Temporal Analysis of Embryonic Epidermal Morphogenesis in Caenorhabditis elegans
Abstract
1. Introduction
2. Results
2.1. Embryo Segmentation and Classification Using ResU-Net and ResNet
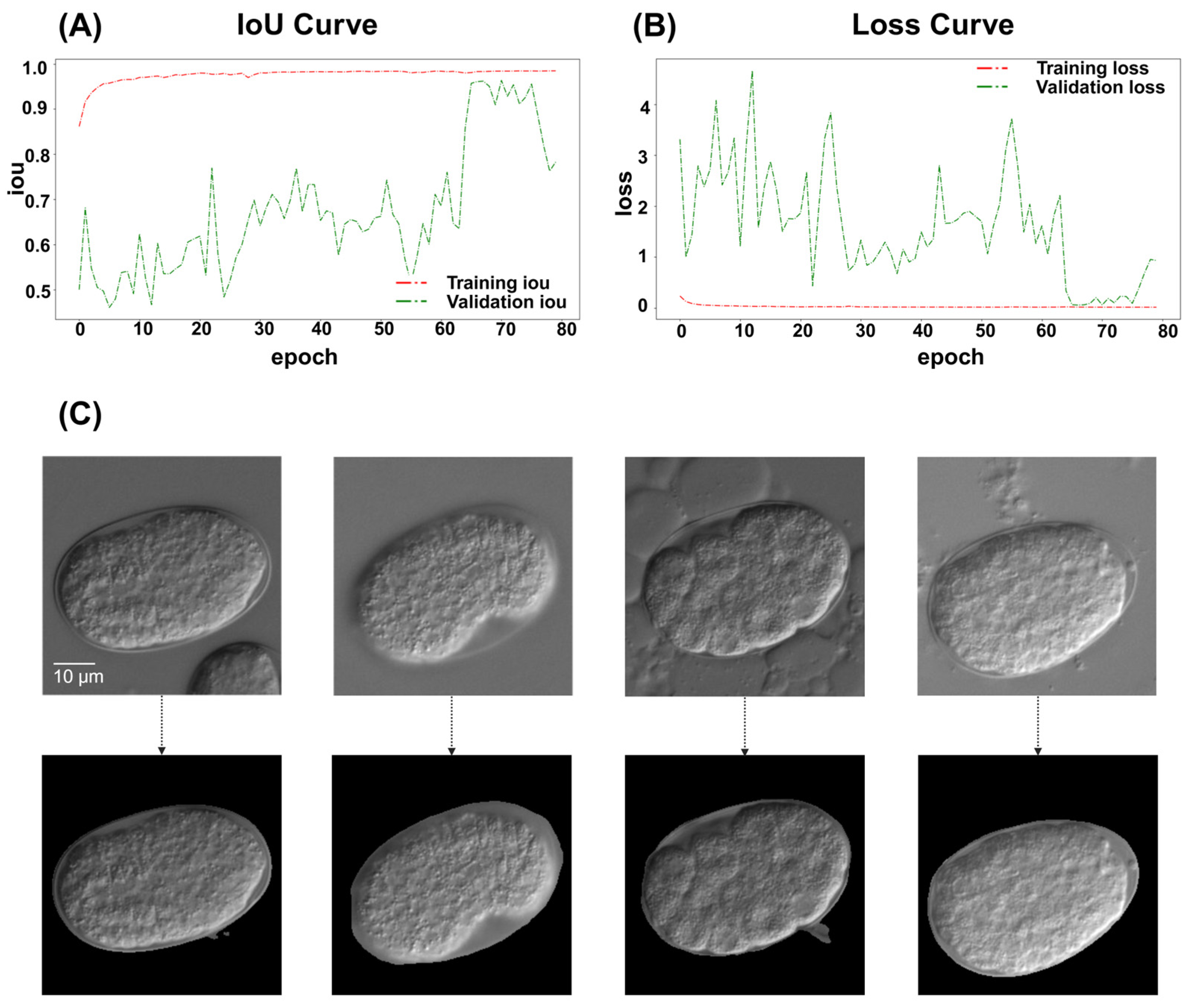
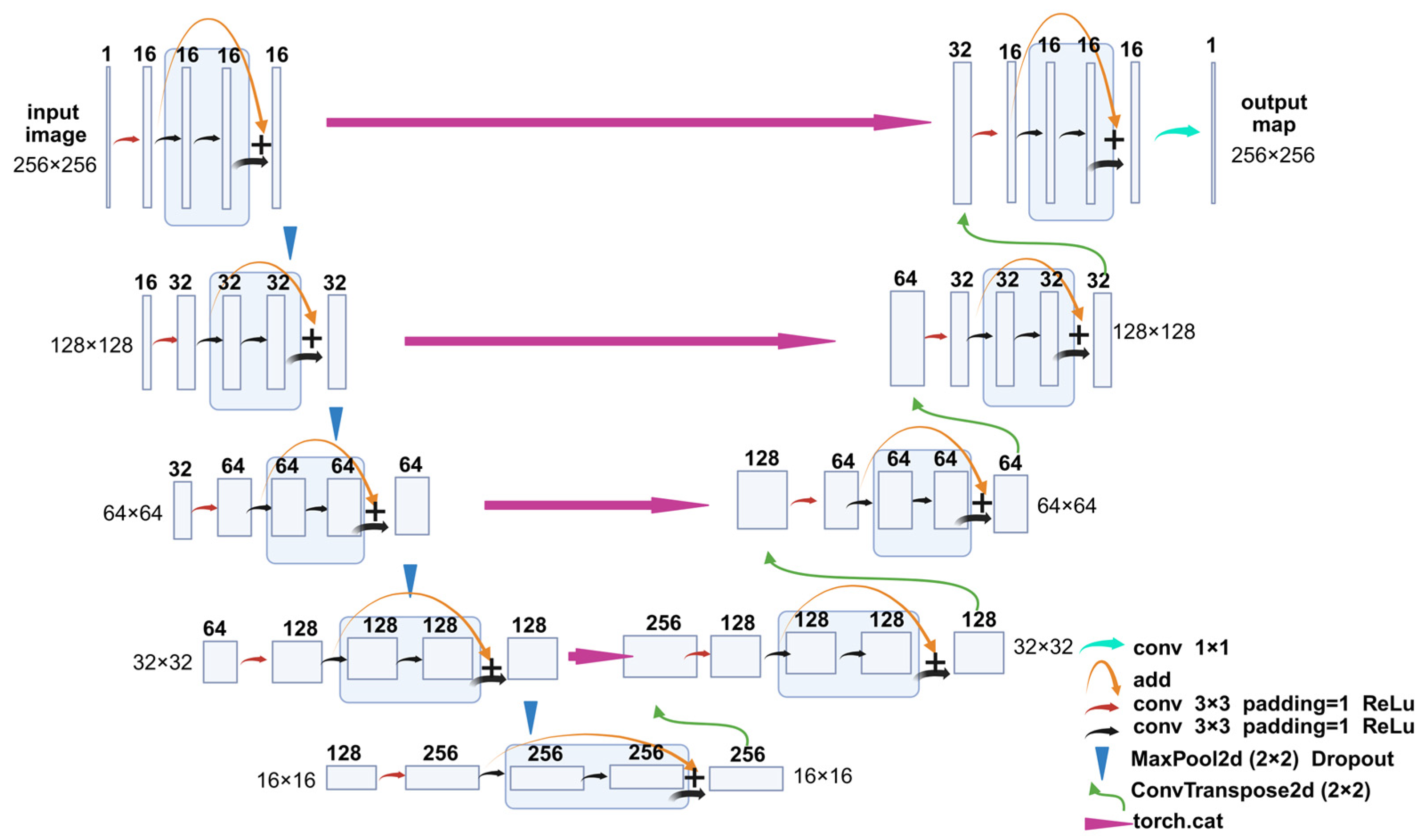
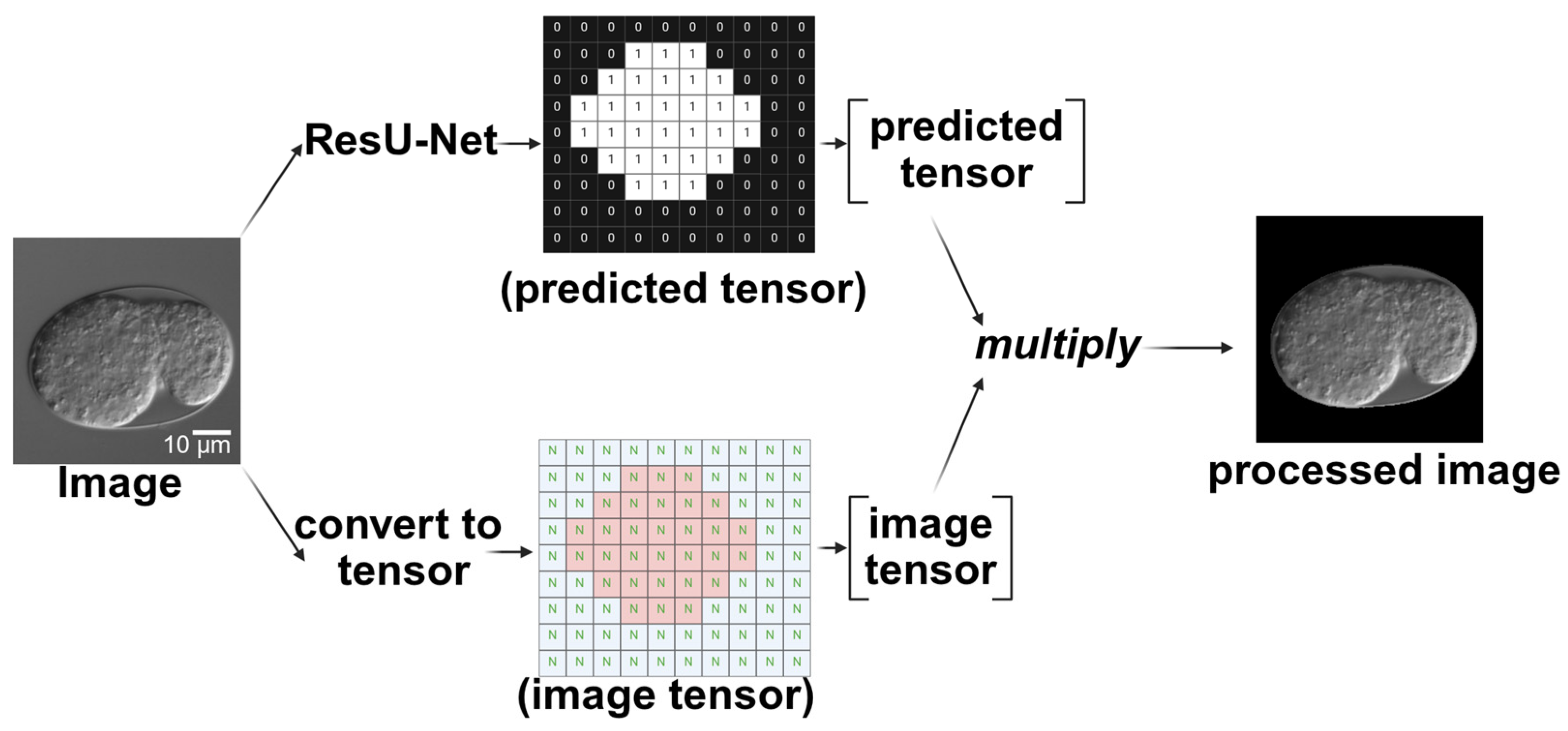
2.1.1. Image Segmentation Using ResU-Net
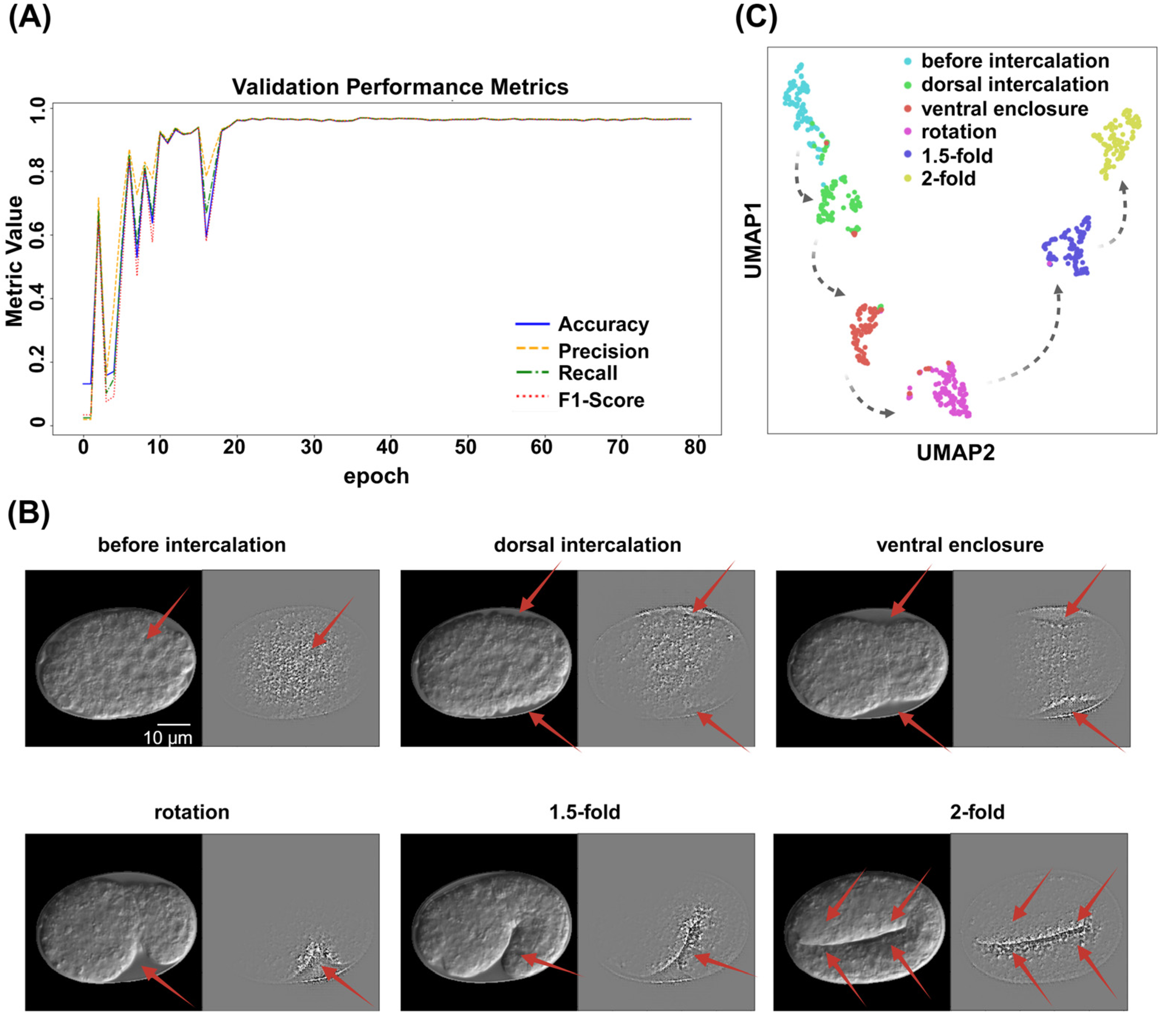
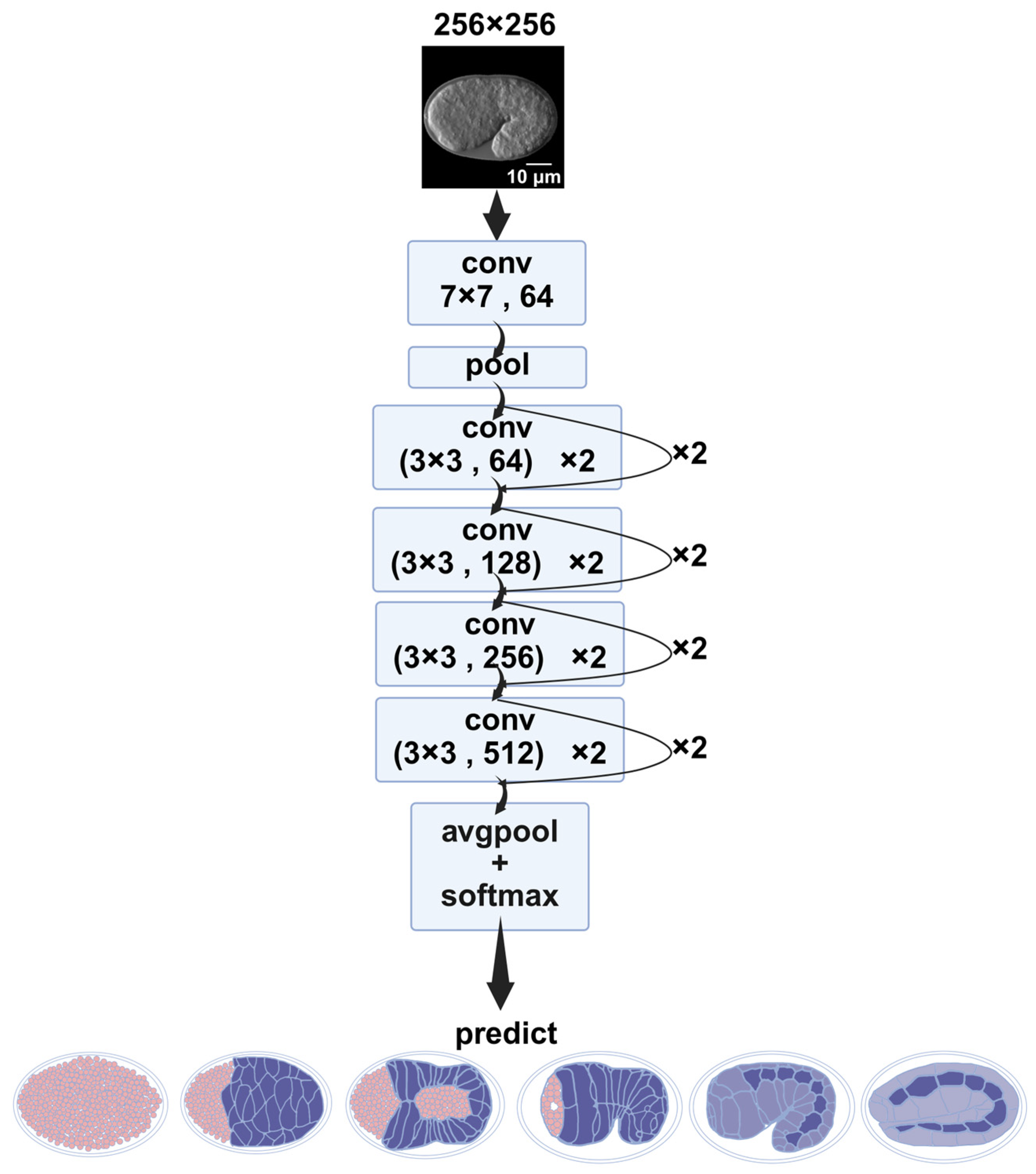
2.1.2. Image Classification Using ResNet
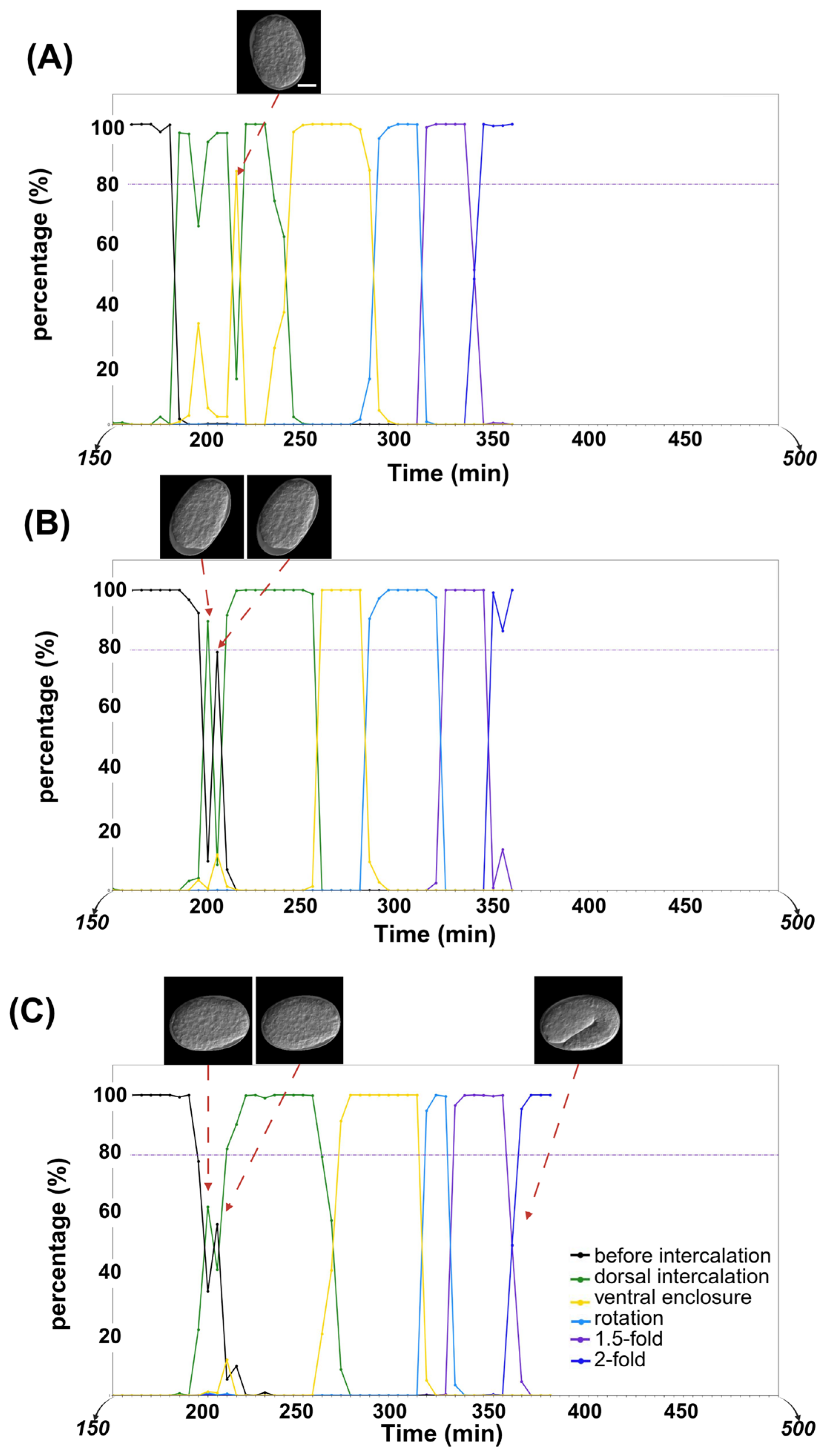
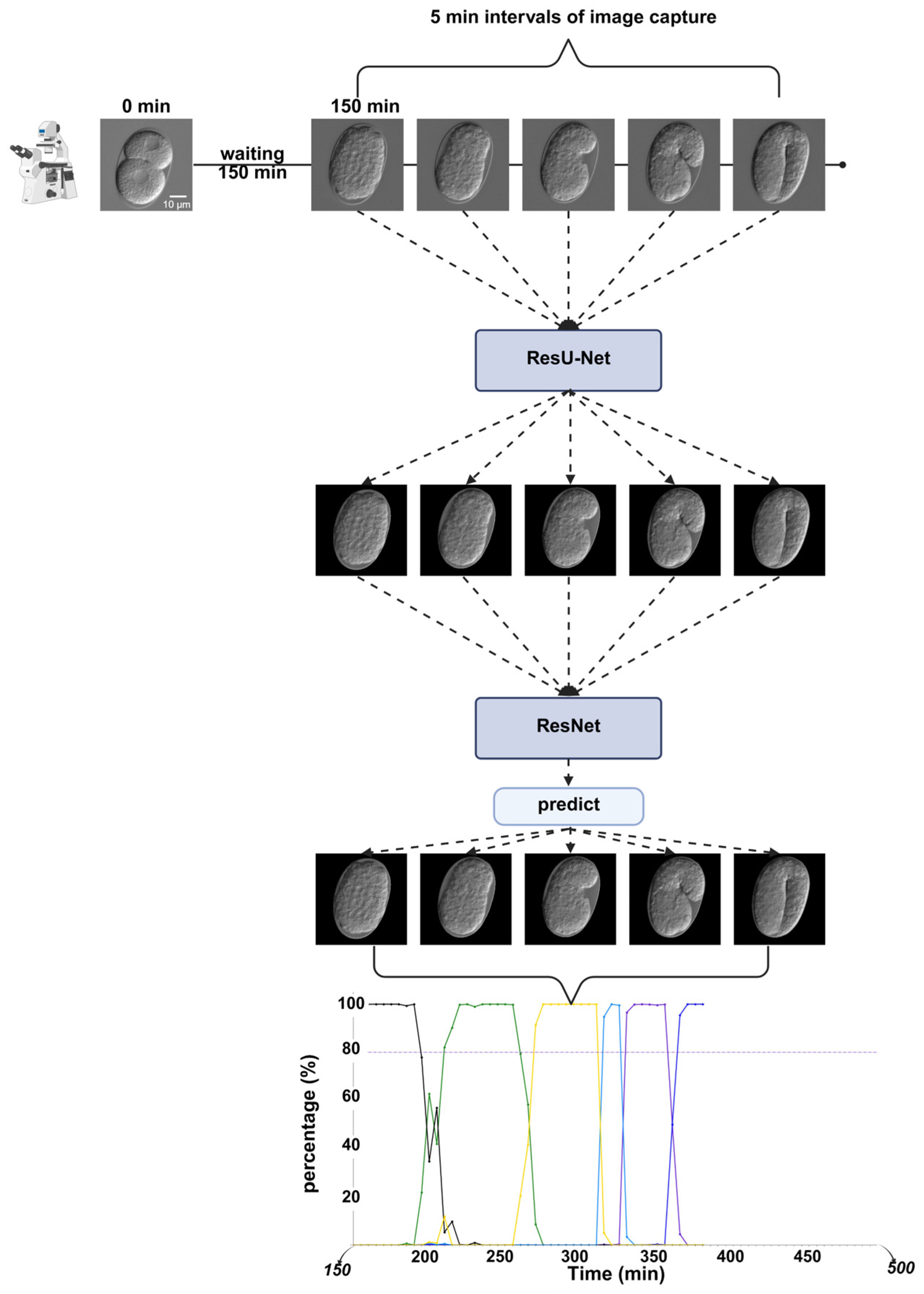
2.2. Analysis of Temporal Prediction Accuracy in RNAi Time-Lapse Data
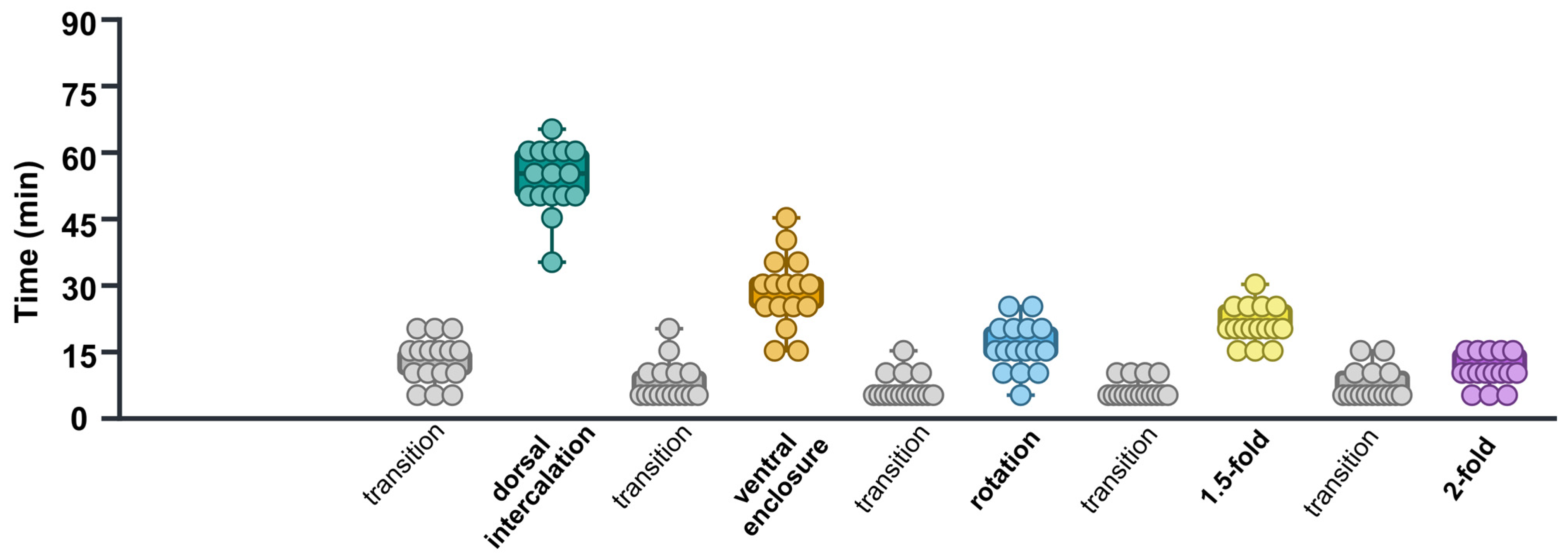
2.3. Analysis of Embryonic Stage Durations in control(RNAi) Time-Lapse Data
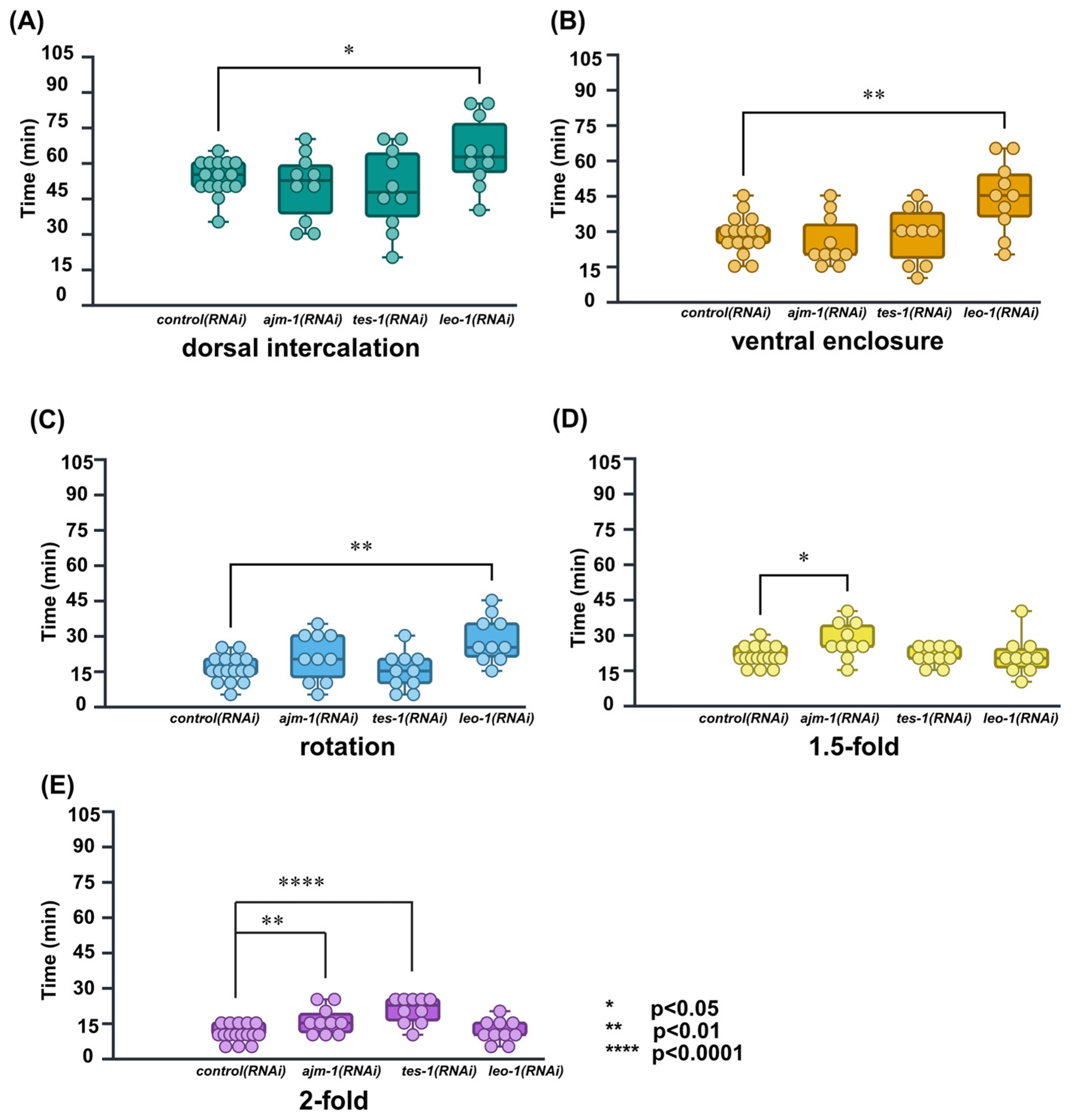
2.4. Application in RNAi Knockdown Time-Lapse Data
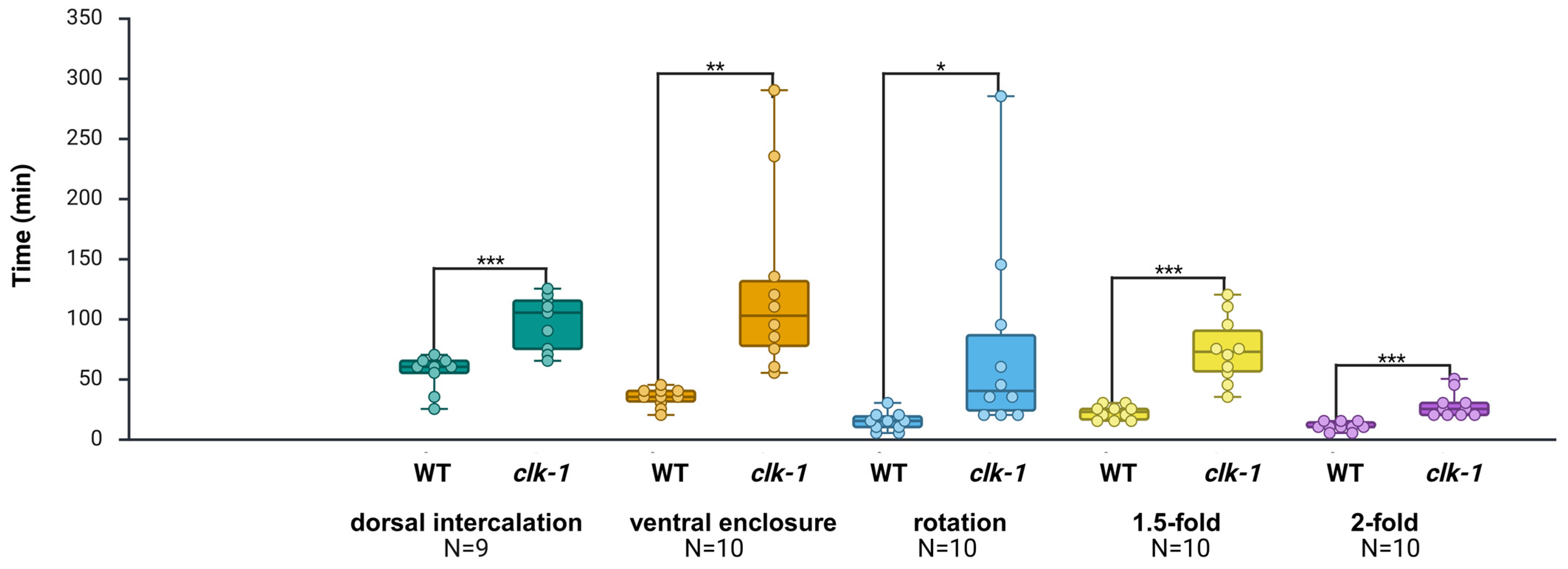
2.5. Expanding Analysis of Temporal Prediction Accuracy in Developmentally Slow clk-1(e2519) Mutant Embryos
2.6. Expanding Application to Developmentally Slow clk-1(e2519) Mutant Embryos
3. Discussion
3.1. Deep Learning-Based Diagnostic Tool for Temporal Analysis
3.2. Calculations of Time Required for Each Stage in control(RNAi) Group
3.3. Trial to Temporal Analysis of the Gene Function in RNAi Knockdown Animals
3.4. Temporal Analysis Reveals That the Developmental Time Course Is Comparable Between control(RNAi) and WT Embryos
3.5. Extended Trial to Temporal Analysis of Developmentally Slow clk-1(e2519) Mutant Embryos
3.6. Contributions and Limitations of the Current Approach
4. Materials and Methods
4.1. Caenorhabditis elegans Strains
4.2. RNA Interference Assay
4.3. Epidermal Morphogenesis Stages
4.3.1. Before Intercalation
