Abstract
Cardiometabolic diseases, including cardiovascular disorders and type 2 diabetes mellitus, are the leading cause of morbidity and mortality worldwide, placing a significant burden on healthcare systems. Although advances in imaging and risk stratification have improved disease management, conventional diagnostic and prognostic tools often lack the requisite sensitivity and specificity for early and precise risk stratification. This limitation stems from their poor ability to capture the full molecular complexity of these conditions, underscoring an urgent need for innovative biomarkers to bridge these gaps. MicroRNAs, small non-coding RNAs that regulate gene expression post-transcriptionally, have emerged as promising candidates. Their characteristics offer several advantages over traditional methods, including exceptional stability in biological fluids, strong tissue and disease specificity, and the ability to reflect dynamic pathological changes. These unique features enable miRNAs to detect subtle molecular alterations that may precede clinical symptoms, thereby overcoming key limitations of current diagnostic approaches. Altered circulating miRNA profiles have been linked to pathological processes such as endothelial dysfunction, inflammation, oxidative stress, and maladaptive cardiac remodeling. This review provides a comprehensive overview of the current evidence supporting the diagnostic and prognostic role of circulating miRNAs in cardiometabolic disease. We highlight their potential as early detection biomarkers, tools for patient stratification, and indicators of therapeutic response. Furthermore, we discuss key limitations to clinical translation, including methodological variability, challenges in sample handling, differences in normalization strategies, and platform-dependent quantification inconsistencies. Overcoming these obstacles and achieving robust large-scale clinical validation will be essential to fully harness the potential of miRNAs as next-generation molecular signatures in precision medicine.
1. Introduction
The global impact of cardiometabolic disorders extends beyond morbidity and mortality widely, shaping public health priorities and straining healthcare systems [1]. Although substantial progress has been made in refining diagnostic imaging and clinical risk scores, current strategies still fall short in detecting disease at its earliest, subclinical stages. This gap highlights the need for innovative biomarkers capable of capturing the underlying molecular disturbances that precede overt clinical manifestations. While valuable, conventional protein biomarkers such as troponins, B-type natriuretic peptide (BNP), C-reactive protein (CRP), and glycated hemoglobin (HbA1c) often lack the sensitivity and specificity required to detect disease at its earliest stages or to provide information about how the disease progresses and how well the treatment is working [2]. These traditional markers often reflect damage after it has occurred, which highlights the critical need for a new generation of biomarkers that can offer earlier, more precise and mechanistically relevant insights.
Over the last decade, microRNAs (miRNAs) have emerged as a novel class of regulatory molecules with significant diagnostic potential, transforming our understanding of disease pathophysiology [3,4]. These small non-coding RNAs act as master regulators of gene expression at the post-transcriptional level, affecting a wide range of essential biological processes. miRNAs exert their effects through the RNA-induced silencing complex (RISC), which binds to complementary sequences within the 3′ untranslated regions (3′UTRs) of target mRNAs, resulting in translational repression or mRNA degradation [5]. They have a significant impact on cardiometabolic pathophysiology, regulating important processes such as inflammation, fibrosis, angiogenesis, and lipid metabolism. Dysregulation of specific miRNA profiles directly contributes to the development and progression of cardiovascular diseases such as atherosclerosis, myocardial infarction and stroke. The intricate regulatory networks governed by miRNAs have led to the identification of specific miRNA signatures, distinct patterns of up- or down-regulated miRNAs that are uniquely associated with particular conditions. For instance, in atherosclerosis, these signatures can differentiate between stable and unstable plaques, which could enable the prediction of rupture before a severe stroke or heart attack. Studies have revealed that miRNAs such as miR-33 [6], which regulates cholesterol homeostasis, and miR-145 [7,8] which modulates vascular smooth muscle cell proliferation, play a pivotal role in this process. In acute ischemic stroke, changes in circulating miRNAs can be detected within hours of the event, providing a more effective diagnostic tool than traditional imaging [9]. Other studies have shown that altered levels of miRNAs, such as miR-124 [10] and miR-155 [11,12], are associated with neuronal damage and neuroinflammation following an ischemic event. Furthermore, specific miRNAs, such as miR-21, have been shown to be involved in a wide range of pathological processes, including fibrosis and inflammation, making them promising theranostic targets as well as diagnostic markers [13,14].
The complexity arises from the fact that a single miRNA can target hundreds of genes, and a single gene can be regulated by multiple miRNAs, creating an interconnected regulatory network that mirrors the intricate nature of these diseases [15,16].
This review critically examines the current understanding of circulating miRNA signatures in cardiometabolic disorders, highlighting their mechanistic relevance, diagnostic potential, and translational challenges. Furthermore, emerging detection technologies and future directions for clinical integration are discussed.
2. Biology of microRNAs
2.1. Biogenesis
MicroRNAs are small non-coding RNAs of ~22 nucleotides that regulate gene expression post-transcriptionally. They are transcribed by RNA polymerase II as primary transcripts (pri-miRNAs) containing stem-loop hairpins [17]. In the nucleus, Drosha and its cofactor DGCR8 process pri-miRNAs into ~65–70 nucleotide precursor miRNAs (pre-miRNAs) with a 2-nucleotide 3′ overhang [17]. Pre-miRNAs are exported to the cytoplasm, where Dicer, with cofactors such as TRBP or PACT, cleaves the terminal loop to produce a ~21–23 nt miRNA duplex. The guide strand is loaded into Argonaute proteins to form the RNA-induced silencing complex (RISC), which mediates gene silencing via mRNA cleavage or translational repression and decay (Figure 1) [18]. Non-canonical pathways, like mirtrons, bypass Drosha processing through spliced introns that fold into pre-miRNA-like hairpins, highlighting the flexibility and precision of miRNA biogenesis [19]. Dysregulation at any stage of miRNA biogenesis, such as transcriptional control, Drosha/DGCR8 processing, nuclear export, Dicer activity, and RISC loading, has been implicated in pathological processes, including cardiovascular disease, immune dysfunction, and neurodegeneration [17]. For instance, impaired Dicer activity directly inhibits miRNA maturation and is closely linked to cardiac hypertrophy and heart failure. Recent studies demonstrate that disruptive modifications of Dicer, such as through oxidative by-products like 4-hydroxynonenal (4-HNE), suppress its function and lead to reduced miRNA biogenesis, defective cardiac remodeling, and progressive heart failure [20,21]. Furthermore, aberrant Drosha expression or function contributes to endothelial dysfunction and metabolic imbalance. Evidence shows that endothelial-specific deletion of Dicer impairs angiogenesis and promotes heart failure, while altered Drosha/Dicer activity increases expression of the anti-angiogenic factor TSP-1, exacerbating vascular complications observed in diabetes and cardiovascular disease [22,23]. These findings emphasize the pivotal role of precise Dicer and Drosha regulation in maintaining cardiovascular and metabolic health, and underscore their relevance in the pathogenesis of cardiometabolic disorders.
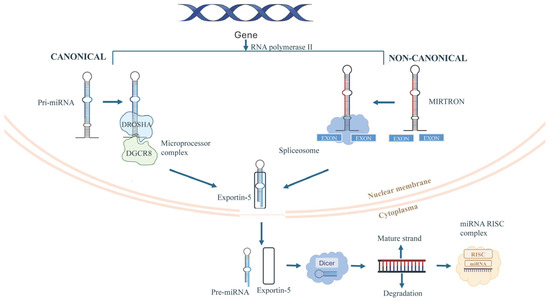
Figure 1.
Schematic representation of canonical and non-canonical miRNA biogenesis. Canonical miRNAs are generated from pri-miRNAs processed by the DROSHA–DGCR8 complex, while non-canonical mirtrons arise from spliced introns. Both pathways produce pre-miRNAs that are exported by exportin-5 from nucleus and cleaved by Dicer in cytoplasm, generating mature miRNAs loaded into the RISC complex.
This makes miRNA biogenesis an important key for precision medicine, both in terms of therapeutic intervention and biomarker discovery.
