Differences in the Gene Regulatory Network for Floral Induction in Two Camellia Species
Abstract
1. Introduction
2. Results
2.1. Annual Distribution of Flower Bud Formation Rates in C. azalea and C. japonica
2.2. Transcriptome Analysis of C. azalea and C. japonica Leaves Throughout the Year
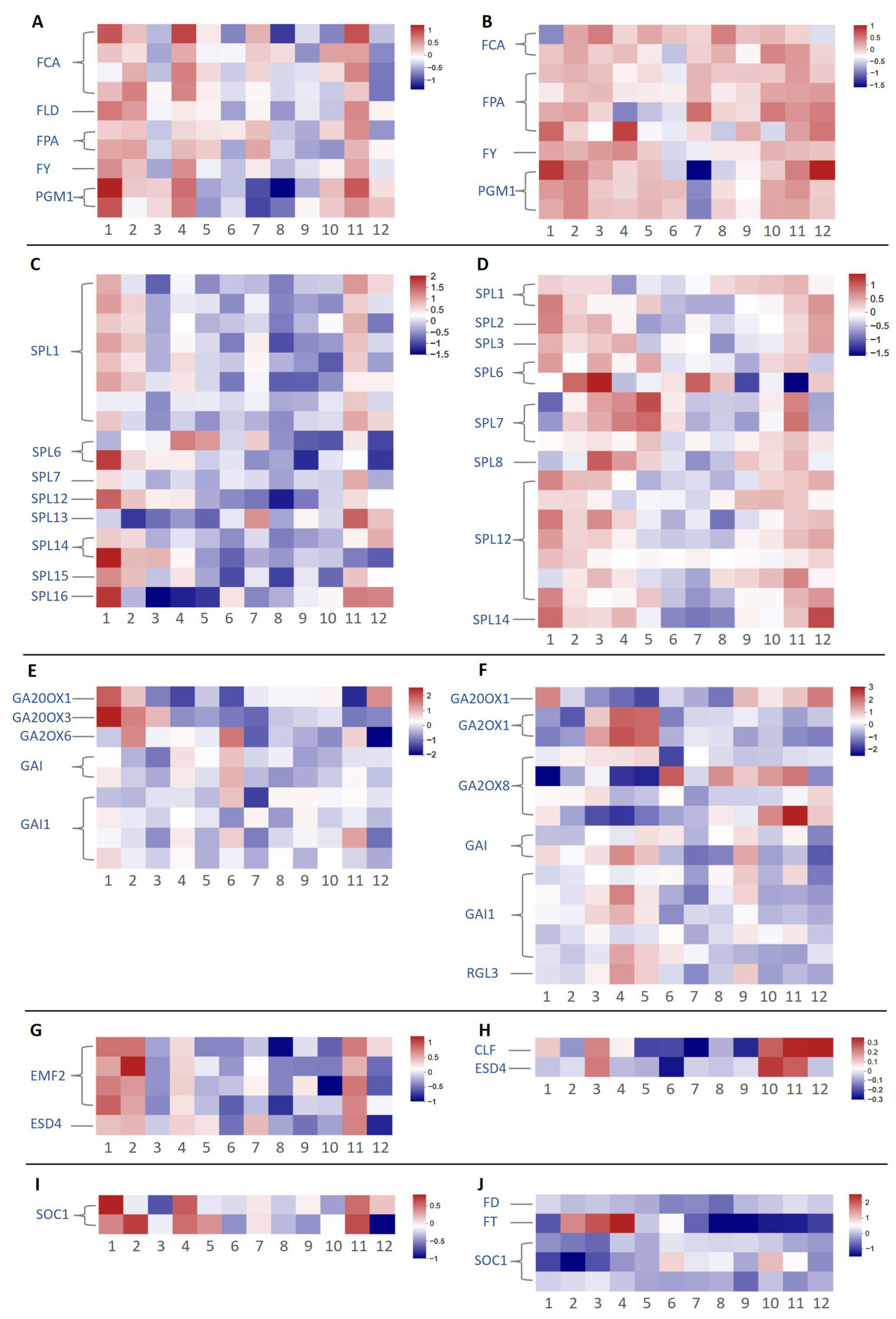
2.3. Analysis of Flowering-Related Genes in Different Pathways Based on Transcriptome Data
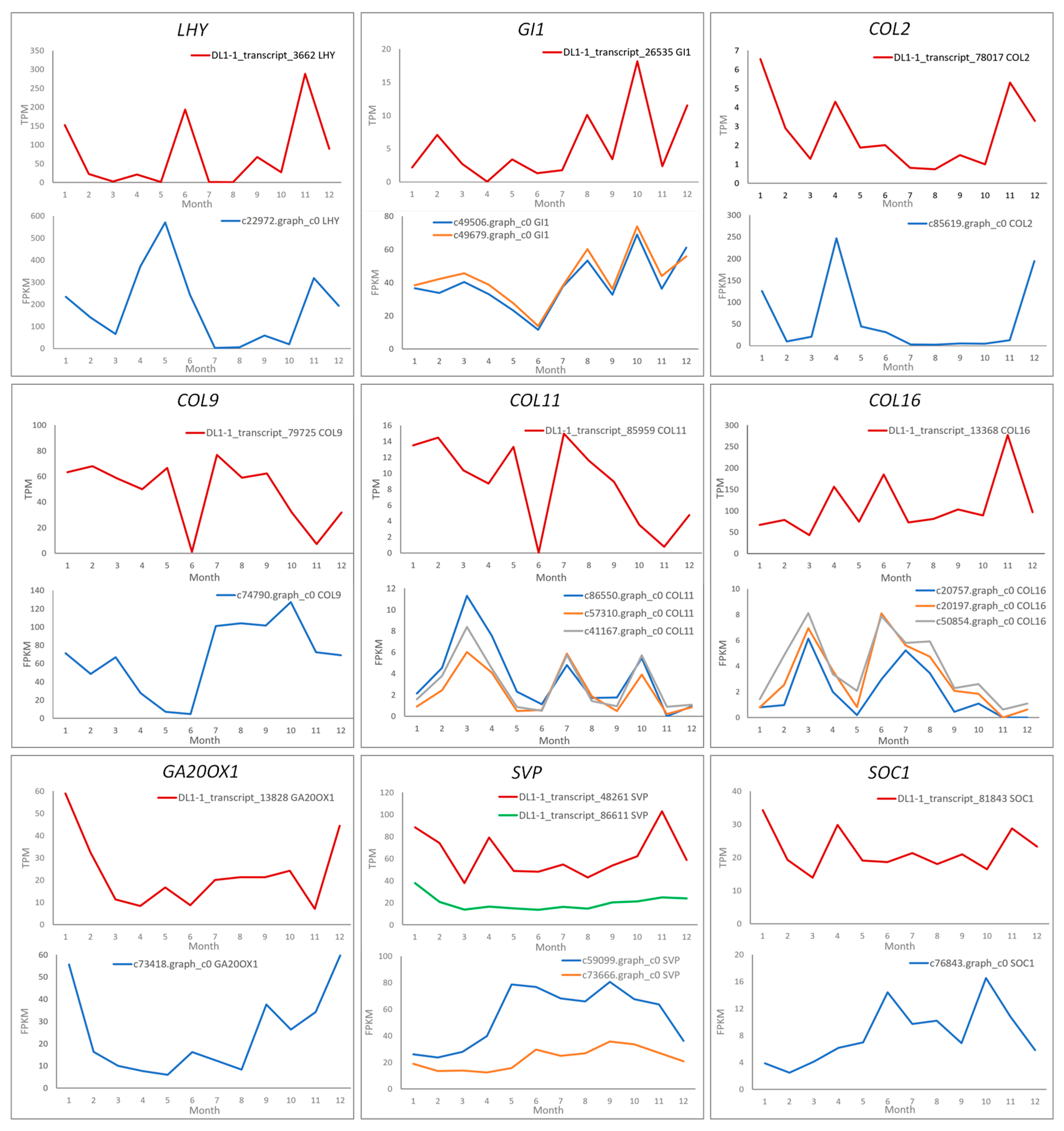
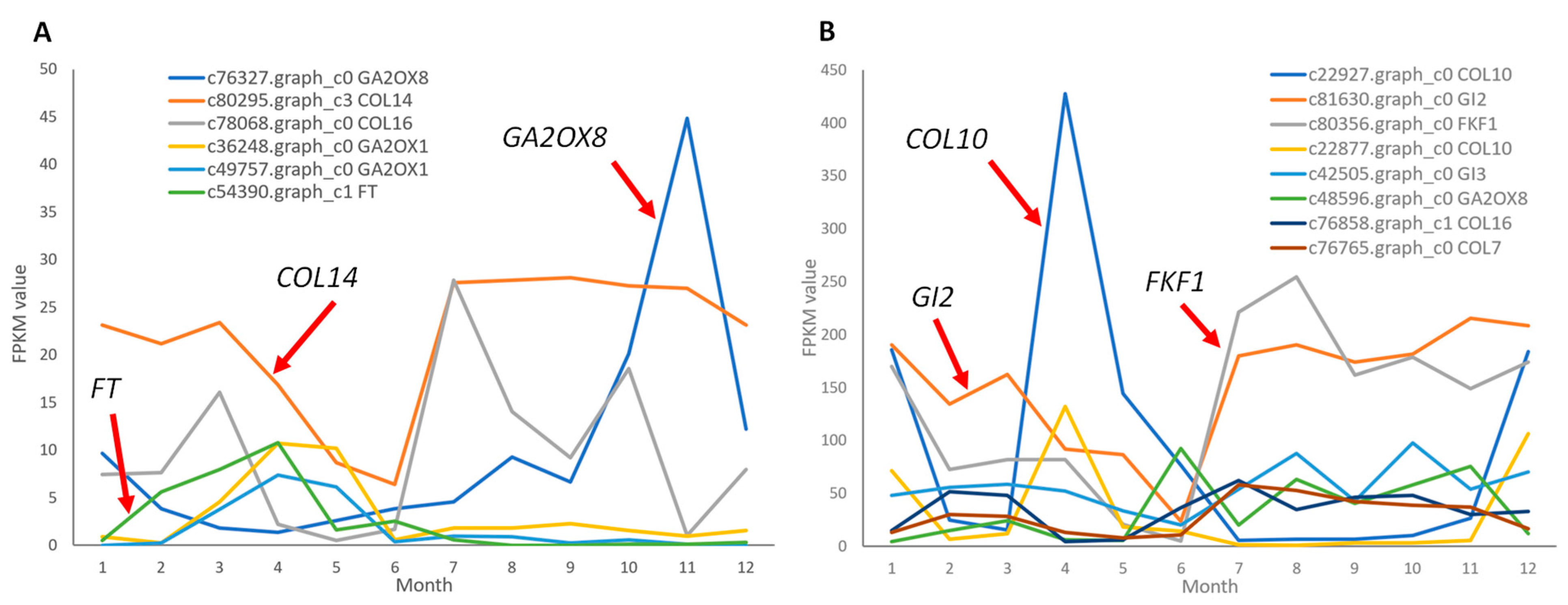
2.4. Differential Expression of Key Floral Induction Genes in Two Camellia Species
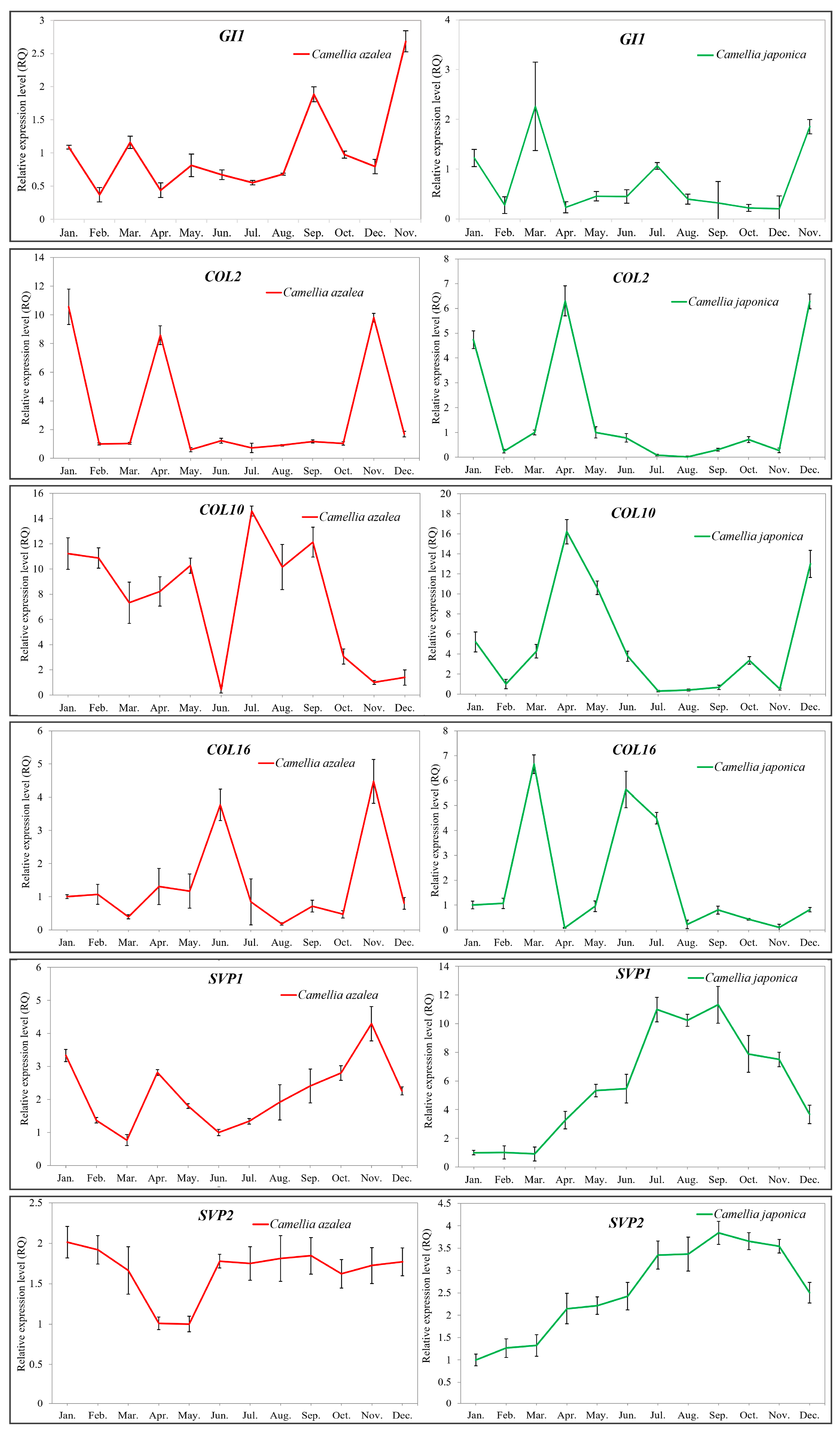
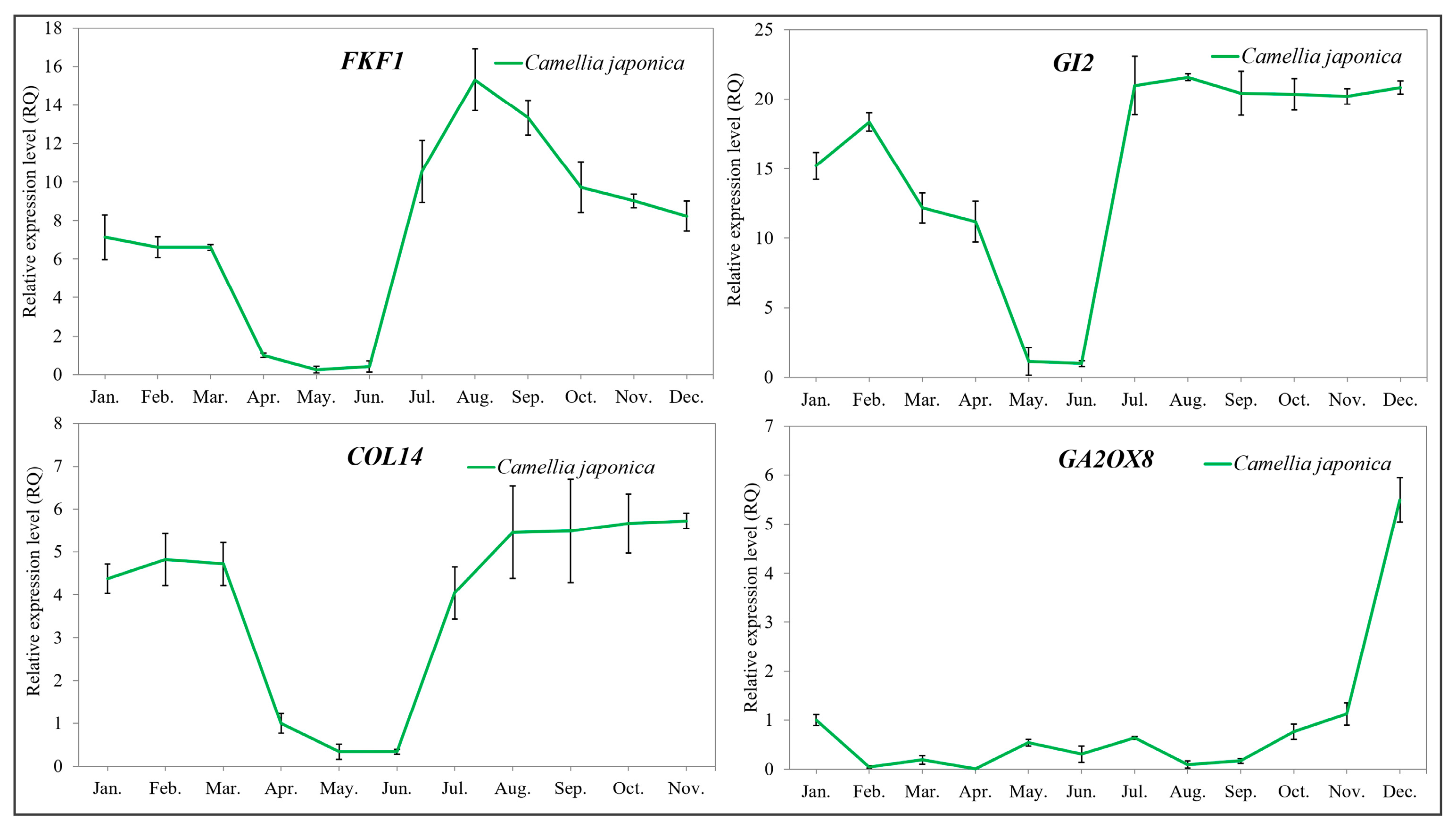
2.5. Validation of the Key Flowering-Related Genes by qRT-PCR
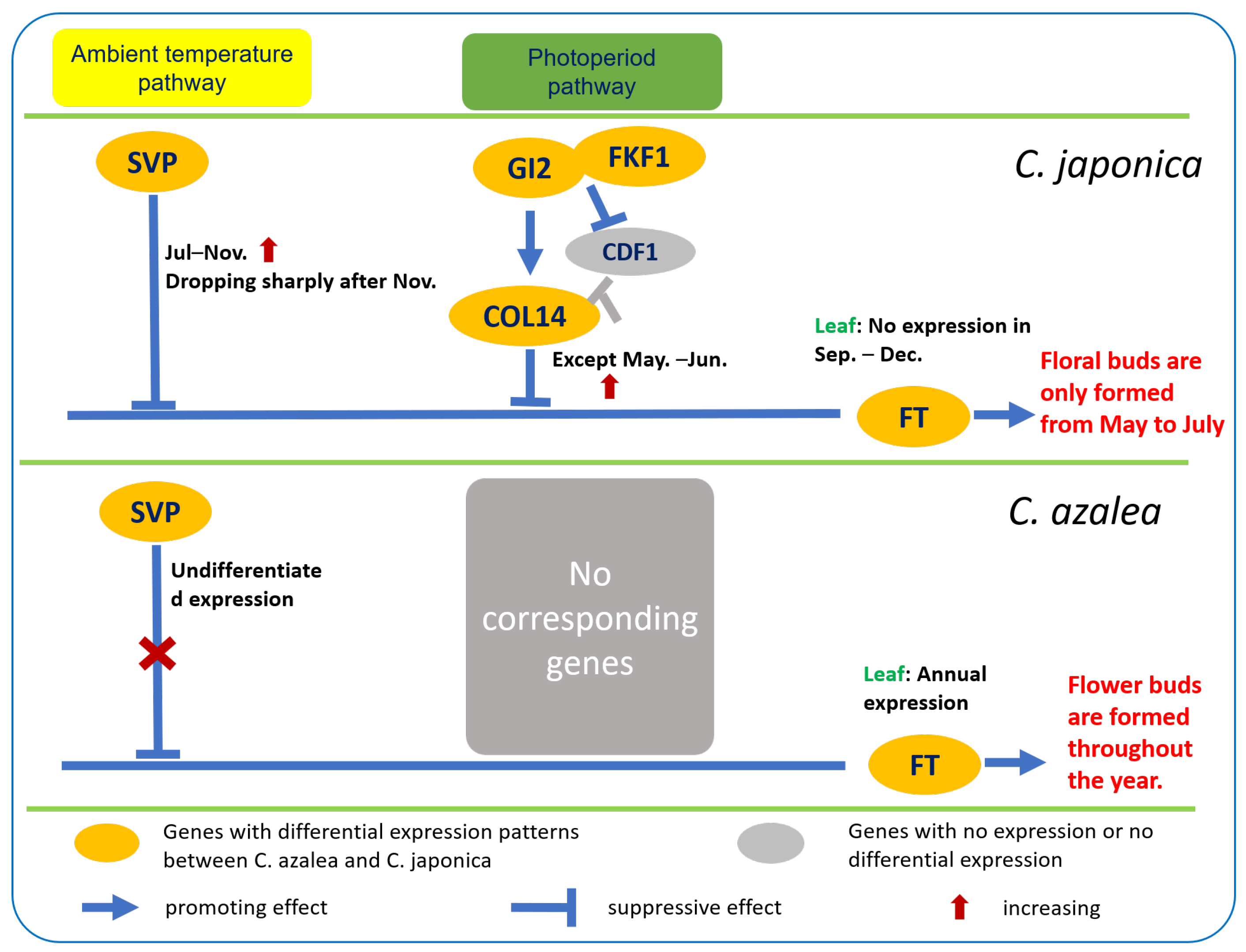
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Investigation of the Annual Bud Formation Pattern
4.3. Transcriptome Sequencing and Gene Expression Analysis of Leaves Throughout the Year
4.4. Validation of the Key Genes by qRT-PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wellmer, F.; Riechmann, J.L. Gene networks controlling the initiation of flower development. Trends Genet. 2010, 26, 519–527. [Google Scholar] [CrossRef]