4.3.2. Dorsal Intercalation
4.3.3. Ventral Enclosure
4.3.4. Rotation
4.3.5. 1.5-Fold and 2-Fold
4.4. Microscope and Image Acquisition
4.5. Models Training and Evaluation
4.5.1. ResU-Net
4.5.2. ResNet
4.6. Timeline
4.7. Image Interpretability Analysis
4.7.1. Grad-CAM
4.7.2. UMAP
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vuong-Brender, T.T.K.; Yang, X.; Labouesse, M.C. elegans Embryonic Morphogenesis. Curr. Top. Dev. Biol. 2016, 116, 597–616. [Google Scholar] [CrossRef]
- Chisholm, A.D.; Hardin, J. Epidermal morphogenesis. In WormBook: The Online Review of C. elegans Biology; WormBook: Pasadena, CA, USA, 2005. [Google Scholar]
- Apfeld, J.; Alper, S. What Can We Learn About Human Disease from the Nematode C. elegans? In Disease Gene Identification: Methods and Protocols; DiStefano, J.K., Ed.; Springer: New York, NY, USA, 2018; pp. 53–75. ISBN 978-1-4939-7471-9. [Google Scholar]
- Sulston, J.E.; Schierenberg, E.; White, J.G.; Thomson, J.N. The Embryonic Cell Lineage of the Nematode Caenorhabditis elegans. Dev. Biol. 1983, 100, 64–119. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Gönczy, P.; Echeverri, C.; Oegema, K.; Coulson, A.; Jones, S.J.; Copley, R.R.; Duperon, J.; Oegema, J.; Brehm, M.; Cassin, E.; et al. Functional Genomic Analysis of Cell Division in C. elegans Using RNAi of Genes on Chromosome III. Nature 2000, 408, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Fraser, A.G.; Kamath, R.S.; Zipperlen, P.; Martinez-Campos, M.; Sohrmann, M.; Ahringer, J. Functional Genomic Analysis of C. elegans Chromosome I by Systematic RNA Interference. Nature 2000, 408, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Williams-Masson, E.M.; Malik, A.N.; Hardin, J. An Actin-Mediated Two-Step Mechanism Is Required for Ventral Enclosure of the C. elegans Hypodermis. Development 1997, 124, 2889–2901. [Google Scholar] [CrossRef]