2.2. Biological Basis for Diagnostic Utility
MicroRNAs have emerged as clinically relevant biomarkers in cardiometabolic disease, offering distinct advantages over conventional diagnostic tools due to their unique biological characteristics. Their expression profiles exhibit remarkable tissue specificity, which enables the precise identification of the cellular origin of circulating signals. For instance, miR-208a and miR-499 are almost exclusively expressed in cardiomyocytes, serving as highly specific markers of myocardial injury [17,18]. Similarly, miR-126 reflects the status of endothelial cells, miR-122 is a strong indicator of hepatic function, and miR-375 is linked to pancreatic β-cell activity [19,20,21]. This selective enrichment allows for the attribution of organ-specific injury, a level of diagnostic resolution rarely attainable with traditional serum proteins. Furthermore, miRNAs display exceptional temporal sensitivity [22]. They are released into the circulation within hours of cellular stress or injury, often preceding the elevation of conventional biomarkers. In acute myocardial infarction, for example, circulating miR-1 levels can increase as early as two hours after onset, providing crucial diagnostic information before cardiac troponins reach their peak [23]. Finally, the exceptional biochemical stability of circulating miRNAs makes them highly promising as biomarkers. Unlike many other RNA species that degrade rapidly, miRNAs are resistant to RNase activity and harsh conditions like multiple freeze–thaw cycles and pH changes [24]. This protection is afforded by their enclosure within extracellular vesicles such as exosomes and microvesicles or their association with RNA-binding proteins, such as Argonaute-2 [25]. This molecular integrity allows for reliable detection and quantification using established techniques like qRT-PCR. Collectively, this intrinsic durability, along with their tissue specificity and ability to reflect subtle cellular pathology, positions circulating miRNAs as ideal, non-invasive biomarkers for liquid biopsy-based diagnostics (Table 1). Nevertheless, despite their remarkable analytical advantages, several technical and pre-analytical challenges still limit their routine clinical implementation. Variability in sample collection, processing, and storage conditions can markedly influence miRNA yield and integrity. Additionally, hemolysis can introduce confounding signals, particularly for erythrocyte-enriched miRNAs such as miR-451 or miR-16. Moreover, the absence of universally accepted normalization strategies and differences among quantification platforms—such as qRT-PCR, microarray, and next-generation sequencing—further complicate data comparability across studies. Addressing these methodological inconsistencies through standardized protocols and robust reference controls will be crucial to harness the diagnostic potential of circulating miRNAs fully.

Table 1.
Key Properties of Circulating miRNAs as Cardiometabolic Biomarkers.
3. Diagnostic Insights from Cardiovascular miRNAs
Given the complexity of cardiovascular disease, a new generation of diagnostic tools is needed to offer a deeper, more specific understanding of a patient’s condition. miRNAs have emerged as a powerful solution, acting as molecular messengers that not only signal the presence of disease but also reveal its unique signature at the cellular and tissue level. Their expression profiles provide a detailed insight into the heart and vasculature, offering an advanced level of information that is expected to transform the approach to diagnosing, categorizing, and monitoring cardiovascular pathology.
3.1. Acute Myocardial Infarction (AMI)
The accurate and timely diagnosis of acute myocardial infarction (AMI) remains a cornerstone of clinical cardiology, where rapid identification directly impacts survival and recovery. While high-sensitivity cardiac troponins (hs-cTn) are the current gold-standard biomarkers, their diagnostic performance can be limited in the very early hours after symptom onset [24]. Circulating miRNAs (c-miRNAs) have therefore emerged as powerful complementary tools, with cardiac-enriched species such as miR-208a, miR-499, miR-1, and miR-133a consistently validated as highly specific indicators of myocardial injury [25]. These miRNAs are rapidly released into the bloodstream, often within 2–4 h of ischemia, preceding detectable rises in hs-cTn, and their levels correlate with infarct size and ventricular dysfunction. Novel candidates such as miR-296-5p, miR-660-3p, miR-107, and miR-101-3p have also demonstrated strong diagnostic power [26]. Their performance is particularly enhanced when combined in multi-marker panels, underscoring the clinical potential of miRNA-based signatures for early “rule-in/rule-out” strategies in patients presenting with chest pain [26]. Beyond these classical markers, new regulatory mechanisms are being uncovered. For instance, the circular RNA circ_0091761 has been shown to be significantly upregulated in AMI patients, promoting endothelial injury by sponging miR-1278, whereas its suppression protects against apoptosis and inflammation, suggesting the circ_0091761/miR-1278 axis as a promising diagnostic and therapeutic target [27]. Similarly, miR-210-3p and miR-582-5p were found downregulated in AMI, with their loss leading to overexpression of MXD1 and the promotion of ferroptosis in hypoxic cardiomyocytes [28]. Their restoration suppressed cell death and provided robust diagnostic accuracy, with combined panels achieving AUCs above 0.80, supporting their role as classifiers of AMI and as predictors of coronary artery disease progression [28]. Elevated levels of miR-142 have been associated with AMI and correlate strongly with cardiac troponin I (r = 0.707), serving as an independent risk factor for the condition.
Additionally, miR-142 predicts worse long-term outcomes and reduced survival in affected patients [29]. Exosomal miRNAs are gaining attention as well, with studies identifying time-dependent increases in panels including miR-3473, miR-504, miR-490-5p, and miR-218a-2-3p in animal and human AMI, enabling machine learning-based diagnostic models with high precision and even accurate estimations of infarction onset time [30]. Among individual biomarkers, miR-1 has been extensively validated through meta-analysis, showing pooled sensitivity and specificity values of 78% and 85%, respectively, and robust diagnostic value across populations and detection platforms [31]. Importantly, miRNAs are not only diagnostic but also therapeutic. Stem cell-derived extracellular vesicles (EVs) and EV-mimetic nanovesicles were shown to confer cardioprotection via delivery of miR-24-3p, which activates the Nrf2 pathway and suppresses cardiomyocyte apoptosis, highlighting translational opportunities for miRNA-based therapies [32]. Finally, novel targets such as miR-409-5p, which is downregulated in AMI and inversely correlated with USP7 expression and cTnI levels, add another promising biomarker to the growing repertoire, with potential applications for both diagnosis and treatment [33].
3.2. Coronary Artery Disease and Atherosclerosis
Emerging evidence consistently highlights the diagnostic potential of c-miRNAs and exosomal miRNA profiles as minimally invasive biomarkers across different manifestations of coronary artery disease (CAD) and related cardiovascular conditions such as atherosclerosis [34]. Several studies have shown their ability to capture disease-specific signatures with high accuracy. For instance, exosomal miR-432-5p and miR-382-3p were found upregulated in patients with impaired myocardial perfusion, suggesting their role in linking functional myocardial alterations to molecular signaling pathways [35]. Similarly, the development of advanced biosensors, such as gold metallene-based electrochemiluminescence platforms, has enabled highly sensitive detection of miR-126-3p.
This confirms its downregulation in coronary artery calcification and validates its utility for early screening [36]. Beyond single markers, novel studies employing both conventional qRT-PCR and machine learning approaches have identified miR-140-3p as a robust diagnostic biomarker, able to improve prediction models when integrated with established risk factors [37]. Expanding this concept, multi-miRNA panels, such as a seven-miRNA signature including miR-10b-5p, miR-29c-3p, and miR-486-3p, have demonstrated outstanding diagnostic power in discriminating late-onset CAD (AUC 0.9924), reinforcing the added value of combinatorial biomarker strategies [38]. Population-specific studies further support the role of lipid-related miRNAs. For instance, miR-128-3p is significantly reduced in Indian CAD patients and shows independent predictive capacity even when adjusted for confounders [39], while miR-106b-5p has emerged as a reliable candidate with strong ROC performance (AUC 0.8975) [40]. Beyond canonical miRNAs, isoform microRNA (isomiR) profiling revealed even stronger correlations with coronary calcium burden in NAFLD patients, indicating that sequence variants may surpass conventional miRNAs in diagnostic accuracy [41]. Additional mechanistic insights come from studies linking specific miRNAs to CAD phenotypes: miR-34a, miR-145, and miR-222 can distinguish between obstructive CAD and INOCA, with miR-145 as an independent predictor of non-obstructive forms [42], while EV-derived miR-146b-5p, miR-4701-3p, and miR-1180-3p showed high specificity for subclinical atherosclerosis [43]. Broadly, different works confirm the consistency of findings across contexts, underscoring miRNAs such as miR-19b, miR-186-5p, miR-331, and miR-638 as promising biomarkers for fibrosis, plaque rupture, myocardial infarction, and vascular calcification, particularly when used in multiplex panels [44,45,46]. More recently, specific miRNAs have been tied to acute and prognostic endpoints, such as miR-139-5p in acute coronary syndrome with correlation to troponin and major adverse cardiovascular events [47], miR-210 in the formation of coronary collaterals via VEGF-A/EphrinA3 pathways [48], and exosomal miR-16-2-3p in diabetic coronary microvascular dysfunction through regulation of fatty acid metabolism [49]. In the diabetic context, miR-375 also emerged as a predictor of CAD with fair diagnostic performance (AUC 0.74) [50]. Finally, expanding beyond miRNAs, the interplay between lncRNAs and miRNAs also shows diagnostic promise, as highlighted by the LINC00426/miR-873-5p/SRRM2 axis in atherosclerosis [51]. Taken together, these findings provide a coherent framework in which miRNAs either individually, in panels, or through regulatory networks emerge as powerful, non-invasive biomarkers for CAD and related vascular conditions. They also show strong potential for integration into clinical diagnostics and personalized risk stratification.
