Rv1899c, an HDAC1–ZBTB25-Interacting Protein of Mycobacterium tuberculosis, Promotes Stress Resistance and Immune Evasion in Infected Macrophages
Abstract
1. Introduction
2. Results
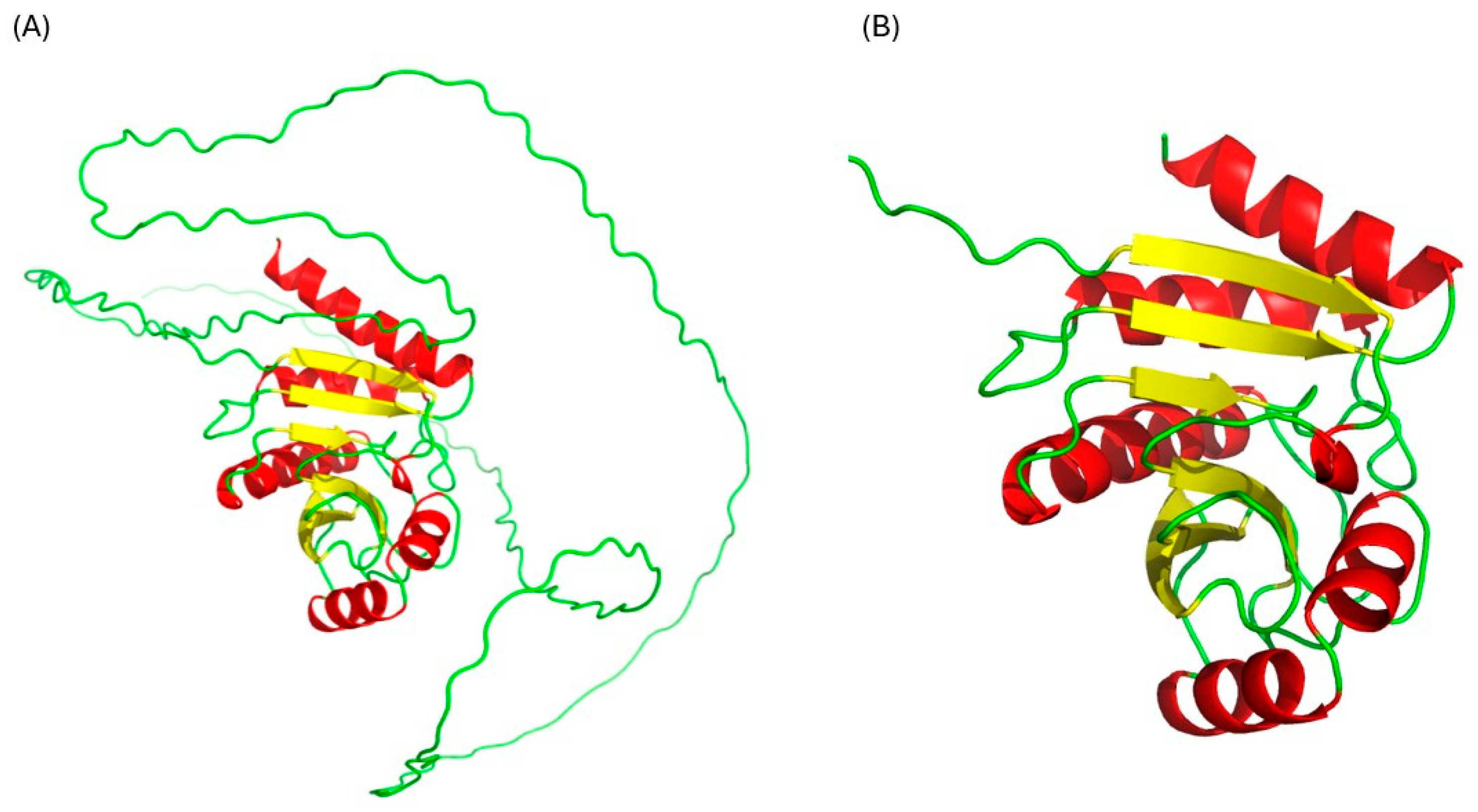
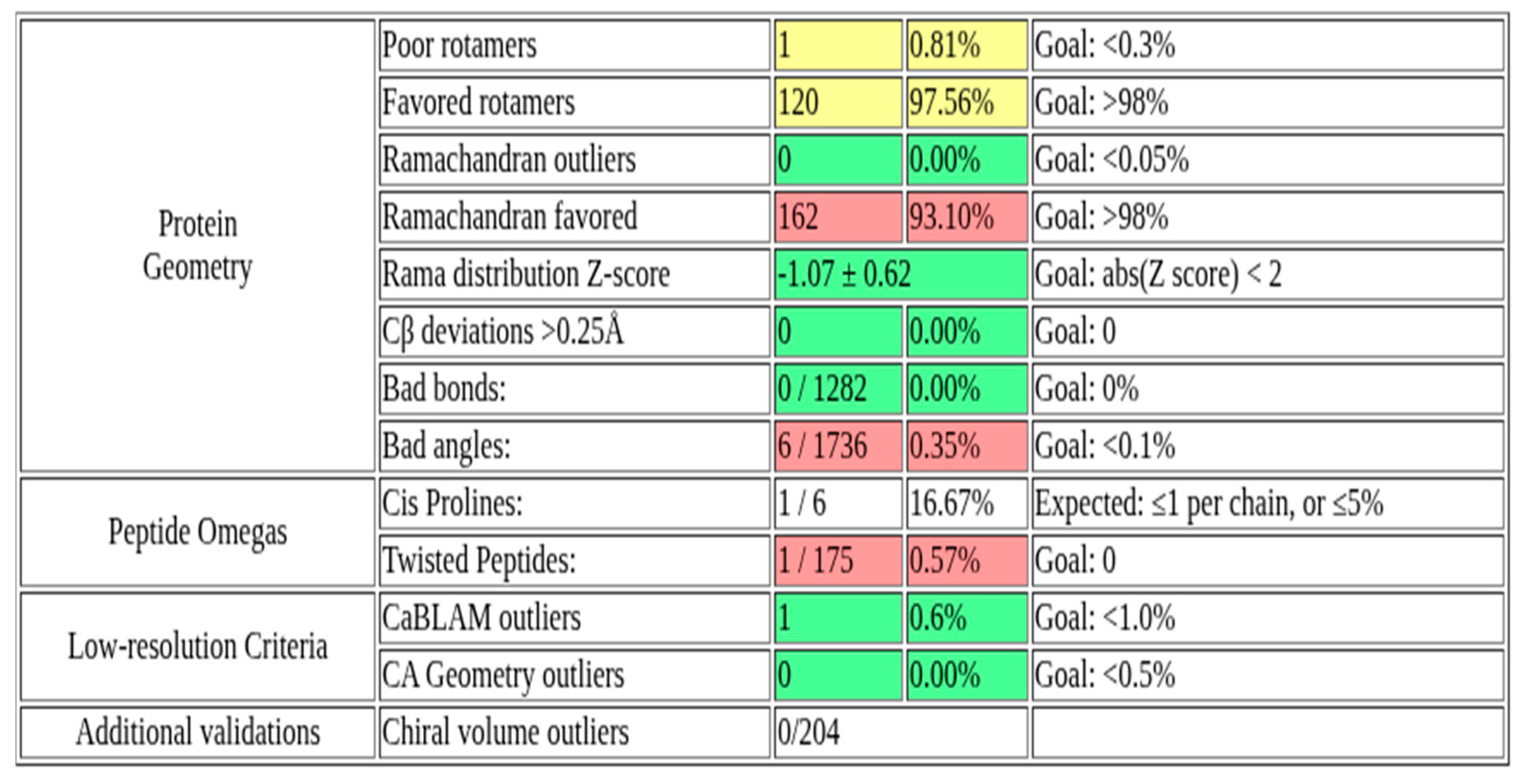
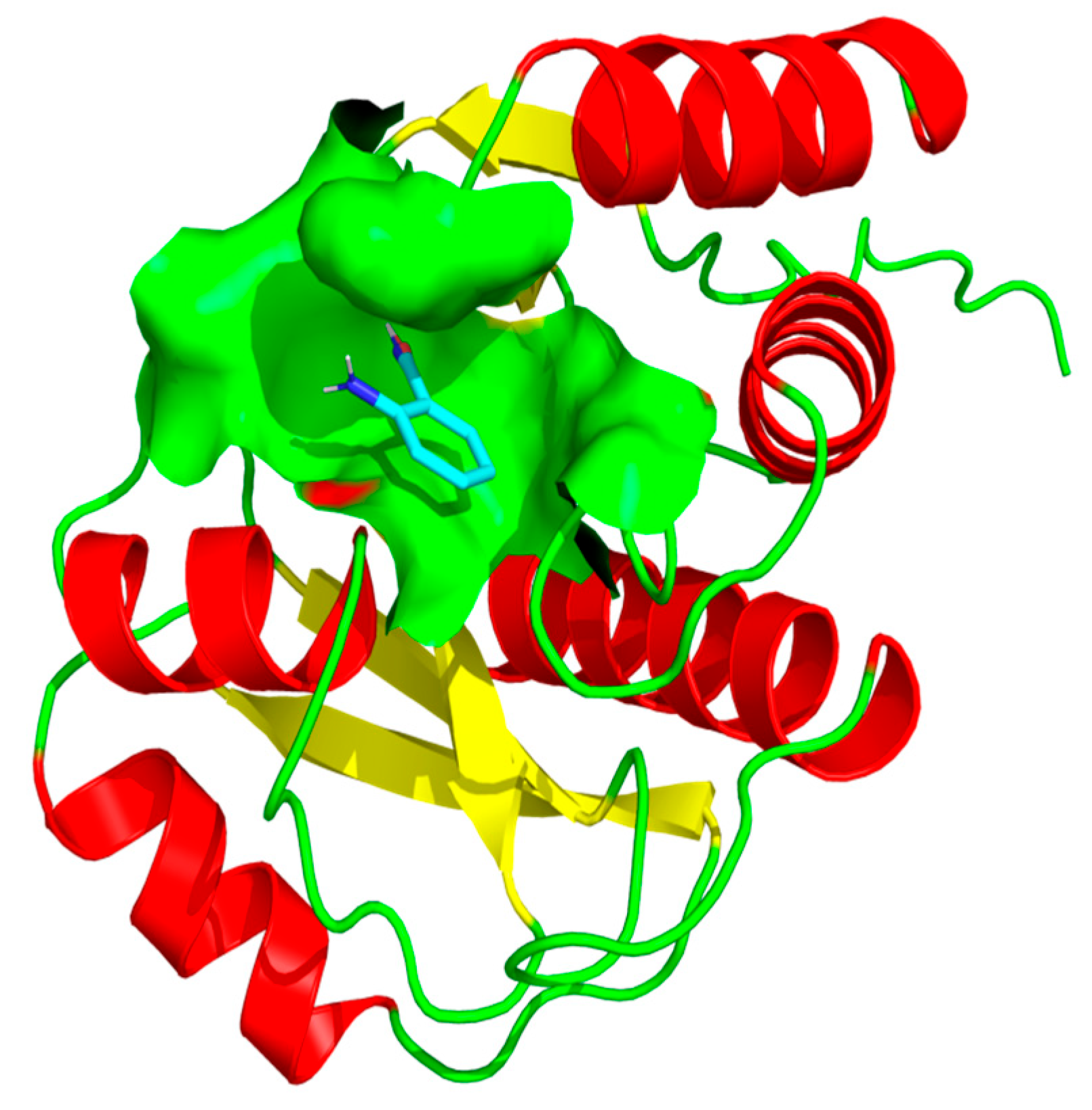
2.1. Structural Validation of the Rv1899c a Macrodomain-Containing Protein