- Quintin, S.; Michaux, G.; McMahon, L.; Gansmuller, A.; Labouesse, M. The Caenorhabditis elegans Gene Lin-26 Can Trigger Epithelial Differentiation without Conferring Tissue Specificity. Dev. Biol. 2001, 235, 410–421. [Google Scholar] [CrossRef]
- Labouesse, M.; Sookhareea, S.; Horvitz, H.R. The Caenorhabditis elegans Gene Lin-26 Is Required to Specify the Fates of Hypodermal Cells and Encodes a Presumptive Zinc-Finger Transcription Factor. Development 1994, 120, 2359–2368. [Google Scholar] [CrossRef]
- Pettitt, J.; Cox, E.A.; Broadbent, I.D.; Flett, A.; Hardin, J. The Caenorhabditis elegans P120 Catenin Homologue, JAC-1, Modulates Cadherin-Catenin Function during Epidermal Morphogenesis. J. Cell Biol. 2003, 162, 15–22. [Google Scholar] [CrossRef]
- George, S.E.; Simokat, K.; Hardin, J.; Chisholm, A.D. The VAB-1 Eph Receptor Tyrosine Kinase Functions in Neural and Epithelial Morphogenesis in C. elegans. Cell 1998, 92, 633–643. [Google Scholar] [CrossRef]
- Meeuse, M.W.; Hauser, Y.P.; Morales Moya, L.J.; Hendriks, G.; Eglinger, J.; Bogaarts, G.; Tsiairis, C.; Großhans, H. Developmental Function and State Transitions of a Gene Expression Oscillator in Caenorhabditis elegans. Mol. Syst. Biol. 2020, 16, e9975, Erratum in Mol. Syst. Biol. 2020, 16, e9498. [Google Scholar] [CrossRef]
- Green, R.A.; Khaliullin, R.N.; Zhao, Z.; Ochoa, S.D.; Hendel, J.M.; Chow, T.-L.; Moon, H.; Biggs, R.J.; Desai, A.; Oegema, K. Automated Profiling of Gene Function during Embryonic Development. Cell 2024, 187, 3141–3160.e23. [Google Scholar] [CrossRef]
