; Hu, Y.X.; Zhang, X. CREB3 protein family: The promising therapeutic targets for cardiovascular and metabolic diseases. Cell Biol. Toxicol. 2024, 40, 103. [Google Scholar] [CrossRef] [PubMed]
- Jessen, U.; Novitskaya, V.; Pedersen, N.; Serup, P.; Berezin, V.; Bock, E. The transcription factors CREB and c-Fos play key roles in NCAM-mediated neuritogenesis in PC12-E2 cells. J. Neurochem. 2001, 79, 1149–1160. [Google Scholar] [CrossRef] [PubMed]
- Delghandi, M.P.; Johannessen, M.; Moens, U. The cAMP signalling pathway activates CREB through PKA, p38 and MSK1 in NIH 3T3 cells. Cell Signal. 2005, 17, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Delmas, V.; Laoide, B.M.; Masquilier, D.; de Groot, R.P.; Foulkes, N.S.; Sassone-Corsi, P. Alternative usage of initiation codons in mRNA encoding the cAMP-responsive-element modulator generates regulators with opposite functions. Proc. Natl. Acad. Sci. USA 1992, 89, 4226–4230. [Google Scholar] [CrossRef] [PubMed]
- de Groot, R.P.; Delmas, V.; Sassone-Corsi, P. DNA bending by transcription factors CREM and CREB. Oncogene 1994, 9, 463–468. [Google Scholar]
- Don, J.; Stelzer, G. The expanding family of CREB/CREM transcription factors that are involved with spermatogenesis. Mol. Cell Endocrinol. 2002, 187, 115–124. [Google Scholar] [CrossRef]
- Dragan, A.I.; Liu, Y.; Makeyeva, E.N.; Privalov, P.L. DNA-binding domain of GCN4 induces bending of both the ATF/CREB and AP-1 binding sites of DNA. Nucleic Acids Res. 2004, 32, 5192–5197. [Google Scholar] [CrossRef][Green Version]
- Chytil, M.; Peterson, B.R.; Erlanson, D.A.; Verdine, G.L. The orientation of the AP-1 heterodimer on DNA strongly affects transcriptional potency. Proc. Natl. Acad. Sci. USA 1998, 95, 14076–14081. [Google Scholar] [CrossRef]
- Papavassiliou, A.G.; Musti, A.M. The Multifaceted Output of c-Jun Biological Activity: Focus at the Junction of CD8 T Cell Activation and Exhaustion. Cells 2020, 9, 2470. [Google Scholar] [CrossRef]
- Kurlishchuk, Y.; Cindric Vranesic, A.; Jessen, M.; Kipping, A.; Ritter, C.; Kim, K.; Cramer, P.; von Eyss, B. A non-canonical repressor function of JUN restrains YAP activity and liver cancer growth. EMBO J. 2024, 43, 4578–4603. [Google Scholar] [CrossRef]
- Li, W.; Yu, S.; Liu, T.; Kim, J.H.; Blank, V.; Li, H.; Kong, A.N. Heterodimerization with small Maf proteins enhances nuclear retention of Nrf2 via masking the NESzip motif. Biochim. Biophys. Acta 2008, 1783, 1847–1856. [Google Scholar] [CrossRef]
- Chowdhury, M.A.R.; An, J.; Jeong, S. The Pleiotropic Face of CREB Family Transcription Factors. Mol. Cells 2023, 46, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, B.R.; Comaills, V. Nuclear Envelope Integrity in Health and Disease: Consequences on Genome Instability and Inflammation. Int. J. Mol. Sci. 2021, 22, 7281. [Google Scholar] [CrossRef] [PubMed]
- Casolari, J.M.; Brown, C.R.; Komili, S.; West, J.; Hieronymus, H.; Silver, P.A. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 2004, 117, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Garcia, P.; Capelson, M. The nuclear pore complex and the genome: Organizing and regulatory principles. Curr. Opin. Genet. Dev. 2021, 67, 142–150. [Google Scholar] [CrossRef]