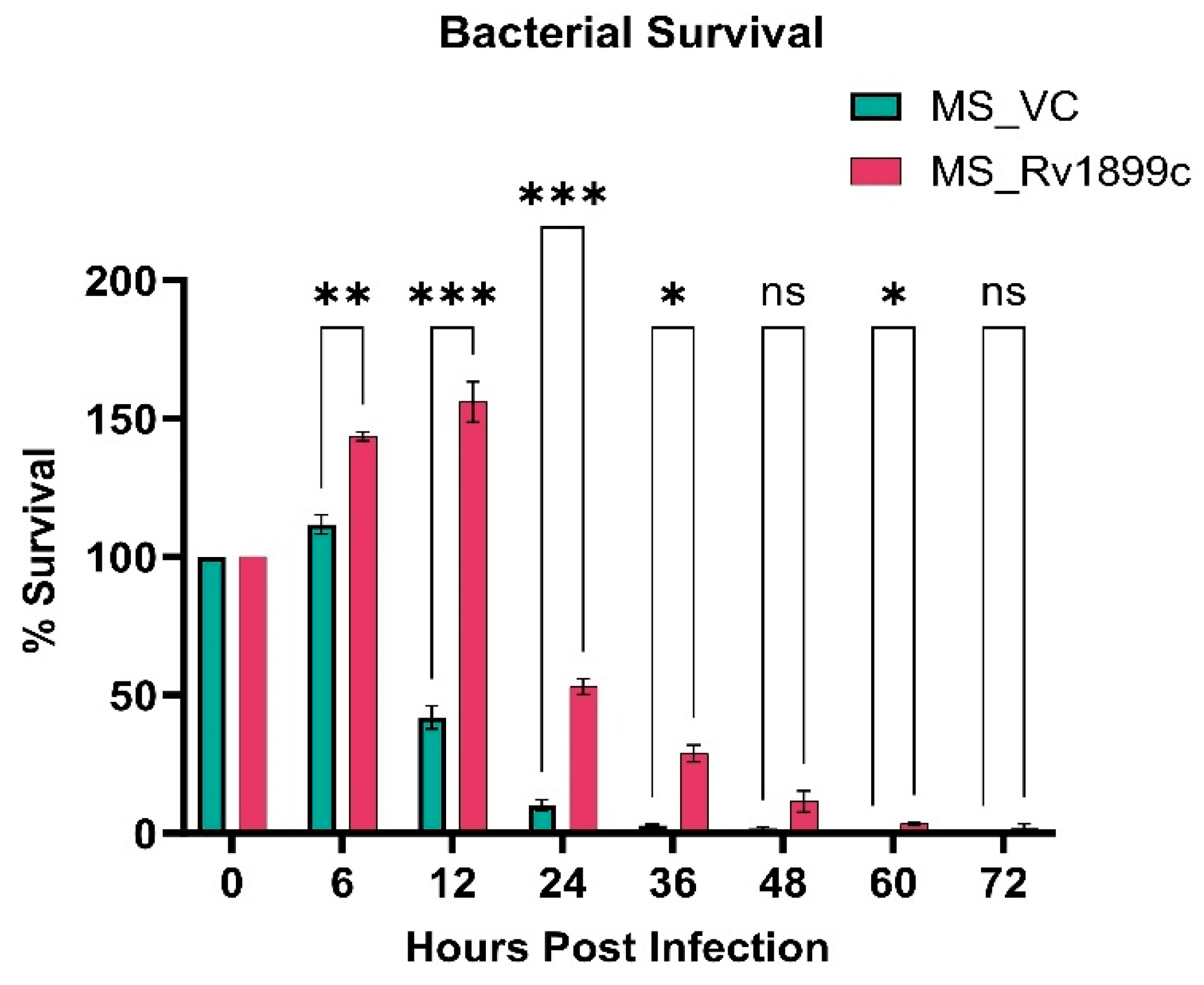
2.2. Rv1899c Expression Enhances the Intracellular Multiplication of Recombinant M. smegmatis
2.3. Rv1899c Enhances the Resistance of M. smegmatis to Various Adverse Environments
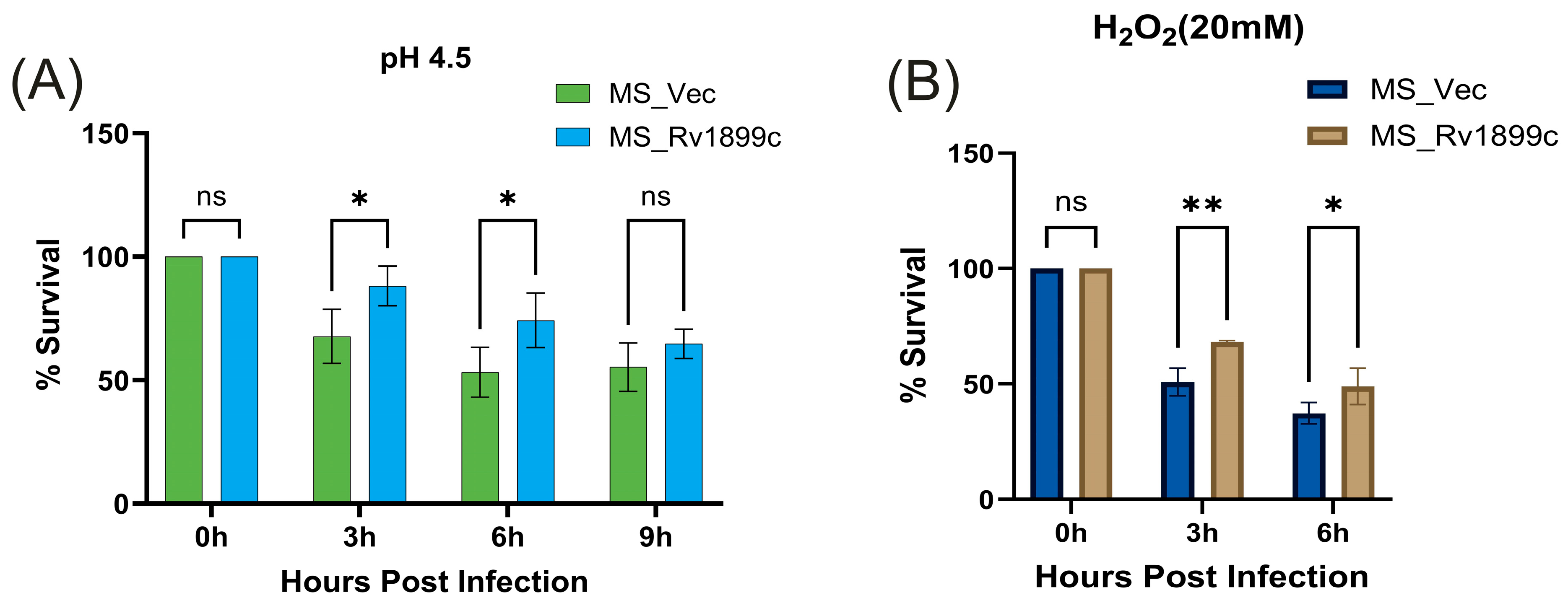
2.3.1. Response to Acidic and Oxidative Stress