- Shaikh, M.A.; Al-Rawashdeh, H.S.; Sait, A.R.W. A Review of Artificial Intelligence-Based Down Syndrome Detection Techniques. Life 2025, 15, 390, Correction in Life 2025, 15, 390. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-H.; Lin, C.-H.; Lane, H.-Y. Machine Learning and Novel Biomarkers for the Diagnosis of Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 2761. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.; Lin, C.-H.; Lane, H.-Y. Deep Learning with Neuroimaging and Genomics in Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 7911. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zheng, S.; Li, L.; Zhang, X.; Zhang, X.; Huang, Z.; Chen, J.; Wang, R.; Zhao, H.; Chong, Y.; et al. Deep Learning Enables Accurate Diagnosis of Novel Coronavirus (COVID-19) With CT Images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 18, 2775–2780. [Google Scholar] [CrossRef]
- Reddy, P.; J, A. Diagnosis of Autism in Children Using Deep Learning Techniques by Analyzing Facial Features. Eng. Proc. 2023, 59, 198. [Google Scholar] [CrossRef]
- Azuma, Y.; Okada, H.; Onami, S. Systematic Analysis of Cell Morphodynamics in C. elegans Early Embryogenesis. Front. Bioinform. 2023, 3, 1082531. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, S.; Yuan, H.; Yong, R.; Duan, S.; Li, Y.; Spencer, J.; Lim, E.G.; Yu, L.; Song, P. Deep Learning for Microfluidic-Assisted Caenorhabditis elegans Multi-Parameter Identification Using YOLOv7. Micromachines 2023, 14, 1339. [Google Scholar] [CrossRef]
- Bates, K.; Le, K.N.; Lu, H. Deep Learning for Robust and Flexible Tracking in Behavioral Studies for C. elegans. PLoS Comput. Biol. 2022, 18, e1009942. [Google Scholar] [CrossRef]