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of flowering in Arabidopsis. Cell 2010, 141, 550. [Google Scholar] [CrossRef]
- Bäurle, I.; Dean, C. The timing of developmental transitions in plants. Cell 2006, 125, 655–664. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Kaya, H.; Goto, K.; Iwabuchi, M.; Araki, T. A pair of related genes with antagonistic roles in mediating flowering signals. Science 1999, 286, 1960–1962. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, M.A.; Dodueva, I.E.; Gancheva, M.S.; Tvorogova, V.E.; Kuznetsova, K.A.; Lutova, L.A. The evolutionary aspects of flowering control: Florigens and anti-florigens. Russ. J. Genet. 2020, 56, 1323–1344. [Google Scholar] [CrossRef]
- Sheppard, T.L. Flowers from a sweetheart. Nat. Chem. Biol. 2013, 9, 215–216. [Google Scholar] [CrossRef]
- Park, D.H.; Somers, D.E.; Kim, Y.S.; Choy, Y.H.; Lim, H.K.; Soh, M.S.; Kim, H.J.; Kay, S.A. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 1999, 285, 1579–1582. [Google Scholar] [CrossRef]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef]
- Xu, X.; Jiang, W.; Chen, Y.; Tian, H.; Yang, Z.; Liu, S.; Li, X.; Song, C.; Ye, Z.; Guo, W.; et al. BRASSINAZOLE RESISTANT 1 delays photoperiodic flowering by repressing CONSTANS transcription. Plant Physiol. 2025, 197, kiaf032. [Google Scholar] [CrossRef]
- Wu, F.; Kang, Y.; Liu, L.; Lei, J.; He, B.; He, Y.; Li, J.; Liu, F.; Du, Q.; Zhang, X.; et al. The blue light receptor ZmFKF1a recruits ZmGI1 to the nucleus to accelerate shoot apex development and flowering in maize. Plant Cell 2025, 37, koaf199. [Google Scholar] [CrossRef] [PubMed]
- Hayama, R.; Yokoi, S.; Tamaki, S.; Yano, M.; Shimamoto, K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 2003, 422, 719–722. [Google Scholar] [CrossRef]
- Robson, F.; Costa, M.M.R.; Hepworth, S.R.; Vizir, I.; Piñeiro, M.; Reeves, P.H.; Putterill, J.; Coupland, G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. the Plant J. 2001, 28, 619–631. [Google Scholar] [CrossRef]
- Valverde, F. CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J. Exp. Bot. 2011, 62, 2453–2463. [Google Scholar] [CrossRef] [PubMed]
- Ledger, S.; Strayer, C.; Ashton, F.; Kay, S.A.; Putterill, J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 2001, 26, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.F.; Wang, Z.Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. the Plant J. 2005, 43, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Wu, F.; Wan, J. Flowering time regulation by the CONSTANS-Like gene OsCOL10. Plant Signal. Behav. 2017, 12, 143–147. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, L.; Yu, X.; Chen, D.; Huang, C.; Li, Y. Cloning and functional analysis of ClCOL16 from Chrysanthemum lavandulifolium. Acta Bot. Boreali-Occident. Sin. 2023, 43, 366–373. [Google Scholar]
- Kinmonth-Schultz, H.; Lewandowska-Sabat, A.; Imaizumi, T.; Ward, J.K.; Rognli, O.A.; Fjellheim, S. Flowering times of wild Arabidopsis accessions from across Norway correlate with expression levels of FT, CO, and FLC genes. Front. Plant Sci. 2021, 12, 747740. [Google Scholar] [CrossRef]
- Angel, A.; Song, J.; Yang, H.; Questa, J.I.; Dean, C.; Howard, M. Vernalizing cold is registered digitally at FLC. Proc. Natl. Acad. Sci. USA 2015, 112, 4146–4151. [Google Scholar] [CrossRef]
- Vidal, A.M.; Ben-Cheikh, W.; Talón, M. Regulation of gibberellin 20-oxidase gene expression and gibberellin content in citrus by temperature and citrus exocortis viroid. Planta 2003, 217, 442–448. [Google Scholar] [CrossRef]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef]