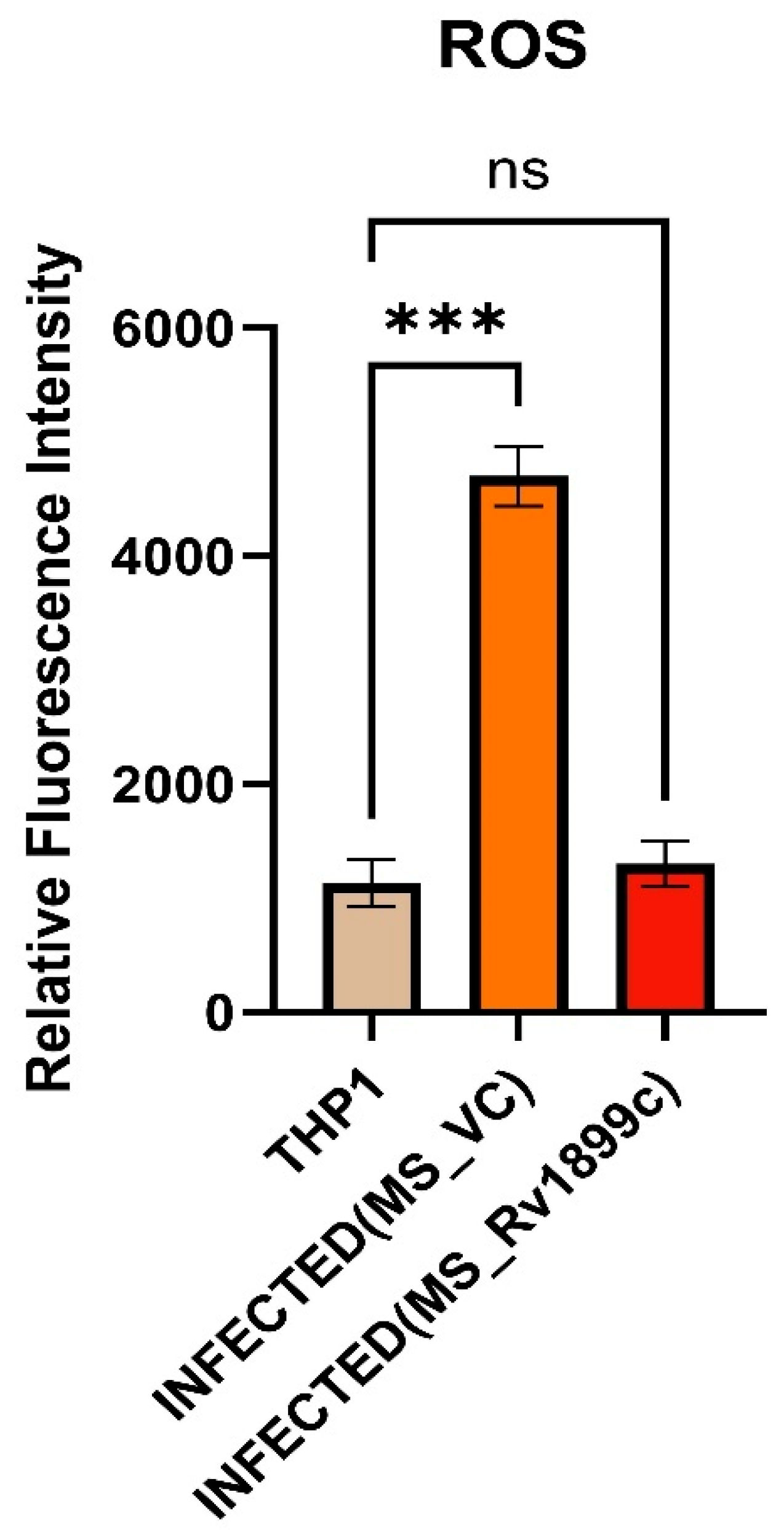
2.3.2. ROS Measurement in Macrophages Infected with M. smegmatis Vector Control and Rv1899c-Overexpressing M. smegmatis
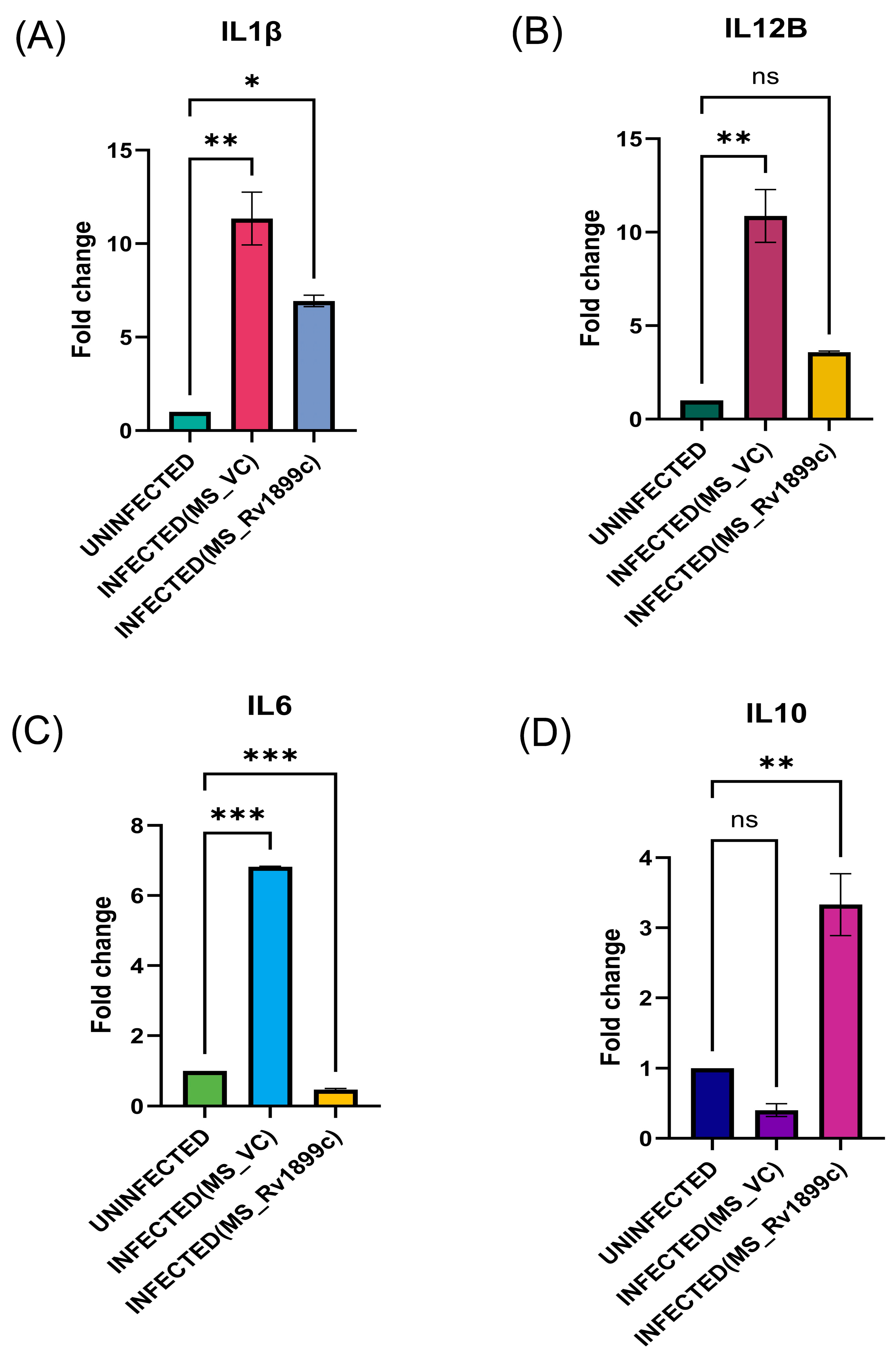
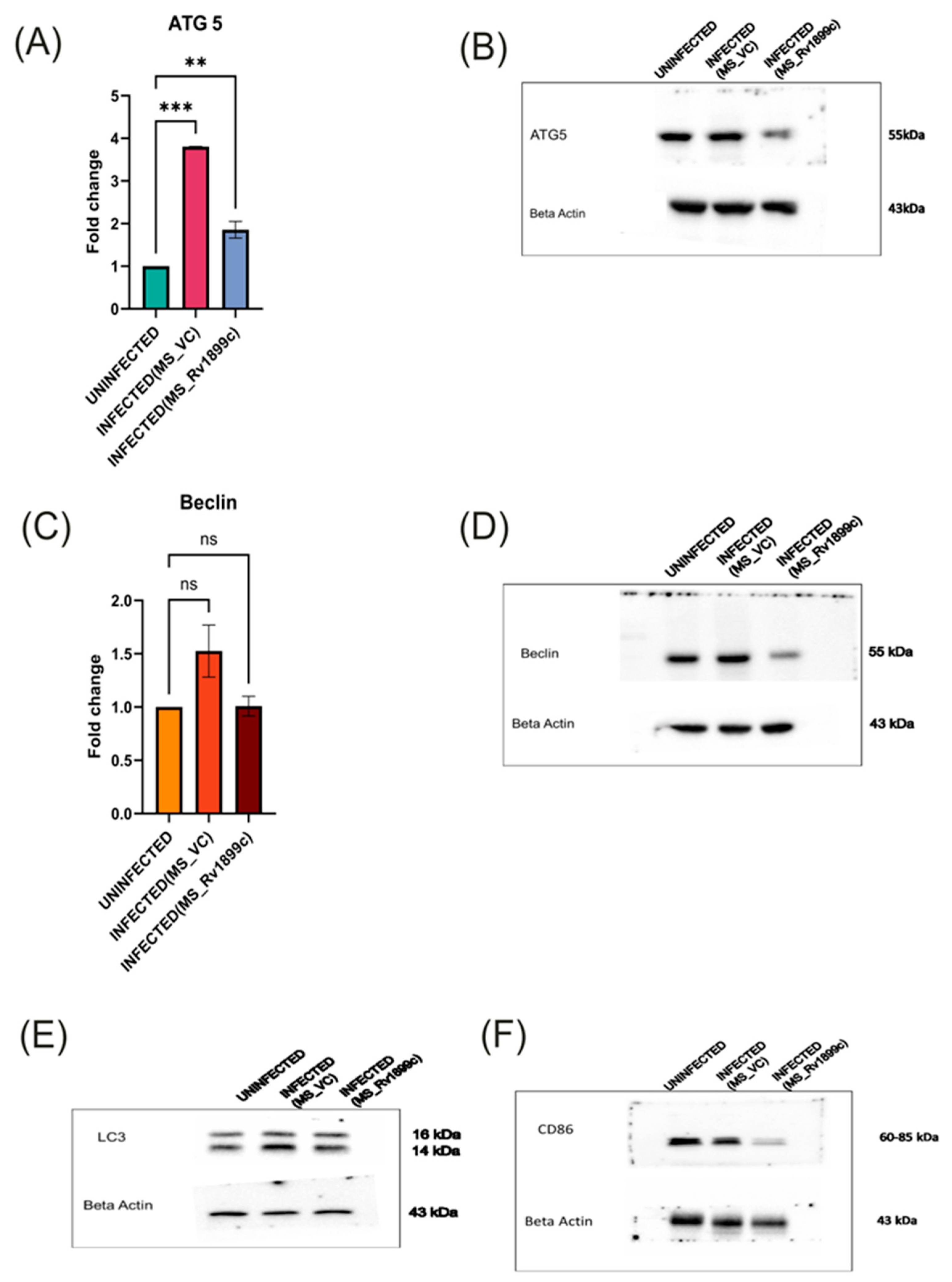
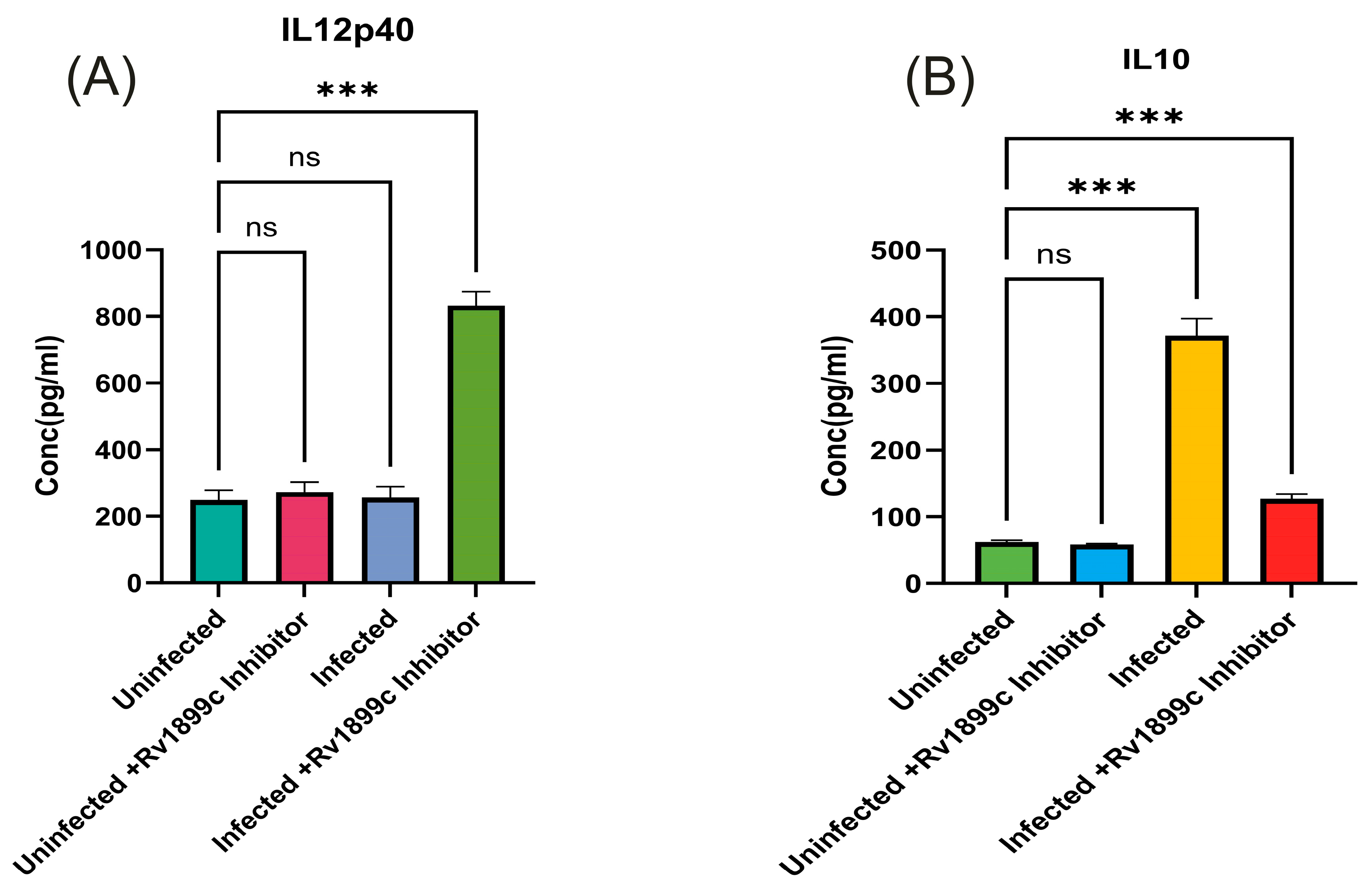
2.4. Rv1899c Suppresses the Secretion of Pro-Inflammatory