Assessing the Functional Significance of Novel and Rare Variants of the SLC26A4 Gene Found in Patients with Hearing Loss by Minigene Assay
Abstract
1. Introduction
2. Results
2.1. In Silico Analysis of the Potential Effects of SLC26A4 Variants on Splicing
2.2. In Vitro Analysis of the SLC26A4 Variants
3. Discussion
4. Materials and Methods
4.1. In Silico Analysis of the SLC26A4 Variants
4.2. Generation of Minigene Constructs
4.3. Cell Lines
4.4. Minigene Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- López-Bigas, N.; Audit, B.; Ouzounis, C.; Parra, G.; Guigó, R. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 2005, 579, 1900–1903. [Google Scholar] [CrossRef] [PubMed]
- Rhine, C.L.; Neil, C.; Glidden, D.T.; Cygan, K.J.; Fredericks, A.M.; Wang, J.; Walton, N.A.; Fairbrother, W.G. Future directions for high-throughput splicing assays in precision medicine. Hum. Mutat. 2019, 40, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Wai, H.; Douglas, A.G.L.; Baralle, D. RNA splicing analysis in genomic medicine. Int. J. Biochem. Cell. Biol. 2019, 108, 61–71. [Google Scholar] [CrossRef]
- Truty, R.; Ouyang, K.; Rojahn, S.; Garcia, S.; Colavin, A.; Hamlington, B.; Freivogel, M.; Nussbaum, R.L.; Nykamp, K.; Aradhya, S. Spectrum of splicing variants in disease genes and the ability of RNA analysis to reduce uncertainty in clinical interpretation. Am. J. Hum. Genet. 2021, 108, 696–708. [Google Scholar] [CrossRef]
- Cartegni, L.; Chew, S.L.; Krainer, A.R. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002, 3, 285–298. [Google Scholar] [CrossRef]
- Baralle, D.; Baralle, M. Splicing in action: Assessing disease causing sequence changes. J. Med. Genet. 2005, 42, 737–748. [Google Scholar] [CrossRef]
- Krawczak, M.; Thomas, N.S.; Hundrieser, B.; Mort, M.; Wittig, M.; Hampe, J.; Cooper, D.N. Single base-pair substitutions in exon-intron junctions of human genes: Nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 2007, 28, 150–158. [Google Scholar] [CrossRef]
- Wang, G.S.; Cooper, T.A. Splicing in disease: Disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007, 8, 749–761. [Google Scholar] [CrossRef]
- Vaz-Drago, R.; Custódio, N.; Carmo-Fonseca, M. Deep intronic mutations and human disease. Hum. Genet. 2017, 136, 1093–1111. [Google Scholar] [CrossRef]
- Abramowicz, A.; Gos, M. Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J. Appl. Genet. 2018, 59, 253–268, Erratum in J. Appl. Genet. 2019, 60, 231. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Riolo, G.; Cantara, S.; Ricci, C. What’s Wrong in a Jump? Prediction and Validation of Splice Site Variants. Methods Protoc. 2021, 4, 62. [Google Scholar] [CrossRef] [PubMed]
- Everett, L.A.; Glaser, B.; Beck, J.C.; Idol, J.R.; Buchs, A.; Heyman, M.; Adawi, F.; Hazani, E.; Nassir, E.; Baxevanis, A.D.; et al. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS). Nat. Genet. 1997, 17, 411–422. [Google Scholar] [CrossRef]
- Everett, L.A.; Morsli, H.; Wu, D.K.; Green, E.D. Expression pattern of the mouse ortholog of the Pendred’s syndrome gene (Pds) suggests a key role for pendrin in the inner ear. Proc. Natl. Acad. Sci. USA 1999, 96, 9727–9732. [Google Scholar] [CrossRef]