- Toulany, N.; Morales-Navarrete, H.; Čapek, D.; Grathwohl, J.; Ünalan, M.; Müller, P. Uncovering Developmental Time and Tempo Using Deep Learning. Nat. Methods 2023, 20, 2000–2010. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Huang, Z.; Cai, H.; Li, Z.; Zhu, J.; Wu, D.; Xu, W.; Qiu, H.; Zhang, N.; Li, G.; et al. WormCNN-Assisted Establishment and Analysis of Glycation Stress Models in C. elegans: Insights into Disease and Healthy Aging. Int. J. Mol. Sci. 2024, 25, 9675. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.I.; Bao, Z.; Boyle, T.J.; Waterston, R.H. The Lineaging of Fluorescently-Labeled Caenorhabditis elegans Embryos with StarryNite and AceTree. Nat. Protoc. 2006, 1, 1468–1476. [Google Scholar] [CrossRef] [PubMed]
- Katzman, B.; Tang, D.; Santella, A.; Bao, Z. AceTree: A Major Update and Case Study in the Long Term Maintenance of Open-Source Scientific Software. BMC Bioinform. 2018, 19, 121. [Google Scholar] [CrossRef]
- Cao, J.; Guan, G.; Ho, V.W.S.; Wong, M.-K.; Chan, L.-Y.; Tang, C.; Zhao, Z.; Yan, H. Establishment of a Morphological Atlas of the Caenorhabditis elegans Embryo Using Deep-Learning-Based 4D Segmentation. Nat. Commun. 2020, 11, 6254. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Spring: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Diakogiannis, F.I.; Waldner, F.; Caccetta, P.; Wu, C. ResUNet-a: A Deep Learning Framework for Semantic Segmentation of Remotely Sensed Data. ISPRS J. Photogramm. Remote Sens. 2020, 162, 94–114. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Kubota, Y.; Tsuyama, K.; Takabayashi, Y.; Haruta, N.; Maruyama, R.; Iida, N.; Sugimoto, A. The PAF1 Complex Is Involved in Embryonic Epidermal Morphogenesis in Caenorhabditis elegans. Dev. Biol. 2014, 391, 43–53. [Google Scholar] [CrossRef]