Cytokines, Autophagy Gene ATG5, Baclin, LC3 and Reduces the Populations of M1 Macrophages upon Infection
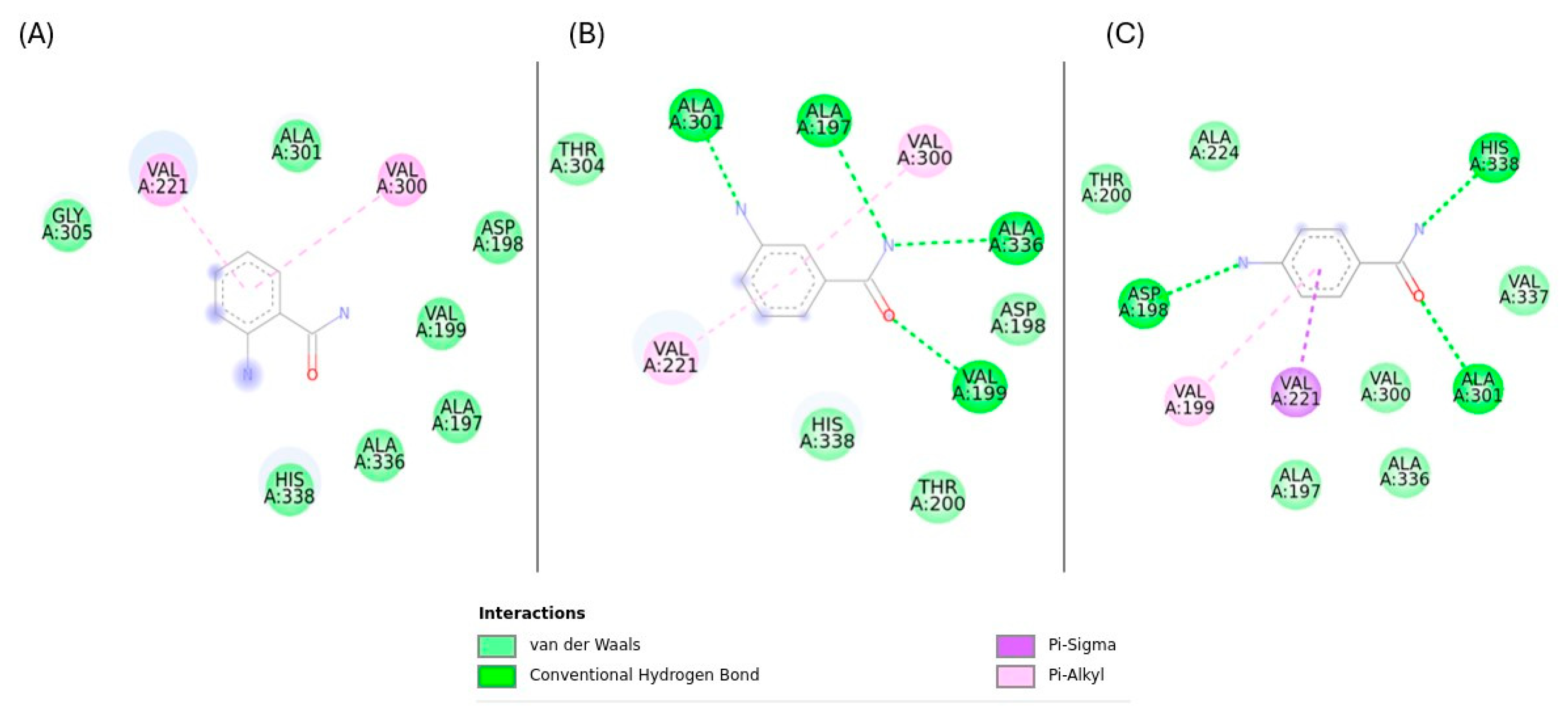
2.5. Binding Affinity and Interaction Analysis of Rv1899c with 3-Aminobenzamide
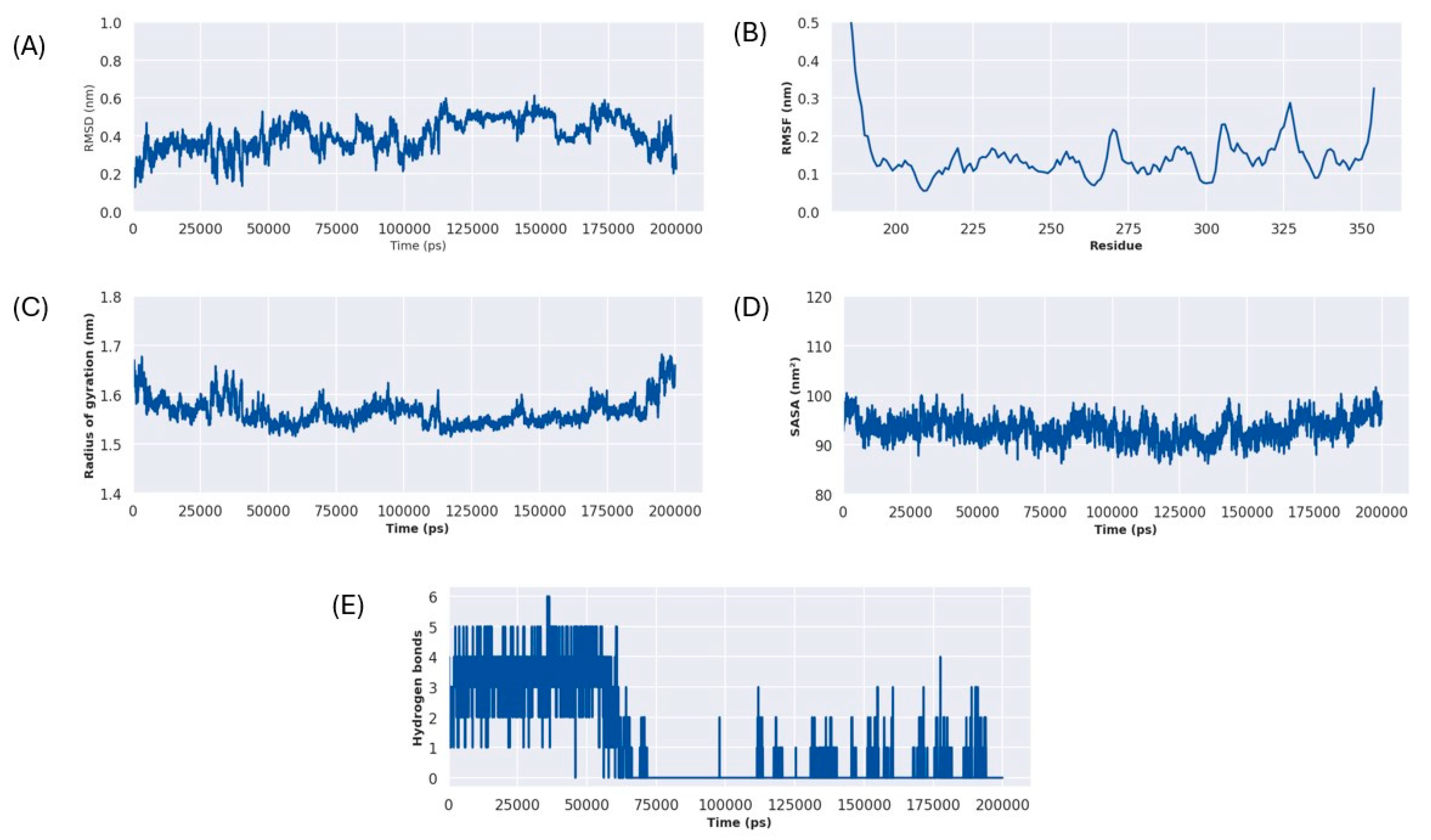
2.6. Molecular Dynamics Simulation Analysis
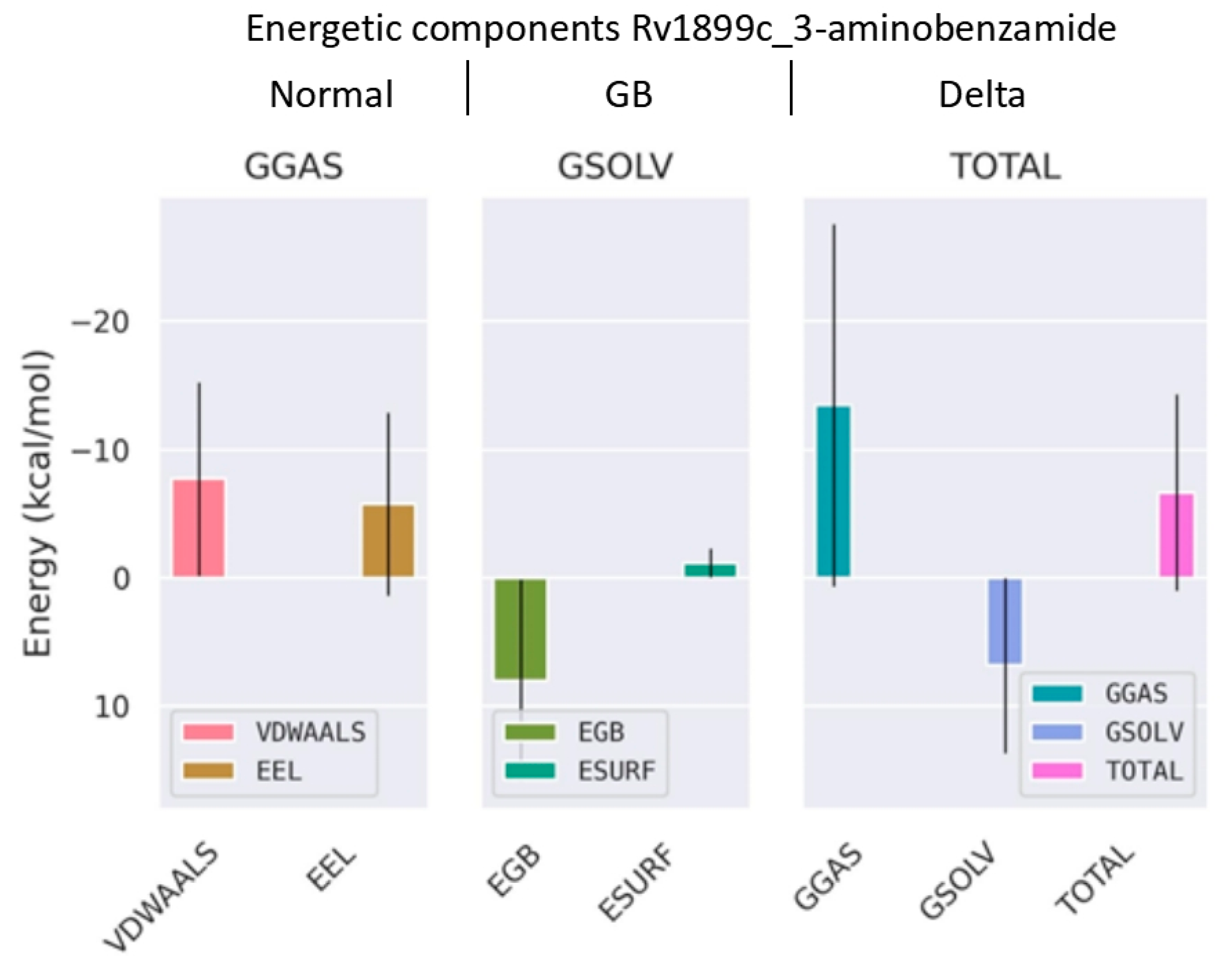