- Hu, Q.; Jin, Y.; Shi, H.; Yang, W. GmFLD, a soybean homolog of the autonomous pathway gene FLOWERING LOCUS D, promotes flowering in Arabidopsis thaliana. BMC Plant Biol. 2014, 14, 263. [Google Scholar] [CrossRef] [PubMed]
- Eom, H.; Park, S.J.; Kim, M.K.; Kim, H.; Kang, H.; Lee, I. TAF15b, involved in the autonomous pathway for flowering, represses transcription of FLOWERING LOCUS C. Plant J. 2018, 93, 79–91. [Google Scholar] [CrossRef]
- Wang, J.; Czech, B.; Weigel, D. MiR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, Y.; Yu, H. Florigen trafficking integrates photoperiod and temperature signals in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 1385–1398. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.C.; Youn, Y.; Duarte, M.I.; Harmon, F.G. The time of day effects of warm temperature on flowering time involve PIF4 and PIF5. J. Exp. Bot. 2014, 65, 1141–1151. [Google Scholar] [CrossRef]
- Wu, R.; Walton, E.F.; Richardson, A.C.; Wood, M.; Hellens, R.P.; Varkonyi-Gasic, E. Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J. Exp. Bot. 2012, 63, 797–807. [Google Scholar] [CrossRef]