3.3. Heart Failure
The quest for superior non-invasive diagnostic and prognostic tools in Human Heart Failure (HF) has positioned circulating miRNAs as indispensable molecular components. Evidence from large-scale meta-analyses and human cohort studies highlights the emergence of multi-miRNA panels for accurate patient stratification across the HF spectrum. For instance, a recently validated diagnostic panel discriminates HFpEF patients from both healthy controls and HFrEF with high specificity [52], while another panel comprising eight miRNAs (miR-17-5p, miR-20a-5p, miR-21, miR-23, miR-27, miR-106b-5p, miR-210, and miR-221) has been linked to overall HF incidence, suggesting its utility for early molecular detection prior to clinical onset [53]. In prognostic stratification, the combination of miR-27a-3p, miR-129-5p, miR-145-5p, and miR-590-3p robustly predicts all-cause mortality in HFrEF, yielding a striking hazard ratio (HR) of 4.26 across thousands of participants [53]. Additionally, miR-122-5p and miR-423-5p have been specifically associated with cardiovascular death. Conversely, in HFpEF, circulating miR-19a-3p independently predicts all-cause mortality. Beyond panels, specific miRNAs act as sentinels of key pathological processes. Cardiac remodeling and fibrosis are consistently reflected by elevated miR-21-5p, miR-23a-3p, miR-142-5p, and miR-126-3p [54], with miR-21 and miR-29a levels in hypertrophic cardiomyopathy directly correlating with myocardial fibrosis burden on imaging [55]. Notably, recurrent dysregulation of miR-126, miR-21, miR-145, miR-92a, and miR-155 delineates a vascular injury and inflammation signature in ischemic heart disease [56], underscoring their overlapping roles across cardiovascular phenotypes. Finally, endocrine involvement in HF has been revealed by a panel of miR-10b-5p, miR-193a-3p, and miR-1-3p, which strongly correlates with multiple hormonal deficiencies [57]. Collectively, these validated miRNAs, whether considered as panels or individual biomarkers, demonstrate their multifaceted and recurring involvement in HF pathophysiology, establishing them as essential tools for implementing precision medicine strategies in cardiology.
4. Metabolic miRNAs in Diagnosis of Diabetes
Circulating miRNAs are increasingly recognized as pivotal molecular regulators and promising biomarkers across the diabetes–cardiovascular axis, offering unprecedented opportunities for early diagnosis, risk prediction, and therapy personalization. In nutritional intervention research, the CORDIOPREV trial demonstrated that baseline plasma miRNA profiles could stratify patients with newly diagnosed Type 2 Diabetes Mellitus (T2DM) according to their likelihood of remission after five years of dietary intervention: low miR-let7b-3p predicted greater benefit from a low-fat diet, whereas elevated miR-141-5p, miR-182, and miR-192 favored remission under a Mediterranean diet, with composite miRNA–clinical scores enhancing predictive accuracy [58]. Beyond dietary responsiveness, diagnostic applications have been exemplified in T2DM-associated coronary artery disease (CAD), where a three-miRNA panel (hsa-miR-4505, hsa-miR-4743-5p, hsa-miR-4750-3p) achieved outstanding diagnostic performance (AUC = 0.959), surpassing individual miRNAs and offering a robust non-invasive tool [59]. Similar vascular implications were reported in INOCA patients, where upregulation of miR-92a-3p and downregulation of miR-363-5p in diabetic patients provided mechanistic insight into endothelial dysfunction and identified potential therapeutic targets [60].
Extending beyond cardiovascular complications, circulating vesicle-derived miR-378a-3p has been validated in multiple mouse models as a surrogate marker of pancreatic β-cell mass, paving the way for earlier detection of β-cell dysfunction [61]. Similarly, clinical studies identified miR-4454 as significantly reduced in T2DM, correlating inversely with HbA1c, LDL-C, and the presence of nephropathy and hypertension, suggesting both diagnostic and prognostic roles [62]. Consolidating such evidence, a systematic review encompassing 71 studies highlighted 79 dysregulated miRNAs, most notably miR-126 and miR-192, across diabetic complications, reinforcing their consistency as molecular hallmarks and revealing convergence on key pathogenic pathways [63]. Genetic variants within non-coding RNA loci further can modulate disease susceptibility, for example MALAT1 rs619586 exerted protective effects against Type 1 Diabetes Mellitus (T1DM), whereas miR-146a rs57095329 variants increased disease risk and metabolic impairment in pediatric patients [64]. On a broader scale, a large international study integrating 50 miRNAs into a machine learning-derived dynamic risk score (DRS) achieved high predictive power for T1DM progression (AUC = 0.84), with added value in stratifying therapeutic responsiveness, particularly through miR-27b-3p, in clinical trials of imatinib [65]. Earlier CORDIOPREV findings similarly identified a predictive four-miRNA signature (high miR-150 and miR-30a-5p, low miR-15a and miR-375) that anticipated T2DM onset years before diagnosis, providing a practical alternative to burdensome glucose tolerance tests [66]. Finally, pediatric investigations in T1DM revealed 28 dysregulated miRNAs. Most notably, there was upregulation of hsa-miR-101-3p, miR-135a-5p, miR-143-3p, miR-223-3p, and miR-410-3p, and downregulation of miR-495-3p. These miRNAs target VEGFA, IGF-1, and AKT1 pathways, thereby linking altered circulating signatures to vascular and metabolic complications [67].
Together, these findings underscore the multifaceted potential of miRNAs. They function not only as static biomarkers but as dynamic molecular signatures capable of guiding nutrition-based interventions, refining cardiovascular and metabolic diagnostics, anticipating complications, and tailoring therapeutic strategies, thus establishing them as a cornerstone in the evolution of precision medicine for diabetes and its cardiovascular sequelae (Table 2).

Table 2.
Schematic summary of miRNA and their diagnostic performance in cardiometabolic disorders.
6. Clinical Translation and Diagnostic Platforms of Circulating miRNAs
The clinical translation of circulating microRNAs (miRNAs) as cardiovascular biomarkers is still at an early stage, mainly due to both technical and validation hurdles. From a diagnostic standpoint, several analytical platforms are available, each with distinct advantages and limitations. Real-time quantitative PCR (qRT-PCR) is still considered the gold standard for miRNA detection, offering high sensitivity and specificity, but its limited multiplexing capacity hampers its application to broad biomarker panels [97]. Microarrays allow the simultaneous profiling of hundreds of miRNAs and are widely used in discovery studies. However, their semi-quantitative performance and poor reproducibility across platforms remain critical drawbacks. Next-Generation Sequencing (NGS) represents the most comprehensive and unbiased tool, enabling the discovery of novel and low-abundance miRNAs, but its cost, analytical complexity, and bioinformatic demands prevent routine use in clinical laboratories [98]. Digital PCR (dPCR) has recently emerged as a promising alternative, providing absolute quantification and improved reproducibility compared with qRT-PCR; however, the lack of standardized protocols and limited diffusion in hospital laboratories still restricts its translational potential [99].