2.7. Binding Free Energy Calculation Using MM/GBSA
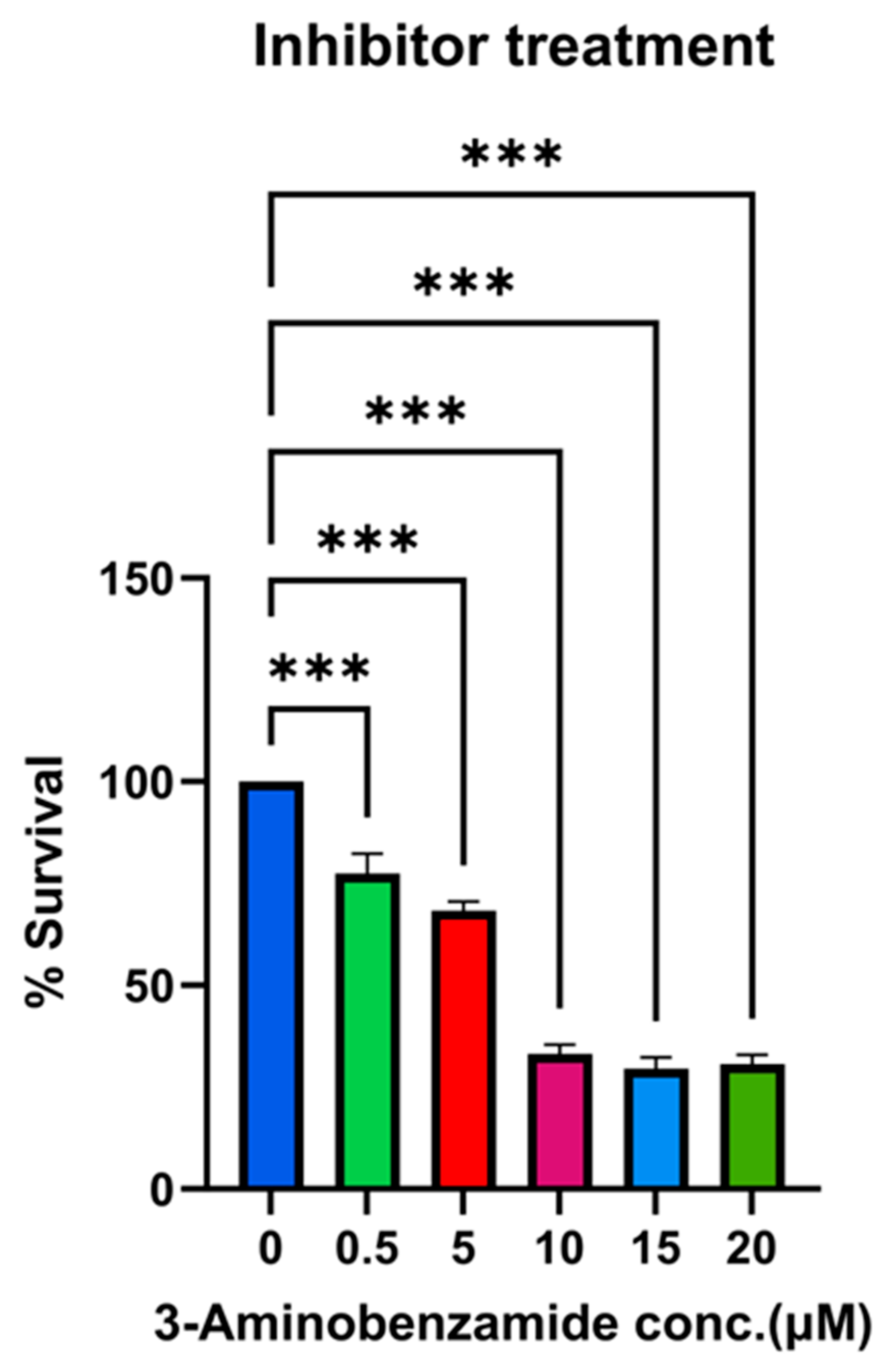
2.8. Inhibitor Treatment for Rv1899c and Survival Assays
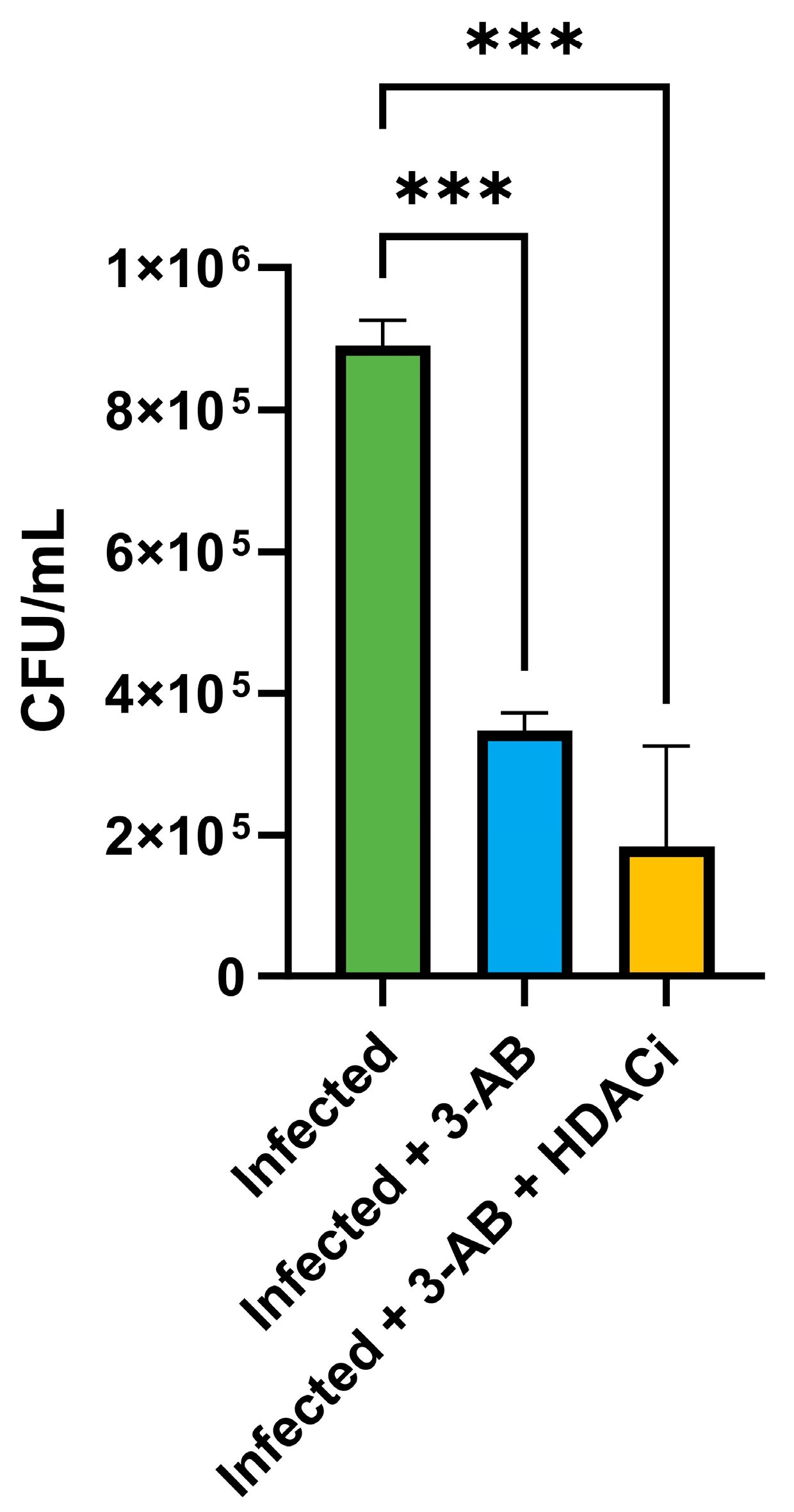
2.9. Combinatorial Treatment for Rv1899c Inhibitor and HDAC1 Inhibitor
3. Discussion
4. Materials and Methods
4.1. Processing and Validation of Protein Structure
4.2. Molecular Docking
4.3. Molecular Dynamics Simulations
4.4. MM/GBSA Analysis
4.5. Cloning and Expression of Rv1899c
4.6. Cell Culture and Differentiation of THP-1 Monocytes
4.7. Infection of THP-1-Derived Macrophage Cells with M. smegmatis
4.8. Measurement of Intracellular ROS
4.9. Intracellular Survival Assays for M. smegmatis
4.10. Total Protein Extraction and Quantification
4.11. Western Blotting
4.12. Quantitative Real-Time PCR (qPCR)
4.13. Cytokine Quantification by ELISA
4.14. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tuberculosis. Available online: https://www.who.int/news-room/fact-sheets/detail/tuberculosis (accessed on 21 September 2025).