- Mount, D.B.; Romero, M.F. The SLC26 gene family of multifunctional anion exchangers. Pflug. Arch. 2004, 447, 710–721. [Google Scholar] [CrossRef]
- Dossena, S.; Nofziger, C.; Tamma, G.; Bernardinelli, E.; Vanoni, S.; Nowak, C.; Grabmayer, E.; Kössler, S.; Stephan, S.; Patsch, W.; et al. Molecular and functional characterization of human pendrin and its allelic variants. Cell Physiol. Biochem. 2011, 28, 451–466. [Google Scholar] [CrossRef]
- Griffith, A.J.; Wangemann, P. Hearing loss associated with enlargement of the vestibular aqueduct: Mechanistic insights from clinical phenotypes, genotypes, and mouse models. Hear. Res. 2011, 281, 11–17. [Google Scholar] [CrossRef]
- Wilcox, E.R.; Everett, L.A.; Li, X.C.; Lalwani, A.K.; Green, E.D. The PDS gene, Pendred syndrome and non-syndromic deafness DFNB4. Adv. Otorhinolaryngol. 2000, 56, 145–151. [Google Scholar] [CrossRef]
- Pryor, S.P.; Madeo, A.C.; Reynolds, J.C.; Sarlis, N.J.; Arnos, K.S.; Nance, W.E.; Yang, Y.; Zalewski, C.K.; Brewer, C.C.; Butman, J.A.; et al. SLC26A4/PDS genotype-phenotype correlation in hearing loss with enlargement of the vestibular aqueduct (EVA): Evidence that Pendred syndrome and non-syndromic EVA are distinct clinical and genetic entities. J. Med. Genet. 2005, 42, 159–165. [Google Scholar] [CrossRef]
- Azaiez, H.; Yang, T.; Prasad, S.; Sorensen, J.L.; Nishimura, C.J.; Kimberling, W.J.; Smith, R.J. Genotype-phenotype correlations for SLC26A4-related deafness. Hum. Genet. 2007, 122, 451–457. [Google Scholar] [CrossRef]
- Lee, H.J.; Jung, J.; Shin, J.W.; Song, M.H.; Kim, S.H.; Lee, J.H.; Lee, K.A.; Shin, S.; Kim, U.K.; Bok, J.; et al. Correlation between genotype and phenotype in patients with bi-allelic SLC26A4 mutations. Clin. Genet. 2014, 86, 270–275. [Google Scholar] [CrossRef]
- Mey, K.; Muhamad, A.A.; Tranebjaerg, L.; Rendtorff, N.D.; Rasmussen, S.H.; Bille, M.; Cayé-Thomasen, P. Association of SLC26A4 mutations, morphology, and hearing in pendred syndrome and NSEVA. Laryngoscope 2019, 129, 2574–2579. [Google Scholar] [CrossRef]
- Roesch, S.; Rasp, G.; Sarikas, A.; Dossena, S. Genetic Determinants of Non-Syndromic Enlarged Vestibular Aqueduct: A Review. Audiol. Res. 2021, 11, 423–442. [Google Scholar] [CrossRef]
- Suzuki, H.; Oshima, A.; Tsukamoto, K.; Abe, S.; Kumakawa, K.; Nagai, K.; Satoh, H.; Kanda, Y.; Iwasaki, S.; Usami, S. Clinical characteristics and genotype-phenotype correlation of hearing loss patients with SLC26A4 mutations. Acta Otolaryngol. 2007, 127, 1292–1297. [Google Scholar] [CrossRef]
- Miyagawa, M.; Nishio, S.Y.; Usami, S. Deafness Gene Study Consortium. Mutation spectrum and genotype-phenotype correlation of hearing loss patients caused by SLC26A4 mutations in the Japanese: A large cohort study. J. Hum. Genet. 2014, 59, 262–268. [Google Scholar] [CrossRef]
- Rose, J.; Muskett, J.A.; King, K.A.; Zalewski, C.K.; Chattaraj, P.; Butman, J.A.; Kenna, M.A.; Chien, W.W.; Brewer, C.C.; Griffith, A.J. Hearing loss associated with enlarged vestibular aqueduct and zero or one mutant allele of SLC26A4. Laryngoscope 2017, 127, E238–E243. [Google Scholar] [CrossRef]