- Köppen, M.; Simske, J.S.; Sims, P.A.; Firestein, B.L.; Hall, D.H.; Radice, A.D.; Rongo, C.; Hardin, J.D. Cooperative Regulation of AJM-1 Controls Junctional Integrity in Caenorhabditis elegans Epithelia. Nat. Cell Biol. 2001, 3, 983–991. [Google Scholar] [CrossRef]
- Lynch, A.M.; Zhu, Y.; Lucas, B.G.; Winkelman, J.D.; Bai, K.; Martin, S.C.T.; Block, S.; Slabodnick, M.M.; Audhya, A.; Goldstein, B.; et al. TES-1/Tes and ZYX-1/Zyxin Protect Junctional Actin Networks under Tension during Epidermal Morphogenesis in the C. elegans Embryo. Curr. Biol. 2022, 32, 5189–5199.e6. [Google Scholar] [CrossRef]
- Wong, A.; Boutis, P.; Hekimi, S. Mutations in the Clk-1 Gene of Caenorhabditis elegans Affect Developmental and Behavioral Timing. Genetics 1995, 139, 1247–1259. [Google Scholar] [CrossRef]
- Chen, J.; Carey, J.R.; Ferris, H. Comparative Demography of Isogenic Populations of Caenorhabditis elegans. Exp. Gerontol. 2001, 36, 431–440. [Google Scholar] [CrossRef]
- Reiss, A.L. Childhood Developmental Disorders: An Academic and Clinical Convergence Point for Psychiatry, Neurology, Psychology and Pediatrics. J. Child Psychol. Psychiatry 2009, 50, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S. The Genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef] [PubMed]
- Maeda, I.; Kohara, Y.; Yamamoto, M.; Sugimoto, A. Large-Scale Analysis of Gene Function in Caenorhabditis elegans by High-Throughput RNAi. Curr. Biol. 2001, 11, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Ota, N.; Takatsuka, H.; Unno, T.; Onami, S.; Sugimoto, A.; Ito, M. The PAF1 Complex Cell Autonomously Promotes Oogenesis in Caenorhabditis elegans. Genes Cells 2022, 27, 409–420. [Google Scholar] [CrossRef]