- Wu, R.; Tomes, S.; Karunairetnam, S.; Tustin, S.D.; Hellens, R.P.; Allan, A.C.; Macknight, R.C.; Varkonyi-Gasic, E. SVP-like MADS box genes control dormancy and budbreak in apple. Front. Plant Sci. 2017, 8, 477. [Google Scholar] [CrossRef]
- Mo, X.; Mo, W.; Jiang, T.; He, S.; Liu, Y.; Xie, F.; Luo, C.; He, X. Functional characterization and regulatory mechanism of mango SHORT VEGETATIVE PHASE (SVP) gene family in response to temperature regulation of flowering. Plant Physiol. Biochem. 2025, 228, 110278. [Google Scholar] [CrossRef]
- Mo, X.; Luo, C.; Xia, L.; Mo, W.; Zhu, J.; Zhang, Y.; Hu, W.; Liu, Y.; Xie, F.; He, X. Overexpression of mango MiSVP3 and Mi-SVP4 delays flowering time in transgenic Arabidopsis. Sci. Hortic. 2023, 317, 112021. [Google Scholar] [CrossRef]
- Wang, X.; Li, J.; Yin, H.; Li, X.; Liu, W.; Fan, Z. Function of FT in flowering induction in two Camellia species. Plants 2024, 13, 784. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
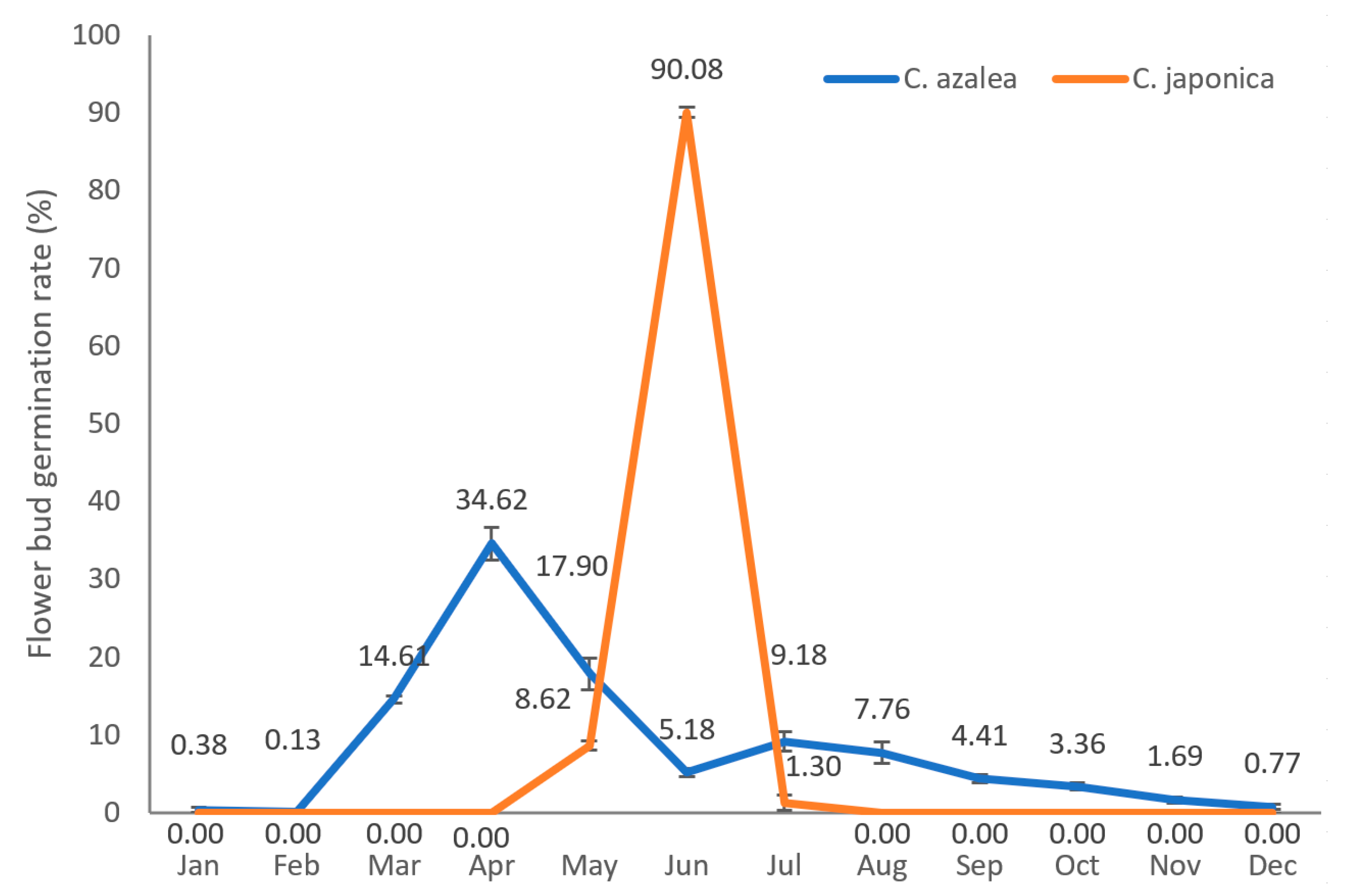
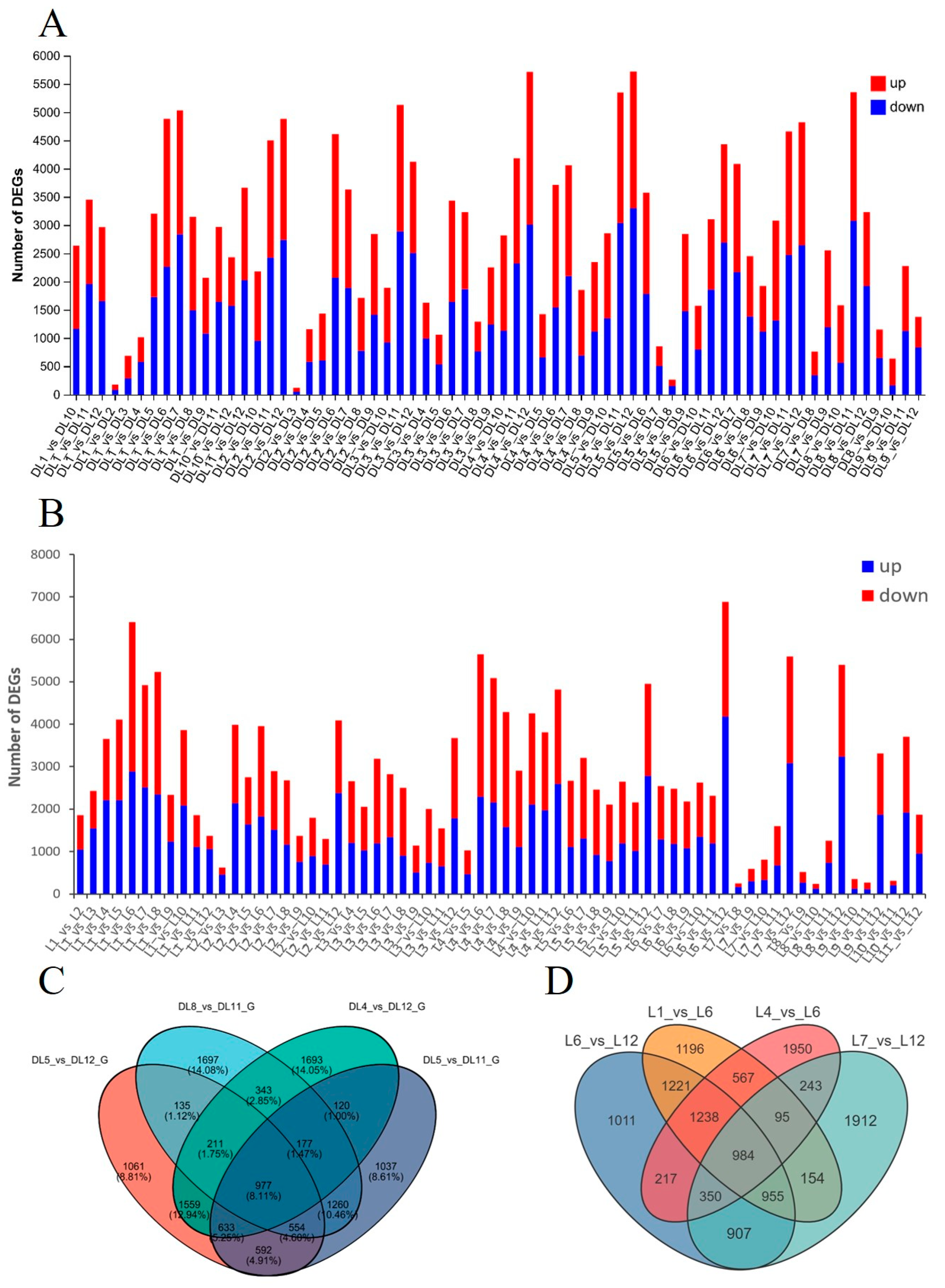
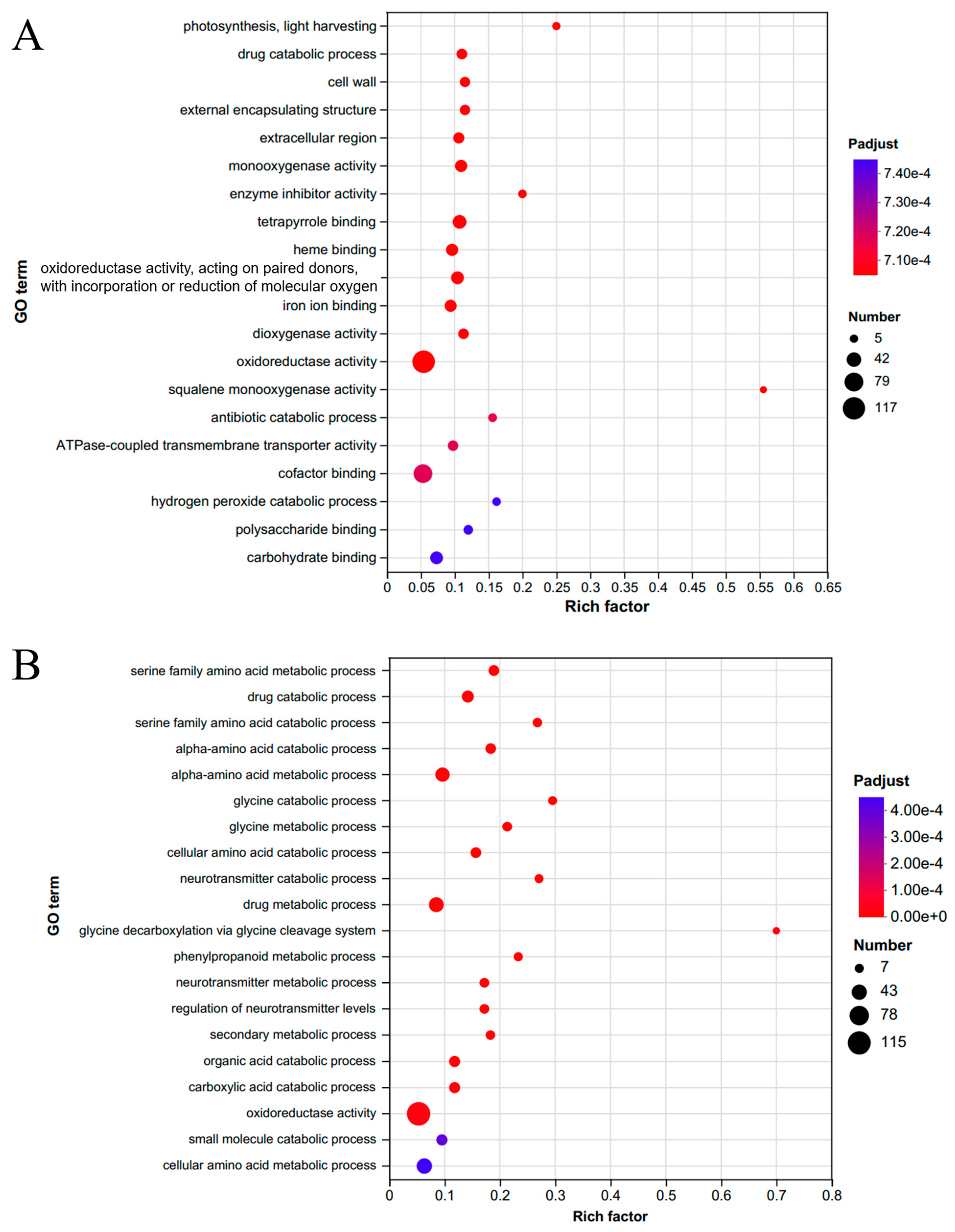
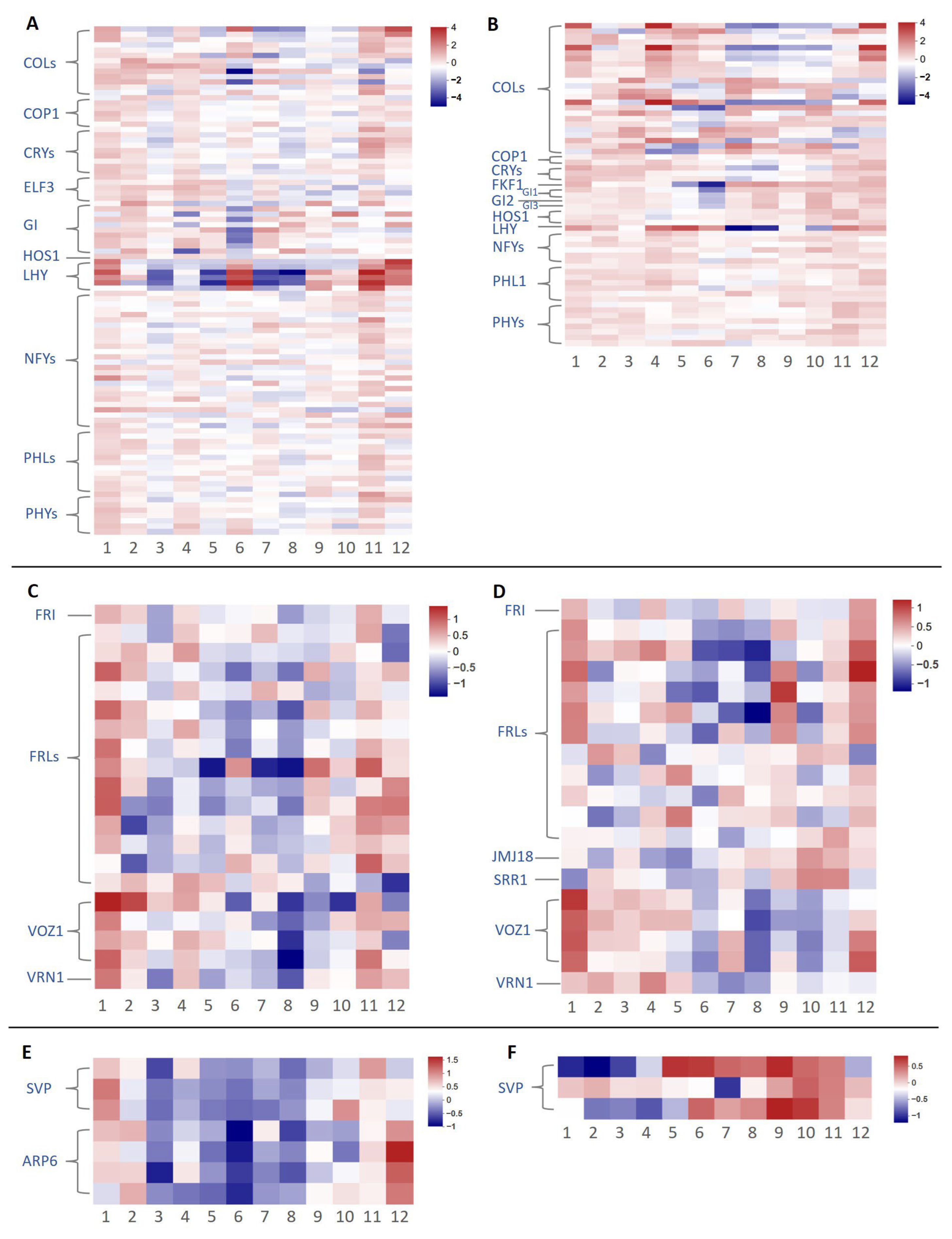