Equally important are pre-analytical variables, such as sample type (plasma vs. serum), hemolysis, RNA extraction, and normalization strategies, that strongly influence results and contribute to inconsistencies across studies [100]. Even in conditions where miRNA panels have shown diagnostic potential, such as acute myocardial infarction or chronic heart failure, their incremental value over established biomarkers and clinical scores remains to be convincingly demonstrated [101]. Machine learning approaches, by integrating complex miRNA datasets, offer the possibility of developing more accurate diagnostic and prognostic models, but their robustness depends on large, externally validated cohorts, which are still scarce [102]. Looking forward, incremental progress is expected from two directions: technical innovation aimed at simplifying and standardizing detection platforms, and clinical studies designed as multicenter, prospective validations. In the near future, the integration of dPCR and targeted NGS into routine workflows, combined with bioinformatic tools optimized for reproducibility, could accelerate the path from bench to bedside. While circulating miRNAs represent one of the most promising classes of liquid biopsy biomarkers, their full clinical adoption will ultimately depend on demonstrating clear superiority, or complementary value, to current diagnostic standards.
7. Conclusions
Circulating microRNAs represent a new frontier in the diagnosis and management of cardiometabolic diseases. Thanks to their high tissue specificity, rapid response to pathological stress and remarkable stability in body fluids, they have the potential to complement or even surpass conventional biomarkers. There is an increasing body of evidence that highlights their ability to detect early subclinical alterations and to provide valuable information on disease phenotypes, progression and therapeutic response in conditions such as atherosclerosis, acute myocardial infarction, heart failure and diabetes. Their dual nature, acting both as molecular effectors of disease pathways and as measurable diagnostic indicators, places them at the intersection of mechanistic biology and clinical practice, offering an opportunity for precision medicine. By capturing dynamic processes such as endothelial dysfunction, vascular smooth muscle cell remodeling, metabolic imbalance, and inflammatory activation, miRNAs provide a multidimensional picture that extends beyond traditional biomarkers. Particularly relevant is the immuno-inflammatory dimension, where miRNAs orchestrate the crosstalk between innate and adaptive immune responses, amplifying or resolving vascular injury and metabolic stress. This adds an additional layer of diagnostic and prognostic value, as immune-related miRNA signatures may distinguish subclinical from overt disease and even predict therapeutic responsiveness.
Across cardiometabolic and vascular disorders, the evidence supporting specific miRNA signatures varies. Among many candidates, miR-126 and miR-21 show the most consistent and reproducible links with endothelial health, oxidative stress regulation, and cardiovascular outcomes across different groups and analyses. miR-210 also has strong prognostic value in conditions related to hypoxia, such as myocardial ischemia and stroke. In contrast, miR-155 and miR-34a display variable regulation, often depending on the inflammatory stage or tissue-specific context. Conflicting results are especially noticeable for miR-146a, whose regulation differs between acute and chronic vascular injuries. The differences in analytical methods, normalization techniques, and sample types (serum vs. plasma vs. tissue) further complicate comparisons across studies. Overall, endothelial-derived miRNAs (miR-126, miR-21, miR-210) stand out as the most reliable and clinically relevant biomarkers, while inflammatory miRNAs (miR-155, miR-34a, miR-132) tend to be more context-dependent, functioning better as indicators of disease activity rather than long-term prognosis. However, despite these promising findings, several limitations continue to hinder their routine application in clinical settings. Current challenges include significant heterogeneity in pre-analytical variables such as sample collection, storage, and RNA isolation; variability in detection methods across laboratories; and a lack of universally accepted normalization strategies.
Furthermore, many studies are still limited by small sample sizes, single-center designs and population-specific biases, which restrict the generalizability of findings. Another critical issue is the lack of large-scale, prospective validation studies that can demonstrate actual clinical utility in real-world practice as well as statistical significance.
Future research should focus on overcoming these barriers by standardizing analytical pipelines internationally, carrying out rigorous multicenter validation and incorporating cutting-edge technologies such as next-generation sequencing, digital PCR and isomiR profiling. In parallel, integrating multi-miRNA panels with advanced computational approaches, including machine learning and artificial intelligence, could enhance predictive accuracy further and enable the development of robust diagnostic and prognostic algorithms. Furthermore, translational efforts should explore the therapeutic potential of miRNAs by investigating whether modulating their expression could benefit patients with cardiometabolic disorders.
Overall, while the field is still in a transitional phase between the laboratory and clinical practice, circulating microRNAs have the potential to transform the diagnostic landscape of cardiometabolic diseases. By enabling earlier detection, more refined risk stratification, and personalized therapeutic monitoring, these molecular signatures could ultimately help reduce the global burden of cardiovascular and metabolic diseases on patients and healthcare systems.
One of the most provocative questions is whether circulating miRNAs, given their early and dynamic expression changes, could one day be monitored in real time outside the laboratory, perhaps even in home-based settings through miniaturized biosensors or wearable technologies. Such an advance would shift miRNAs from being static diagnostic markers to becoming continuous indicators of gene expression dynamics, enabling pre-symptomatic detection, personalized risk forecasting, and adaptive treatment adjustment. While this scenario remains speculative, it underscores the transformative potential of miRNAs as true “sentinels” of cardiometabolic health and raises the challenge of bridging molecular biology with next-generation digital health solutions.
Author Contributions