- Hardin, J.; Serre, J.; King, R.; Walck-Shannon, E.; Reiner, D. Imaging Epidermal Cell Rearrangement in the C. elegans Embryo. Methods Mol. Biol. 2022, 2438, 345–376. [Google Scholar] [CrossRef]
- Oegema, K.; Hyman, T. Cell Division. In WormBook: The Online Review of C. elegans Biology; WormBook: Pasadena, CA, USA, 2006. [Google Scholar]

| Metric | Value |
|---|---|
| True Positive (TP) | 519,293 |
| False Positive (FP) | 14,760 |
| True Negative (TN) | 714,840 |
| False Negative (FN) | 4483 |
| Sensitivity (TPR) | 99.1% |
| Specificity (TNR) | 98.0% |
| Overall Accuracy | 98.5% |
| Precision (PPV) | 97.2% |
| F1-Score | 98.2% |
| Intersection over Union (IoU) | 96.4% |
| Metric | Value |
|---|---|
| Sensitivity (TPR) | 96.9% |
| Specificity (TNR) | 98.0% |
| Overall Accuracy | 96.9% |
| Precision (PPV) | 96.9% |
| F1-Score | 96.8% |
| RNAi | Number of Timelines | Timeline Number with Misclassification | Total Images | Images Number with Misclassification | Continuous Misclassification |
|---|---|---|---|---|---|
| control(RNAi) | 16 | 1 | 681 | 0.2% (n= 1) | 0 |
| leo-1(RNAi) | 10 | 2 | 541 | 0.4% (n = 2) | 0 |
| ajm-1(RNAi) | 10 | 2 | 467 | 0.6% (n = 3) | 0 |
| tes-1(RNAi) | 10 | 3 | 457 | 0.7% (n = 3) | 0 |
| RNAi | Dorsal Intercalation (min) | Ventral Enclosure (min) | Rotation (min) | 1.5-Fold (min) | 2-Fold (min) |
|---|---|---|---|---|---|
| control(RNAi) | 53.75 ± 1.85 | 28.43 ± 2.02 | 15.93 ± 1.38 | 20.93 ± 1.04 | 10.62 ± 0.89 |
| leo-1(RNAi) | 64.50 ± 4.74 * | 44.50 ± 4.80 ** | 28.50 ± 3.08 ** | 21.00 ± 2.56 | 11.50 ± 1.50 |
| ajm-1(RNAi) | 50.50 ± 4.47 | 25.50 ± 3.37 | 21.00 ± 3.23 | 27.50 ± 2.39 * | 16.00 ± 1.80 ** |