| Month | Jan. | Feb. | Mar. | Apr. | May | Jun. | Jul. | Aug | Sep. | Oct. | Nov. | Dec. | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ca | Plant 1 | 1 | 0 | 38 | 90 | 48 | 12 | 28 | 15 | 10 | 7 | 5 | 3 | 257 |
| Plant 2 | 0 | 1 | 35 | 80 | 50 | 15 | 20 | 22 | 13 | 9 | 5 | 1 | 251 | |
| Plant 3 | 2 | 0 | 40 | 98 | 40 | 13 | 23 | 23 | 11 | 10 | 3 | 2 | 265 | |
| Cj | Plant 1 | 0 | 0 | 0 | 0 | 16 | 159 | 3 | 0 | 0 | 0 | 0 | 0 | 178 |
| Plant 2 | 0 | 0 | 0 | 0 | 6 | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 66 | |
| Plant 3 | 0 | 0 | 0 | 0 | 7 | 81 | 2 | 0 | 0 | 0 | 0 | 0 | 90 |
| Species | Target Gene | Forward Primers (5′ to 3′) | Reverse Primers (5′ to 3′) |
|---|---|---|---|
| C. azalea and C. japonica | GAPDH | CTGTCGATGTCTCAGTGGTTGAC | TGATCTCATCATAGGAAGCCTTCTT |
| FKF1 | TTCGAGATCTTCACTGGCTATCG | TCACTGTTCCATCATCACCATGT | |
| GI1 | TAGTCCTCCATTTGCGTCTTTCA | TTTTCCTCTCCTGCTGTACATCC | |
| GI2 | CCCTTGCAACCTCCCATTCT | AGGCAGACCCAACACCAAAA | |
| COL2 | TGACAATGACGACGAGGCTG | TCTCCACACTGAACTGGCAC | |
| COL10 | GGAGGAGGAGGAGATGACCA | CGCTAGGCATTCGAGGAGTT | |
| COL14 | GGGAGGTAGTGGGAGTGGAT | TCGCTACTTTTGGGACCGAC | |
| COL16 | TGTGTGGGGATAGGAGGAGG | AGGAGGAGGACGAGGATGAC | |
| GA2OX8 | TCGCTACTTTTGGGACCGAC | GGCTCTCTTCGACGACTACG | |