Conceptualization, C.I. and A.C.; methodology, C.I., F.P., P.D.P., E.V. and A.C.A.; data curation, C.I.; writing—original draft preparation, C.I., F.P., P.D.P. and A.C.; writing—review and editing, A.D., C.V. and A.C.; writing—review and editing, V.P., S.A. and S.P.; visualization, C.I. and F.P.; supervision, C.V. and A.C. All authors have read and agreed to the published version of the manuscript.
Funding
This project was supported by MASE_Regione Campania, CUP_ B21I22000180001.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Eroglu, T.; Capone, F.; Schiattarella, G.G. The evolving landscape of cardiometabolic diseases. EBioMedicine 2024, 109, 105447. [Google Scholar] [CrossRef]
- Nazir, A.A.-O.; Nazir, A.; Afzaal, U.; Aman, S.; Sadiq, S.U.R.; Akah, O.Z.; Jamal, M.S.W.; Hassan, S.Z. Advancements in Biomarkers for Early Detection and Risk Stratification of Cardiovascular Diseases-A Literature Review. Health Sci. Rep. 2025, 8, e70878. [Google Scholar] [CrossRef]
- Wronska, A. The Role of microRNA in the Development, Diagnosis, and Treatment of Cardiovascular Disease: Recent Developments. J. Pharmacol. Exp. Ther. 2022, 384, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Aleshcheva, G.; Baumeier, C.; Harms, D.; Bock, C.T.; Escher, F.; Schultheiss, H.P. MicroRNAs as novel biomarkers and potential therapeutic options for inflammatory cardiomyopathy. ESC Heart Fail. 2023, 10, 3410–3418. [Google Scholar] [CrossRef]
- Diener, C.A.-O.; Keller, A.A.-O.; Meese, E.A.-O.X. The miRNA-target interactions: An underestimated intricacy. Nucleic Acids Res. 2023, 52, 1544–1557. [Google Scholar] [CrossRef]
- Rayner, K.J.; Moore, K.J. MicroRNA control of high-density lipoprotein metabolism and function. Circ. Res. 2014, 114, 183–192. [Google Scholar] [CrossRef]
- Li, S.; Sun, W.; Zheng, H.; Tian, F. Microrna-145 accelerates the inflammatory reaction through activation of NF-κB signaling in atherosclerosis cells and mice. Biomed. Pharmacother. 2018, 103, 851–857. [Google Scholar] [CrossRef]
- Wang, D.A.-O.; Atanasov, A.A.-O. The microRNAs Regulating Vascular Smooth Muscle Cell Proliferation: A Minireview. Int. J. Mol. Sci. 2019, 20, 324. [Google Scholar] [CrossRef]
- Loggini, A.; Hornik, J.; Hornik, A. The role of microRNAs as super-early biomarkers in acute ischemic stroke: A systematic review. Clin. Neurol. Neurosurg. 2024, 244, 108416. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tian, T.; Cai, E.L.; Yang, C.; Yang, X. miR-214 Alleviates Ischemic Stroke-Induced Neuronal Death by Targeting DAPK1 in Mice. Front. Neurosci. 2021, 15, 649982. [Google Scholar]
- Caballero-Garrido, E.; Pena-Philippides, J.C.; Lordkipanidze, T.A.-O.; Bragin, D.; Yang, Y.; Erhardt, E.B.; Roitbak, T.A.-O. In Vivo Inhibition of miR-155 Promotes Recovery after Experimental Mouse Stroke. J. Neurosci. 2015, 35, 12446–12464. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Stoica, B.A.; Loane, D.J.; Yang, M.; Abulwerdi, G.; Khan, N.; Kumar, A.; Thom, S.R.; Faden, A.I. Microglial-derived microparticles mediate neuroinflammation after traumatic brain injury. J. Neuroinflamm. 2017, 14, 47. [Google Scholar] [CrossRef]
- Hazra, J.; Vijayakumar, A.; Mahapatra, N.R. Emerging role of heat shock proteins in cardiovascular diseases. Adv. Protein Chem. Struct. Biol. 2023, 134, 271–306. [Google Scholar]
- Popat, A.; Jnaneswaran, G.; Yerukala Sathipati, S.; Sharma, P.P. MicroRNAs in cardiac arrhythmias: Mechanisms, biomarkers, and therapeutic frontiers. Heart Rhythm 2025, in press. [Google Scholar] [CrossRef]
- Xu, J.; Shao, T.; Ding, N.; Li, Y.; Li, X. miRNA-miRNA crosstalk: From genomics to phenomics. Brief. Bioinform. 2017, 18, 1002–1011. [Google Scholar] [CrossRef]
- Naderi Yeganeh, P.; Teo, Y.Y.; Karagkouni, D.; Pita-Juárez, Y.; Morgan, S.L.; Slack, F.J.; Vlachos, I.S.; Hide, W.A. PanomiR: A systems biology framework for analysis of multi-pathway targeting by miRNAs. Brief. Bioinform. 2023, 46, bbad418. [Google Scholar] [CrossRef] [PubMed]
- Bofill-De Ros, X.; Vang Ørom, U.A.-O. Recent progress in miRNA biogenesis and decay. RNA Biol. 2023, 21, 36–43. [Google Scholar] [CrossRef]
- Hynes, C.; Kakumani, P.K. Regulatory role of RNA-binding proteins in microRNA biogenesis. Front. Mol. Biosci. 2024, 11, 1374843. [Google Scholar] [CrossRef]
- Salim, U.; Kumar, A.A.-O.; Kulshreshtha, R.; Vivekanandan, P. Biogenesis, characterization, and functions of mirtrons. Wiley Interdiscip. Rev. RNA 2021, 13, e1680. [Google Scholar] [CrossRef]
- Kiyuna, L.A.; Candido, D.S.; Bechara, L.R.G.; Jesus, I.C.G.; Ramalho, L.S.; Krum, B.; Albuquerque, R.P.; Campos, J.C.; Bozi, L.H.M.; Zambelli, V.O.; et al. 4-Hydroxynonenal impairs miRNA maturation in heart failure via Dicer post-translational modification. Eur. Heart J. 2023, 44, 4696–4712. [Google Scholar] [CrossRef] [PubMed]
- Fumarulo, I.A.-O.; De Prisco, A.; Salerno, E.N.M.; Ravenna, S.A.-O.; Vaccarella, M.A.-O.; Garramone, B.A.-O.X.; Burzotta, F.A.-O.; Aspromonte, N. New Frontiers of microRNA in Heart Failure: From Clinical Risk to Therapeutic Applications. J. Clin. Med. 2025, 14, 6361. [Google Scholar] [CrossRef] [PubMed]
- Kuehbacher, A.; Urbich, C.; Fau-Zeiher, A.M.; Zeiher Am Fau-Dimmeler, S.; Dimmeler, S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ. Res. 2007, 101, 59–68. [Google Scholar] [CrossRef]
- Braun, H.; Hauke, M.; Ripperger, A.; Ihling, C.; Fuszard, M.; Eckenstaler, R.A.-O.; Benndorf, R.A.-O. Impact of DICER1 and DROSHA on the Angiogenic Capacity of Human Endothelial Cells. Int. J. Mol. Sci. 2021, 22, 9855. [Google Scholar] [CrossRef]
- Boeddinghaus, J.; Nestelberger, T.; Twerenbold, R.; Koechlin, L.; Meier, M.; Troester, V.; Wussler, D.; Badertscher, P.; Wildi, K.; Puelacher, C.; et al. High-Sensitivity Cardiac Troponin I Assay for Early Diagnosis of Acute Myocardial Infarction. Clin. Chem. 2019, 65, 893–904. [Google Scholar] [CrossRef]
- Melman, Y.F.; Shah, R.; Fau-Das, S.; Das, S. MicroRNAs in heart failure: Is the picture becoming less miRky? Circ. Heart Fail. 2014, 7, 203–214. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Y.; Zhao, R.; Zhu, B.; Liu, J.; Yue, Q.; Wu, R.; Han, S.; Gao, Y.; Chen, J.; et al. Global surveillance of circulating microRNA for diagnostic and prognostic assessment of acute myocardial infarction based on the plasma small RNA sequencing. Biomark. Res. 2024, 12, 143. [Google Scholar] [CrossRef]
- Zhou, B.; Wang, H.; Zhang, K.; Xie, J.; Yuan, C.; Jin, J.; Zhang, J.; Ma, M.; Zhang, Z. Suppression of circ_0091761 ameliorates acute myocardial infarction-induced endothelial injury through regulation of miR-1278. Hereditas 2025, 162, 169. [Google Scholar] [CrossRef]
- Liu, S.; Han, J.; Gao, S. Study on the mechanism and clinical value of miR-210-3p and miR-582-5p in acute myocardial infarction by targeting MXD1. J. Cardiothorac. Surg. 2025, 20, 282. [Google Scholar] [CrossRef]