| tes-1(RNAi) | 49.50 ± 5.46 | 28.50 ± 3.73 | 15.00 ± 2.47 | 21.00 ± 1.25 | 20.50 ± 1.74 **** |
| Strains | Number of Timelines | Timeline Number with Misclassification | Total Images | Images Number with Misclassification | Continuous Misclassification |
|---|---|---|---|---|---|
| WT | 10 | 2 | 383 | 0.5% (n = 2) | 1 |
| clk-1(e2519) | 10 | 7 | 1000 | 2.3% (n = 23) | 1 |
| Strains | Dorsal Intercalation (min) | Ventral Enclosure (min) | Rotation (min) | 1.5-fold (min) | 2-fold (min) |
|---|---|---|---|---|---|
| WT | 55.00 ± 5.00 | 34.50 ± 2.41 | 14.5 ± 2.41 | 22.00 ± 1.86 | 10.50 ± 1.17 |
| clk-1(e2519) | 97.22 ± 7.60 *** | 126.00 ± 24.45 ** | 76.00 ± 26.41 * | 74.00 ± 8.69 *** | 28.50 ± 3.42 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Li, P.; Onishi, M.; Chuen, L.K.; Kubota, Y.; Ito, M. Temporal Analysis of Embryonic Epidermal Morphogenesis in Caenorhabditis elegans. Int. J. Mol. Sci. 2025, 26, 10802. https://doi.org/10.3390/ijms262110802
Li F, Li P, Onishi M, Chuen LK, Kubota Y, Ito M. Temporal Analysis of Embryonic Epidermal Morphogenesis in Caenorhabditis elegans. International Journal of Molecular Sciences. 2025; 26(21):10802. https://doi.org/10.3390/ijms262110802
Chicago/Turabian StyleLi, Fangzheng, Peiyue Li, Mao Onishi, Law King Chuen, Yukihiko Kubota, and Masahiro Ito. 2025. "Temporal Analysis of Embryonic Epidermal Morphogenesis in Caenorhabditis elegans" International Journal of Molecular Sciences 26, no. 21: 10802. https://doi.org/10.3390/ijms262110802
APA StyleLi, F., Li, P., Onishi, M., Chuen, L. K., Kubota, Y., & Ito, M. (2025). Temporal Analysis of Embryonic Epidermal Morphogenesis in Caenorhabditis elegans. International Journal of Molecular Sciences, 26(21), 10802. https://doi.org/10.3390/ijms262110802