- Diriba, G.; Alemu, A.; Yenew, B.; Ayano, B.Z.; Hailu, M.; Buta, B.; Wondimu, A.; Tefera, Z.; Meaza, A.; Seid, G.; et al. Second-line drug resistance among multidrug-resistant tuberculosis patients in Ethiopia: A laboratory-based surveillance. J. Glob. Antimicrob. Resist. 2025, 42, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, J.R.; Boccia, D.; Evans, C.A.; Adato, M.; Petticrew, M.; Porter, J.D.H. The Social Determinants of Tuberculosis: From Evidence to Action. Am. J. Public Health 2011, 101, 654. [Google Scholar] [CrossRef] [PubMed]
- Frota, C.C.; Hunt, D.M.; Buxton, R.S.; Rickman, L.; Hinds, J.; Kremer, K.; van Soolingen, D.; Colston, M.J. Genome structure in the vole bacillus, Mycobacterium microti, a member of the Mycobacterium tuberculosis complex with a low virulence for humans. Microbiology 2004, 150 Pt 5, 1519. [Google Scholar] [CrossRef]
- García, J.; Puentes, A.; Rodríguez, L.; Ocampo, M.; Curtidor, H.; Vera, R.; Lopez, R.; Valbuena, J.; Cortes, J.; Vanegas, M.; et al. Mycobacterium tuberculosis Rv2536 protein implicated in specific binding to human cell lines. Protein Sci. 2005, 14, 2236. [Google Scholar] [CrossRef]
- Manjunath, P.; Ahmad, J.; Samal, J.; Sheikh, J.A.; Arora, S.K.; Khubaib, M.; Aggarwal, H.; Kumari, I.; Luthra, K.; Rahman, S.A.; et al. Mycobacterium tuberculosis Specific Protein Rv1509 Evokes Efficient Innate and Adaptive Immune Response Indicative of Protective Th1 Immune Signature. Front. Immunol. 2021, 12, 706081. [Google Scholar] [CrossRef]
- Kim, K.H.; An, D.R.; Song, J.; Yoon, J.Y.; Kim, H.S.; Yoon, H.J.; Im, H.N.; Kim, J.; Kim, D.J.; Lee, S.J.; et al. Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7. Proc. Natl. Acad. Sci. USA 2012, 109, 7729–7734. [Google Scholar] [CrossRef]
- Shin, D.M.; Jeon, B.-Y.; Lee, H.-M.; Jin, H.S.; Yuk, J.-M.; Song, C.-H.; Lee, S.-H.; Lee, Z.-W.; Cho, S.-N.; Kim, J.-M.; et al. Mycobacterium tuberculosis Eis Regulates Autophagy, Inflammation, and Cell Death through Redox-dependent Signaling. PLoS Pathog. 2010, 6, e1001230. [Google Scholar] [CrossRef]
- Wong, D.; Bach, H.; Sun, J.; Hmama, Z.; Av-Gay, Y. Mycobacterium tuberculosis protein tyrosine phosphatase (PtpA) excludes host vacuolar-H +-ATPase to inhibit phagosome acidification. Proc. Natl. Acad. Sci. USA 2011, 108, 19371–19376. [Google Scholar] [CrossRef]
- Anes, E.; Pires, D.; Mandal, M.; Azevedo-Pereira, J.M. ESAT-6 a Major Virulence Factor of Mycobacterium tuberculosis. Biomolecules 2023, 13, 968. [Google Scholar] [CrossRef]
- Singh, V.; Kaur, C.; Chaudhary, V.K.; Rao, K.V.S.; Chatterjee, S.M. tuberculosis Secretory Protein ESAT-6 Induces Metabolic Flux Perturbations to Drive Foamy Macrophage Differentiation. Sci. Rep. 2015, 5, 12906. [Google Scholar] [CrossRef]
- Madhavan, A.; Arun, K.B.; Pushparajan, A.R.; Balaji, M.; Kumar, R.A. Transcription Repressor Protein ZBTB25 Associates with HDAC1-Sin3a Complex in Mycobacterium tuberculosis-Infected Macrophages, and Its Inhibition Clears Pathogen by Autophagy. mSphere 2021, 6, e00036-21. [Google Scholar] [CrossRef]
- Rankin, P.W.; Jacobson, E.L.; Benjamin, R.C.; Moss, J.; Jacobson, M.K. Quantitative studies of inhibitors of ADP-ribosylation in vitro and in vivo. J. Biol. Chem. 1989, 264, 4312–4317. [Google Scholar] [CrossRef]
- Rastogi, S.; Ellinwood, S.; Augenstreich, J.; Mayer-Barber, K.D.; Briken, V. Mycobacterium tuberculosis inhibits the NLRP3 inflammasome activation via its phosphokinase PknF. PLoS Pathog. 2021, 17, e1009712. [Google Scholar] [CrossRef]
- Simeone, R.; Bobard, A.; Lippmann, J.; Bitter, W.; Majlessi, L.; Brosch, R.; Enninga, J. Phagosomal Rupture by Mycobacterium tuberculosis Results in Toxicity and Host Cell Death. PLoS Pathog. 2012, 8, e1002507. [Google Scholar] [CrossRef] [PubMed]
- Forrellad, M.A.; Klepp, L.I.; Gioffré, A.; García, J.S.Y.; Morbidoni, H.R.; Santangelo, M.d.l.P.; Cataldi, A.A.; Bigi, F. Virulence factors of the Mycobacterium tuberculosis complex. Virulence 2013, 4, 3–66. [Google Scholar] [CrossRef] [PubMed]
- Dorhoi, A.; Nouailles, G.; Jörg, S.; Hagens, K.; Heinemann, E.; Pradl, L.; Oberbeck-Müller, D.; Duque-Correa, M.A.; Reece, S.T.; Ruland, J.; et al. Activation of the NLRP3 inflammasome by Mycobacterium tuberculosis is uncoupled from susceptibility to active tuberculosis. Eur. J. Immunol. 2012, 42, 374–384. [Google Scholar] [CrossRef]
- Ge, P.; Lei, Z.; Yu, Y.; Lu, Z.; Qiang, L.; Chai, Q.; Zhang, Y.; Zhao, D.; Li, B.; Pang, Y.; et al. M. tuberculosis PknG manipulates host autophagy flux to promote pathogen intracellular survival. Autophagy 2022, 18, 576–594. [Google Scholar] [CrossRef]
- Vergne, I.; Chua, J.; Lee, H.H.; Lucas, M.; Belisle, J.; Deretic, V. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2005, 102, 4033–4038. [Google Scholar] [CrossRef]
- Strong, E.J.; Ng, T.W.; Porcelli, S.A.; Lee, S. Mycobacterium tuberculosis PE_PGRS20 and PE_PGRS47 Proteins Inhibit Autophagy by Interaction with Rab1A. mSphere 2021, 6, e0054921. [Google Scholar] [CrossRef]
- Joseph, S.; Yuen, A.; Singh, V.; Hmama, Z. Mycobacterium tuberculosis Cpn60.2 (GroEL2) blocks macrophage apoptosis via interaction with mitochondrial mortalin. Biol. Open 2017, 6, 481–488. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, Version 1.8; Schrödinger LLC: New York, NY, USA, 2015.
- Chen, V.B.; Arendall, W.B., III; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 1, 12–21. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.; Thiessen, P.; Yu, B.; et al. PubChem 2025 update. Nucleic Acids Res. 2025, 53, D1516–D1525. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Croitoru, A.; Park, S.-J.; Kumar, A.; Lee, J.; Im, W.; MacKerell, A.D.; Aleksandrov, A. Additive CHARMM36 Force Field for Nonstandard Amino Acids. J. Chem. Theory Comput. 2021, 17, 3554–3570. [Google Scholar] [CrossRef]
- Huang, J.; Mackerell, A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; MacKerell, A.D. Automation of the CHARMM General Force Field (CGenFF) I: Bond perception and atom typing. J. Chem. Inf. Model. 2012, 52, 3144. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Raman, E.P.; MacKerell, A.D. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of Bonded Parameters and Partial Atomic Charges. J. Chem. Inf. Model. 2012, 52, 3155–3168. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Lin, Y.; Pan, D.; Li, J.; Zhang, L.; Shao, X. Application of Berendsen barostat in dissipative particle dynamics for nonequilibrium dynamic simulation. J. Chem. Phys. 2017, 146, 124108. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 14631472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Cuendet, M.A.; Van Gunsteren, W.F. On the calculation of velocity-dependent properties in molecular dynamics simulations using the leapfrog integration algorithm. J. Chem. Phys. 2007, 127, 184102. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M.L. seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. Gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menon, A.M.; Gopichand, B.; Thomas, S.S.; Abhinand, K.; Nair, B.G.; Kumar, G.B.; Babu, P.; Arun, K.; Edison, L.K.; Madhavan, A. Rv1899c, an HDAC1–ZBTB25-Interacting Protein of Mycobacterium tuberculosis, Promotes Stress Resistance and Immune Evasion in Infected Macrophages. Int. J. Mol. Sci. 2025, 26, 10872. https://doi.org/10.3390/ijms262210872
Menon AM, Gopichand B, Thomas SS, Abhinand K, Nair BG, Kumar GB, Babu P, Arun K, Edison LK, Madhavan A. Rv1899c, an HDAC1–ZBTB25-Interacting Protein of Mycobacterium tuberculosis, Promotes Stress Resistance and Immune Evasion in Infected Macrophages. International Journal of Molecular Sciences. 2025; 26(22):10872. https://doi.org/10.3390/ijms262210872
Chicago/Turabian StyleMenon, Arjun M., Boinapalli Gopichand, Shwetha Susan Thomas, Kuniyil Abhinand, Bipin G. Nair, Geetha B. Kumar, Pradeesh Babu, KB Arun, Lekshmi K. Edison, and Aravind Madhavan. 2025. "Rv1899c, an HDAC1–ZBTB25-Interacting Protein of Mycobacterium tuberculosis, Promotes Stress Resistance and Immune Evasion in Infected Macrophages" International Journal of Molecular Sciences 26, no. 22: 10872. https://doi.org/10.3390/ijms262210872
APA StyleMenon, A. M., Gopichand, B., Thomas, S. S., Abhinand, K., Nair, B. G., Kumar, G. B., Babu, P., Arun, K., Edison, L. K., & Madhavan, A. (2025). Rv1899c, an HDAC1–ZBTB25-Interacting Protein of Mycobacterium tuberculosis, Promotes Stress Resistance and Immune Evasion in Infected Macrophages. International Journal of Molecular Sciences, 26(22), 10872. https://doi.org/10.3390/ijms262210872