- Guo, X.; Chen, Y.; Lu, Y.; Li, P.; Yu, H.; Diao, F.R.; Tang, W.D.; Hou, P.; Zhao, X.X.; Shi, C.A.-O. High level of circulating microRNA-142 is associated with acute myocardial infarction and reduced survival. Ir. J. Med. Sci. 2020, 189, 933–937. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Zhang, J.; Wu, X.; Zhang, L.; Liu, Q.; Xiong, R.; Wang, Y.; Li, M.; Wei, R.; Xu, X.; et al. Differential diagnosis of acute myocardial infarction based on plasma Exosomal MicroRNA. Int. J. Leg. Med. 2025; ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhao, J.; Zhou, Z.; Xu, W.; Wang, S.; Sun, Q. Circulating microRNA-1 as a diagnostic biomarker for acute myocardial infarction: A meta-analysis. Cardiol. J. 2025. ahead of print. [Google Scholar] [CrossRef]
- Huang, C.; Chng, W.H.; Neupane, Y.R.; Lai, Y.; Cui, W.; Yang, M.; Wolfram, J.; Chong, S.Y.; Yu, X.; Zhang, S.; et al. Adipose stem cell-derived nanovesicles for cardioprotection: Production and identification of therapeutic components. J. Control. Release 2025, 385, 113989. [Google Scholar] [CrossRef]
- Fu, X.; Zhang, X.; Wang, L.; Zhang, J.; Li, W.; Qin, S.; Zhang, M.; Zheng, X.; Li, Y.; Yang, S.; et al. The potential value of microRNA-409-5p-mediated negative regulation of USP7 in the diagnosis and treatment of acute myocardial infarction. BMC Cardiovasc. Disord. 2025, 25, 167. [Google Scholar] [CrossRef]
- Zhang, P.; Liang, T.; Chen, Y.; Wang, X.; Wu, T.; Xie, Z.; Luo, J.; Yu, Y.A.-O.X.; Yu, H.A.-O. Circulating Exosomal miRNAs as Novel Biomarkers for Stable Coronary Artery Disease. BioMed Res. Int. 2020, 2020, 3593962. [Google Scholar] [CrossRef]
- Liu, C.J.; Chen, J.J.; Wu, J.H.; Chung, Y.T.; Chen, J.W.; Liu, M.T.; Chiu, C.H.; Chang, Y.C.; Chang, S.N.; Lin, J.W.; et al. Association of exosomes in patients with compromised myocardial perfusion on functional imaging. J. Formos. Med. Assoc. 2024, 123, 968–974. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Chen, W.; Wang, P.; Fan, J.; Si, D.; Ma, Q.; Shi, J.; He, Y. Gold metallene-based ECL biosensor to detect miRNA-126 for coronary artery calcification diagnosis. Biosens. Bioelectron. 2024, 271, 116993. [Google Scholar] [CrossRef]
- Mo, P.; Tian, C.W.; Li, Q.; Teng, M.; Fang, L.; Xiong, Y.; Liu, B. Decreased plasma miR-140-3p is associated with coronary artery disease. Heliyon 2024, 10, e26960. [Google Scholar] [CrossRef] [PubMed]
- Kanašniece, E.; Daukšaite, V.; Vilne, B.; Gailīte, L.; Caunīte, L.; Ērglis, A.; Trušinskis, K. Circulating plasma microRNA expression profile: Potential minimally invasive biomarker for early detection of coronary artery disease development. Mol. Biol. Rep. 2025, 52, 927. [Google Scholar] [CrossRef]
- Choudhury, R.R.; Gupta, H.; Bhushan, S.; Singh, A.; Roy, A.; Saini, N. Role of miR-128-3p and miR-195-5p as biomarkers of coronary artery disease in Indians: A pilot study. Sci. Rep. 2024, 14, 11881. [Google Scholar] [CrossRef] [PubMed]
- Azar Bahadori, R.; Shabani, D.; Arjmandrad, E.; Kazerani, M.; Rohani, M.; Ramazani Karim, Z.; Ali-Kheyl, M.; Nosratabadi, R.; Pourghadamyari, H.; Zaemi, M.A. Circulating miRNA-106b-5p As a Potential Biomarker for Coronary Artery Disease. Int. J. Mol. Cell. Med. 2024, 13, 325. [Google Scholar]
- Makarenkov, N.A.-O.X.; Yoel, U.A.-O.; Haim, Y.; Pincu, Y.A.-O.; Bhandarkar, N.A.-O.; Shalev, A.; Shelef, I.A.-O.; Liberty, I.F.; Ben-Arie, G.; Yardeni, D.; et al. Circulating isomiRs May Be Superior Biomarkers Compared to Their Corresponding miRNAs: A Pilot Biomarker Study of Using isomiR-Ome to Detect Coronary Calcium-Based Cardiovascular Risk in Patients with NAFLD. Int. J. Mol. Sci. 2024, 25, 890. [Google Scholar] [CrossRef]
- Iusupova, A.A.-O.; Pakhtusov, N.A.-O.; Slepova, O.A.-O.; Khabarova, N.V.; Privalova, E.V.; Bure, I.V.; Nemtsova, M.V.; Belenkov, Y.A.-O. MiRNA-34a, miRNA-145, and miRNA-222 Expression, Matrix Metalloproteinases, TNF-α and VEGF in Patients with Different Phenotypes of Coronary Artery Disease. Int. J. Mol. Sci. 2024, 25, 12978. [Google Scholar] [CrossRef] [PubMed]
- Peña-Peña, M.A.-O.; Zepeda-García, Ó.A.-O.; Posadas-Sánchez, R.A.-O.; Sánchez-Muñoz, F.A.-O.; Domínguez-Pérez, M.A.-O.; Martínez-Greene, J.A.-O.X.; López-Bautista, F.A.-O.; Hernández-Díazcouder, A.A.-O.; Jiménez-Ortega, R.F.; Valencia-Cruz, A.I.; et al. A Plasma Extracellular Vesicle-Derived microRNA Signature as a Potential Biomarker for Subclinical Coronary Atherosclerosis. Int. J. Mol. Sci. 2025, 26, 8727. [Google Scholar] [CrossRef] [PubMed]
- Mir, R.A.-O.; Elfaki, I.A.-O.X.; Khullar, N.; Waza, A.A.; Jha, C.A.-O.; Mir, M.A.-O.; Nisa, S.; Mohammad, B.; Mir, T.A.; Maqbool, M.; et al. Role of Selected miRNAs as Diagnostic and Prognostic Biomarkers in Cardiovascular Diseases, Including Coronary Artery Disease, Myocardial Infarction and Atherosclerosis. J. Cardiovasc. Dev. Dis. 2021, 8, 22. [Google Scholar] [CrossRef]
- Murase, H.; Minatoguchi, S.; Heishima, K.; Yasuda, S.; Satake, A.; Yoshizumi, R.; Komaki, H.; Baba, S.; Ojio, S.; Tanaka, T.; et al. Plasma microRNA-143 and microRNA-145 levels are elevated in patients with left ventricular dysfunction. Heart Vessels 2024, 39, 867–876. [Google Scholar] [CrossRef]
- Mansour, R.M.; Hemdan, M.; Moustafa, H.A.M.; Mageed, S.S.A.; Rizk, N.I.; Ali, M.A.; Ashour, M.M.; Ashraf, A.; Doghish, Y.A.; Mohammed, O.A.; et al. Global Perspectives on Coronary Artery Disease: The Emerging Role of miRNAs. Curr. Atheroscler. Rep. 2025, 27, 66. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ma, Q.; Lin, F.A.-O. The Clinical Significance of miR-139-5p in Acute Coronary Syndrome and its Potential Effect on the Progression of Acute Coronary Syndrome. J. Cardiovasc. Transl. Res. 2025, 18, 983–992. [Google Scholar] [CrossRef]
- Zhao, N.; Na, K.; Sun, W.; Shi, H.; Zhang, X.; Liu, B.; Han, Y. Plasma microRNA-210 is associated with VEGF-A and EphrinA3 and relates to coronary collateral circulation in patients with coronary heart disease: A cross-sectional study. BMC Cardiovasc. Disord. 2025, 25, 547. [Google Scholar] [CrossRef]
- Liu, Y.; Zhong, C.; Chen, S.; Xue, Y.; Wei, Z.; Dong, L.; Kang, L. Circulating exosomal mir-16-2-3p is associated with coronary microvascular dysfunction in diabetes through regulating the fatty acid degradation of endothelial cells. Cardiovasc. Diabetol. 2024, 23, 60. [Google Scholar] [CrossRef]
- Sangali, P.; Abdullahi, S.; Nosrati, M.; Khosravi-Asrami, O.F.; Mahrooz, A.A.-O.; Bagheri, A. Altered expression of miR-375 and miR-541 in type 2 diabetes patients with and without coronary artery disease (CAD): The potential of miR-375 as a CAD biomarker. J. Diabetes Metab. Disord. 2024, 23, 1101–1106. [Google Scholar] [CrossRef]
- Zhou, B.; Gan, L.; Zhou, P.; Yang, T.; Tang, F.; Jin, P.; Jin, P.; Chen, J. LINC00426 promotes the progression of atherosclerosis by regulating miR-873-5p/SRRM2 axis. Cytokine 2025, 191, 156938. [Google Scholar] [CrossRef] [PubMed]