| SVP1 | GACGAAGGGTGAGATAATGGAGAA | GCCCTCAAATTCAGTTCTGGAAAA | |
| SVP2 | AAGACAAAGGATGACAGGGTTGA | TTCTGAAGACTGCCCTTGTTCTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Liu, W.; Li, J.; Yin, H.; Li, X.; Wang, M.; Fan, Z. Differences in the Gene Regulatory Network for Floral Induction in Two Camellia Species. Int. J. Mol. Sci. 2025, 26, 10854. https://doi.org/10.3390/ijms262210854
Wang X, Liu W, Li J, Yin H, Li X, Wang M, Fan Z. Differences in the Gene Regulatory Network for Floral Induction in Two Camellia Species. International Journal of Molecular Sciences. 2025; 26(22):10854. https://doi.org/10.3390/ijms262210854
Chicago/Turabian StyleWang, Xiong, Weixin Liu, Jiyuan Li, Hengfu Yin, Xinlei Li, Minyan Wang, and Zhengqi Fan. 2025. "Differences in the Gene Regulatory Network for Floral Induction in Two Camellia Species" International Journal of Molecular Sciences 26, no. 22: 10854. https://doi.org/10.3390/ijms262210854
APA StyleWang, X., Liu, W., Li, J., Yin, H., Li, X., Wang, M., & Fan, Z. (2025). Differences in the Gene Regulatory Network for Floral Induction in Two Camellia Species. International Journal of Molecular Sciences, 26(22), 10854. https://doi.org/10.3390/ijms262210854