- Poleshko, A.; Shah, P.P.; Gupta, M.; Babu, A.; Morley, M.P.; Manderfield, L.J.; Ifkovits, J.L.; Calderon, D.; Aghajanian, H.; Sierra-Pagan, J.E.; et al. Genome-Nuclear Lamina Interactions Regulate Cardiac Stem Cell Lineage Restriction. Cell 2017, 171, 573–587.e14. [Google Scholar] [CrossRef]
- Stephens, A.D.; Liu, P.Z.; Banigan, E.J.; Almassalha, L.M.; Backman, V.; Adam, S.A.; Goldman, R.D.; Marko, J.F. Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell 2018, 29, 220–233. [Google Scholar] [CrossRef]
- Asada, R.; Kanemoto, S.; Kondo, S.; Saito, A.; Imaizumi, K. The signalling from endoplasmic reticulum-resident bZIP transcription factors involved in diverse cellular physiology. J. Biochem. 2011, 149, 507–518. [Google Scholar] [CrossRef]
- Kamikawa, Y.; Saito, A.; Matsuhisa, K.; Kaneko, M.; Asada, R.; Horikoshi, Y.; Tashiro, S.; Imaizumi, K. OASIS/CREB3L1 is a factor that responds to nuclear envelope stress. Cell Death Discov. 2021, 7, 152. [Google Scholar] [CrossRef]
- Thuerauf, D.J.; Morrison, L.; Glembotski, C.C. Opposing roles for ATF6alpha and ATF6beta in endoplasmic reticulum stress response gene induction. J. Biol. Chem. 2004, 279, 21078–21084. [Google Scholar] [CrossRef]
- Maciejowski, J.; Hatch, E.M. Nuclear Membrane Rupture and Its Consequences. Annu. Rev. Cell Dev. Biol. 2020, 36, 85–114. [Google Scholar] [CrossRef] [PubMed]
- Raggo, C.; Rapin, N.; Stirling, J.; Gobeil, P.; Smith-Windsor, E.; O’Hare, P.; Misra, V. Luman, the cellular counterpart of herpes simplex virus VP16, is processed by regulated intramembrane proteolysis. Mol. Cell Biol. 2002, 22, 5639–5649. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, H.; Chen, B.; Kou, H.; Lai, L.; Zhang, X.; Xu, Y.; Sun, Y. Mechanical signal-chromatin interactions: Molecular networks from nuclear membrane force transmission to epigenetic regulation. Front. Med. 2025, 12, 1631645. [Google Scholar] [CrossRef] [PubMed]
- Stephens, A.D.; Banigan, E.J.; Marko, J.F. Chromatin’s physical properties shape the nucleus and its functions. Curr. Opin. Cell Biol. 2019, 58, 76–84. [Google Scholar] [CrossRef]
- Papadopoulos, N.; Lennartsson, J.; Heldin, C.H. PDGFRbeta translocates to the nucleus and regulates chromatin remodeling via TATA element-modifying factor 1. J. Cell Biol. 2018, 217, 1701–1717. [Google Scholar] [CrossRef]
- Vinson, C.; Acharya, A.; Taparowsky, E.J. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim. Biophys. Acta 2006, 1759, 4–12. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeung, D.; Li, X.; Cho, Y.-Y. New Roles of bZIP-Containing Membrane-Bound Transcription Factors in Chromatin Tethering and Karyoptosis. Int. J. Mol. Sci. 2025, 26, 10896. https://doi.org/10.3390/ijms262210896
Jeung D, Li X, Cho Y-Y. New Roles of bZIP-Containing Membrane-Bound Transcription Factors in Chromatin Tethering and Karyoptosis. International Journal of Molecular Sciences. 2025; 26(22):10896. https://doi.org/10.3390/ijms262210896
Chicago/Turabian StyleJeung, Dohyun, Xianzhe Li, and Yong-Yeon Cho. 2025. "New Roles of bZIP-Containing Membrane-Bound Transcription Factors in Chromatin Tethering and Karyoptosis" International Journal of Molecular Sciences 26, no. 22: 10896. https://doi.org/10.3390/ijms262210896
APA StyleJeung, D., Li, X., & Cho, Y.-Y. (2025). New Roles of bZIP-Containing Membrane-Bound Transcription Factors in Chromatin Tethering and Karyoptosis. International Journal of Molecular Sciences, 26(22), 10896. https://doi.org/10.3390/ijms262210896