- Parvan, R.; Rolim, N.; Gevaert, A.B.; Cataliotti, A.; van Craenenbroeck, E.M.; Adams, V.; Wisløff, U.; Silva, G.A.-O. Multi-microRNA diagnostic panel for heart failure with preserved ejection fraction in preclinical and clinical settings. ESC Heart Fail. 2025, 12, 3028–3041. [Google Scholar] [CrossRef] [PubMed]
- Parvan, R.; Becker, V.; Hosseinpour, M.; Moradi, Y.; Louch, W.E.; Cataliotti, A.; Devaux, Y.; Frisk, M.; Silva, G.J.J. Prognostic and predictive microRNA panels for heart failure patients with reduced or preserved ejection fraction: A meta-analysis of Kaplan-Meier-based individual patient data. BMC Med. 2025, 23, 409. [Google Scholar] [CrossRef]
- Evin, L.; Sigutova, R.; Sulc, P.; Kufova, E.; Branny, M.; Vaclavik, J.; Stejskal, D. Serum levels of miR-21, miR-23a, miR-142-5p, and miR-126 in chronic failure with reduced ejection fraction: A case-control study. Front. Cardiovasc. Med. 2025, 12, 1529451. [Google Scholar] [CrossRef]
- Mylavarapu, M.; Kodali, L.S.M.; Vempati, R.; Nagarajan, J.S.; Vyas, A.; Desai, R. Circulating microRNAs in predicting fibrosis in hypertrophic cardiomyopathy: A systematic review. World J. Cardiol. 2025, 17, 106123. [Google Scholar] [CrossRef]
- Alcibahy, Y.; Darwish, R.; Abu-Sharia, G.; Maes, Q.; Elgamassy, O. Circulating microRNAs as biomarkers for ischemic heart disease: A systematic review and gene set enrichment analysis. Front. Med. 2025, 12, 1545023. [Google Scholar] [CrossRef]
- Grimaldi, A.A.-O.; D’Assante, R.A.-O.X.; Fiore, F.A.-O.; Marcella, S.A.-O.; Paolillo, S.; Cacciatore, F.; Mercurio, V.A.-O.; Bossone, E.A.-O.; Cittadini, A.; Tocchetti, C.A.-O.X.; et al. Circulating miR-10b-5p, miR-193a-3p, and miR-1-3p Are Deregulated in Patients with Heart Failure and Correlate with Hormonal Deficiencies. Int. J. Mol. Sci. 2025, 26, 5225. [Google Scholar] [CrossRef]
- Alcala-Diaz, J.F.; Camargo, A.; Vals-Delgado, C.; Leon-Acuña, A.; Garcia-Fernandez, H.; Arenas-de Larriva, A.P.; Perez-Cardelo, M.; Mora-Ortiz, M.A.-O.; Perez-Martinez, P.; Delgado-Lista, J.; et al. MiRNAs as biomarkers of nutritional therapy to achieve T2DM remission in patients with coronary heart disease: From the CORDIOPREV study. Nutr. Diabetes 2025, 15, 7. [Google Scholar] [CrossRef]
- Szydełko, J.; Czop, M.; Petniak, A.; Lenart-Lipińska, M.; Kocki, J.; Zapolski, T.; Matyjaszek-Matuszek, B. Identification of plasma miR-4505, miR-4743-5p and miR-4750-3p as novel diagnostic biomarkers for coronary artery disease in patients with type 2 diabetes mellitus: A case-control study. Cardiovasc. Diabetol. 2024, 23, 278. [Google Scholar] [CrossRef]
- Ferrone, M.; Ciccarelli, M.; Varzideh, F.; Kansakar, U.; Guerra, G.; Cerasuolo, F.A.; Buonaiuto, A.; Fiordelisi, A.; Venga, E.; Esposito, M.; et al. Endothelial microRNAs in INOCA patients with diabetes mellitus. Cardiovasc. Diabetol. 2024, 23, 268. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, A.; Asahara, S.I.; Inoue, H.; Goto, A.; Seike, M.; Kido, N.; Suzuki, H.; Kanno, A.; Kimura-Koyanagi, M.; Uto, K.; et al. miR378a-3p in serum extracellular vesicles is associated with pancreatic beta-cell mass in diabetic states. Res. Commun. 2025, 750, 151367. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, M.; Deng, Q. Circulating miR-4454 as a potential biomarker for the diagnosis of T2DM and the prediction value of comorbidity and complications in T2DM. BMC Endocr. Disord. 2025, 25, 144. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Zhang, Y.; Fang, X.; Zhang, Y.; Miao, R.; Yao, Y.; Guan, H.; Tian, J. Discovering diabetes complications-related microRNAs: Meta-analyses and pathway modeling approach. BMC Med. Genom. 2025, 18, 86. [Google Scholar] [CrossRef]
- Hamdy, S.M.; Ibrahim, H.A.; Abdelaleem, O.A.-O.; Shaker, O.G.; Hussein, S.K.; Massoud, S.; Sayed, O.N. MALAT1 SNP (rs619586) shows a protective effect against type 1 diabetes mellitus, while the miR-146a SNP (rs57095329) is linked to an increased risk of developing the disease. Mol. Biol. Rep. 2025, 52, 340. [Google Scholar] [CrossRef]
- Joglekar, M.A.-O.; Wong, W.K.M.; Kunte, P.S.; Hardikar, H.P.; Kulkarni, R.A.-O.; Ahmed, I.A.-O.; Farr, R.A.-O.; Pham, N.H.T.; Coles, M.; Kaur, S.A.-O.; et al. A microRNA-based dynamic risk score for type 1 diabetes. Nat. Med. 2025, 31, 2622–2631. [Google Scholar] [CrossRef]
- Jiménez-Lucena, R.; Camargo, A.; Alcalá-Diaz, J.F.; Romero-Baldonado, C.; Luque, R.M.; van Ommen, B.; Delgado-Lista, J.; Ordovás, J.M.; Pérez-Martínez, P.; Rangel-Zúñiga, O.A.; et al. A plasma circulating miRNAs profile predicts type 2 diabetes mellitus and prediabetes: From the CORDIOPREV study. Exp. Mol. Med. 2018, 50, 1–12. [Google Scholar] [CrossRef]
- Swolin-Eide, D.A.-O.; Forsander, G.A.-O.; Pundziute Lyckå, A.A.-O.; Novak, D.A.-O.; Grillari, J.A.-O.; Diendorfer, A.A.-O.; Hackl, M.A.-O.; Magnusson, P.A.-O. Circulating microRNAs in young individuals with long-duration type 1 diabetes in comparison with healthy controls. Sci. Rep. 2023, 13, 11634. [Google Scholar] [CrossRef]
- Ait-Aissa, K.A.-O.; Nguyen, Q.M.; Gabani, M.; Kassan, A.; Kumar, S.; Choi, S.K.; Gonzalez, A.A.; Khataei, T.; Sahyoun, A.M.; Chen, C.; et al. MicroRNAs and obesity-induced endothelial dysfunction: Key paradigms in molecular therapy. Cardiovasc. Diabetol. 2020, 19, 136. [Google Scholar] [CrossRef]
- He, F.; Guan, W. The role of miR-21 as a biomarker and therapeutic target in cardiovascular disease. Clin. Chim. Acta 2025, 574, 120304. [Google Scholar] [CrossRef] [PubMed]
- Zapata-Martínez, L.; Águila, S.; de Los Reyes-García, A.M.; Carrillo-Tornel, S.; Lozano, M.L.; González-Conejero, R.; Martínez, C. Inflammatory microRNAs in cardiovascular pathology: Another brick in the wall. Front. Immunol. 2023, 14, 1196104. [Google Scholar] [CrossRef]
- Ardinal, A.A.-O.; Wiyono, A.A.-O.; Estiko, R.A.-O. Unveiling the therapeutic potential of miR-146a: Targeting innate inflammation in atherosclerosis. J. Cell. Mol. Med. 2024, 28, e70121. [Google Scholar] [CrossRef] [PubMed]
- Meng, W.T.; Zhu, J.; Wang, Y.C.; Shao, C.L.; Li, X.Y.; Lu, P.P.; Huang, M.Y.; Mou, F.F.; Guo, H.D.; Ji, G. Targeting delivery of miR-146a via IMTP modified milk exosomes exerted cardioprotective effects by inhibiting NF-κB signaling pathway after myocardial ischemia-reperfusion injury. J. Nanobiotechnol. 2024, 22, 382. [Google Scholar] [CrossRef]
- Wang, Y.A.-O.; Yu, J.; Ou, C.; Zhao, Y.; Chen, L.; Cai, W.; Wang, H.; Huang, S.; Hu, J.; Sun, G.; et al. miRNA-146a-5p Inhibits Hypoxia-Induced Myocardial Fibrosis Through EndMT. Cardiovasc. Toxicol. 2024, 24, 133–145. [Google Scholar] [CrossRef]
- Villagrán-Silva, F.A.-O.; Loren, P.A.-O.; Sandoval, C.A.-O.; Lanas, F.; Salazar, L.A.-O. Circulating microRNAs as Potential Biomarkers of Overweight and Obesity in Adults: A Narrative Review. Genes 2025, 16, 349. [Google Scholar] [CrossRef]
- Al-Rawaf, H.A.; Gabr, S.A.; Alghadir, T.; Alghadir, F.; Iqbal, A.; Alghadir, A.H. Correlation between circulating microRNAs and vascular biomarkers in type 2 diabetes based upon physical activity: A biochemical analytic study. BMC Endocr. Disord. 2025, 25, 55. [Google Scholar] [CrossRef]
- Luo, M.; Zhao, F.; Cheng, H.; Su, M.; Wang, Y. Macrophage polarization: An important role in inflammatory diseases. Front. Immunol. 2024, 15, 1352946. [Google Scholar] [CrossRef]
- Gager, G.M.; Eyileten, C.; Postuła, M.A.-O.; Nowak, A.A.-O.; Gąsecka, A.A.-O.; Jilma, B.A.-O.; Siller-Matula, J.A.-O. Expression Patterns of MiR-125a and MiR-223 and Their Association with Diabetes Mellitus and Survival in Patients with Non-ST-Segment Elevation Acute Coronary Syndrome. Biomedicines 2023, 11, 1118. [Google Scholar] [CrossRef] [PubMed]
- Blevins, H.M.; Xu, Y.; Biby, S.; Zhang, S. The NLRP3 Inflammasome Pathway: A Review of Mechanisms and Inhibitors for the Treatment of Inflammatory Diseases. Front. Aging Neurosci. 2022, 14, 879021. [Google Scholar] [CrossRef]
- Rayat Pisheh, H.A.-O.; Sani, M.A.-O. Mesenchymal stem cells derived exosomes: A new era in cardiac regeneration. Stem Cell Res. Ther. 2025, 16, 16. [Google Scholar] [CrossRef] [PubMed]
- Letonja, J.A.-O.; Petrovič, D.A.-O. A Review of MicroRNAs and lncRNAs in Atherosclerosis as Well as Some Major Inflammatory Conditions Affecting Atherosclerosis. Biomedicines 2024, 12, 1322. [Google Scholar] [CrossRef]
- Zhu, H.; Leung, S.W. MicroRNA biomarkers of type 2 diabetes: Evidence synthesis from meta-analyses and pathway modelling. Diabetologia 2022, 66, 288–299. [Google Scholar] [CrossRef]
- Hu, F.; Liu, L.; Liu, Z.; Cao, M.; Li, G.; Zhang, X. Meta-analysis of the characteristic expression of circulating microRNA in type 2 diabetes mellitus with acute ischemic cerebrovascular disease. Front. Endocrinol. 2023, 14, 1129860. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.; Peng, B.; Zhang, G.; Fu, X.; Tian, J.; Tian, Y. MicroRNAs in diabetic macroangiopathy. Cardiovasc. Diabetol. 2024, 23, 344. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhu, M.; Schober, A. Macrophage MicroRNAs as Therapeutic Targets for Atherosclerosis, Metabolic Syndrome, and Cancer. Int. J. Mol. Sci. 2018, 19, 1756. [Google Scholar] [CrossRef]
- Zhang, R.L.; Wang, W.M.; Li, J.Q.; Li, R.W.; Zhang, J.; Wu, Y.; Liu, Y. The role of miR-155 in cardiovascular diseases: Potential diagnostic and therapeutic targets. Int. J. Cardiol. Cardiovasc. Risk Prev. 2024, 24, 200355. [Google Scholar] [CrossRef]
- Fitzsimons, S.; Oggero, S.; Bruen, R.; McCarthy, C.; Strowitzki, M.J.; Mahon, N.G.; Ryan, N.; Brennan, E.P.; Barry, M.; Perretti, M.; et al. microRNA-155 Is Decreased During Atherosclerosis Regression and Is Increased in Urinary Extracellular Vesicles During Atherosclerosis Progression. Front. Immunol. 2020, 11, 576516. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Hu, J.; Zhang, A.; Li, F.; Li, X. miR-155 induces endothelial cell apoptosis and inflammatory response in atherosclerosis by regulating Bmal1. Exp. Ther. Med. 2020, 20, 128. [Google Scholar] [CrossRef]
- Cerda, A.A.-O.; Amaral, A.A.; de Oliveira, R.; Moraes, T.I.; Braga, A.A.; Graciano-Saldarriaga, M.E.; Fajardo, C.M.; Hirata, T.A.-O.; Bonezi, V.; Campos-Salazar, A.B.; et al. Peripheral Blood miRome Identified miR-155 as Potential Biomarker of MetS and Cardiometabolic Risk in Obese Patients. Int. J. Mol. Sci. 2021, 22, 1468. [Google Scholar] [CrossRef]
- Khalaji, A.A.-O.; Mehrtabar, S.; Jabraeilipour, A.; Doustar, N.; Rahmani Youshanlouei, H.; Tahavvori, A.; Fattahi, P.; Alavi, S.M.A.; Taha, S.R.; Fazlollahpour-Naghibi, A.; et al. Inhibitory effect of microRNA-21 on pathways and mechanisms involved in cardiac fibrosis development. Ther. Adv. Cardiovasc. Dis. 2024, 18, 17539447241253134. [Google Scholar] [CrossRef]
- Li, X.; Meng, C.; Han, F.; Yang, J.; Wang, J.; Zhu, Y.; Cui, X.; Zuo, M.; Xu, J.; Chang, B. Vildagliptin Attenuates Myocardial Dysfunction and Restores Autophagy via miR-21/SPRY1/ERK in Diabetic Mice Heart. Front. Pharmacol. 2021, 12, 634365. [Google Scholar] [CrossRef]
- Xu, X.; Hong, P.; Wang, Z.; Tang, Z.; Li, K. MicroRNAs in Transforming Growth Factor-Beta Signaling Pathway Associated with Fibrosis Involving Different Systems of the Human Body. Front. Mol. Biosci. 2021, 8, 707461. [Google Scholar] [CrossRef]
- Li, D.; Mao, C.; Zhou, E.; You, J.; Gao, E.; Han, Z.; Fan, Y.; He, Q.; Wang, C. MicroRNA-21 Mediates a Positive Feedback on Angiotensin II-Induced Myofibroblast Transformation. J. Inflamm. Res. 2020, 13, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- Wenlan, L.; Zhongyuan, X.; Shaoqing, L.; Liying, Z.; Bo, Z.; Min, L. MiR-34a-5p mediates sevoflurane preconditioning induced inhibition of hypoxia/reoxygenation injury through STX1A in cardiomyocytes. Biomed. Pharmacother. 2018, 102, 153–159. [Google Scholar] [CrossRef]
- Lei, Z.; Fang, J.; Deddens, J.C.; Metz, C.H.G.; van Eeuwijk, E.C.M.; El Azzouzi, H.; Doevendans, P.A.; Sluijter, J.P.G. Loss of miR-132/212 Has No Long-Term Beneficial Effect on Cardiac Function After Permanent Coronary Occlusion in Mice. Front. Physiol. 2020, 11, 590. [Google Scholar] [CrossRef]
- Baker, A.H.; Giacca, M. Antagonism of miRNA in heart failure: First evidence in human. Eur. Heart J. 2021, 42, 189–191. [Google Scholar] [CrossRef]
- Surina; Fontanella, R.A.; Scisciola, L.; Marfella, R.; Paolisso, G.; Barbieri, M. Corrigendum: miR-21 in Human Cardiomyopathies. Front. Cardiovasc. Med. 2021, 8, 767064, Correction in Front. Cardiovasc. Med. 2022, 9, 913429. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Xu, M.; Tian, X.; Cai, S.; Zeng, S. Research advances in the detection of miRNA. J. Pharm. Anal. 2019, 9, 217–226. [Google Scholar] [CrossRef]
- Siddika, T.; Heinemann, I.U. Bringing MicroRNAs to Light: Methods for MicroRNA Quantification and Visualization in Live Cells. Front. Bioeng. Biotechnol. 2021, 8, 619583. [Google Scholar] [CrossRef]
- Bernardi, S.; Cavalleri, A.; Mutti, S.; Garuffo, L.; Farina, M.; Leoni, A.; Iurlo, A.; Bucelli, C.; Toffoletti, E.; Di Giusto, S.; et al. Digital PCR (dPCR) is able to anticipate the achievement of stable deep molecular response in adult chronic myeloid leukemia patients: Results of the DEMONSTRATE study. Ann. Hematol. 2024, 104, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.F.; Cheng, H.; Goh, K.K.; Zou, R. Preanalytic Methodological Considerations and Sample Quality Control of Circulating miRNAs. J. Mol. Diagn. 2023, 25, 438–453. [Google Scholar] [CrossRef]
- Qian, L.; Zhao, Q.; Yu, P.; Lü, J.; Guo, Y.; Gong, X.; Ding, Y.; Yu, S.; Fan, L.; Fan, H.; et al. Diagnostic potential of a circulating miRNA model associated with therapeutic effect in heart failure. J. Transl. Med. 2022, 20, 267. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Pathak, A.K.; Kural, S.; Kumar, L.; Bhardwaj, M.G.; Yadav, M.; Trivedi, S.; Das, P.; Gupta, M.; Jain, G. Integrating miRNA profiling and machine learning for improved prostate cancer diagnosis. Sci. Rep. 2025, 15, 30477. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).