- Honda, K.; Griffith, A.J. Genetic architecture and phenotypic landscape of SLC26A4-related hearing loss. Hum. Genet. 2022, 141, 455–464. [Google Scholar] [CrossRef]
- Zhao, X.; Huang, L.; Wang, X.; Wang, X.; Zhao, L.; Cheng, X.; Ruan, Y. Genotyping and audiological characteristics of infants with a single-allele SLC26A4 mutation. Int. J. Pediatr. Otorhinolaryngol. 2019, 116, 153–158. [Google Scholar] [CrossRef]
- Kallel-Bouattour, R.; Belguith-Maalej, S.; Zouari-Bradai, E.; Mnif, M.; Abid, M.; Hadj Kacem, H. Intronic variants of SLC26A4 gene enhance splicing efficiency in hybrid minigene assay. Gene 2017, 620, 10–14. [Google Scholar] [CrossRef]
- Lee, B.; Kim, Y.R.; Kim, S.J.; Goh, S.H.; Kim, J.H.; Oh, S.K.; Baek, J.I.; Kim, U.K.; Lee, K.Y. Modified U1 snRNA and antisense oligonucleotides rescue splice mutations in SLC26A4 that cause hereditary hearing loss. Hum. Mutat. 2019, 40, 1172–1180. [Google Scholar] [CrossRef]
- Wasano, K.; Takahashi, S.; Rosenberg, S.K.; Kojima, T.; Mutai, H.; Matsunaga, T.; Ogawa, K.; Homma, K. Systematic quantification of the anion transport function of pendrin (SLC26A4) and its disease-associated variants. Hum. Mutat. 2020, 41, 316–331. [Google Scholar] [CrossRef]
- Zhou, K.; Huang, L.; Feng, M.; Li, X.; Zhao, Y.; Liu, F.; Wei, J.; Qin, D.; Lu, Q.; Shi, M.; et al. A novel SLC26A4 splicing mutation identified in two deaf Chinese twin sisters with enlarged vestibular aqueducts. Mol. Genet. Genomic. Med. 2020, 8, e1447. [Google Scholar] [CrossRef]
- Albader, N.; Zou, M.; BinEssa, H.A.; Abdi, S.; Al-Enezi, A.F.; Meyer, B.F.; Alzahrani, A.S.; Shi, Y. Insights of Noncanonical Splice-site Variants on RNA Splicing in Patients with Congenital Hypothyroidism. J. Clin. Endocrinol. Metab. 2022, 107, e1263–e1276. [Google Scholar] [CrossRef] [PubMed]
- Bałdyga, N.; Oziębło, D.; Gan, N.; Furmanek, M.; Leja, M.L.; Skarżyński, H.; Ołdak, M. The Genetic Background of Hearing Loss in Patients with EVA and Cochlear Malformation. Genes 2023, 14, 335. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Liu, M.; Lu, Y.; Zhao, X.; Yan, Z.; Sun, Y.; Ma, J.; Tang, W.; Wang, H.; Xu, H. Exonic Deletions and Deep Intronic Variants of the SLC26A4 Gene Contribute to the Genetic Diagnosis of Unsolved Patients With Enlarged Vestibular Aqueduct. Hum. Mutat. 2024, 2024, 8444122. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhu, Y.M.; Xu, C.; Ding, W.; Jia, H.; Bian, P.; Xu, B.; Guo, Y.; Liu, X. A novel intronic variant causing aberrant splicing identified in two deaf Chinese siblings with enlarged vestibular aqueducts. Mol. Genet. Genomic. Med. 2024, 12, e2361. [Google Scholar] [CrossRef]
- Zhao, Y.; Long, Y.; Shi, T.; Ma, X.; Lian, C.; Wang, H.; Xu, H.; Yu, L.; Zhao, X. Validating the splicing effect of rare variants in the SLC26A4 gene using minigene assay. BMC Med. Genom. 2024, 17, 233. [Google Scholar] [CrossRef]
- Panina, E.A.; Danilchenko, V.Y.; Zytsar, M.V.; Orishchenko, K.E.; Posukh, O.L. Analysis by Minigene Assay of the Splicing Effect of a Novel Variant c.1545T>G in the SLC26A4 Gene Associated with Hearing Loss. Russ. J. Genet. 2025, 61, 602–607. [Google Scholar] [CrossRef]
- Danilchenko, V.Y.; Zytsar, M.V.; Maslova, E.A.; Bady-Khoo, M.S.; Barashkov, N.A.; Morozov, I.V.; Bondar, A.A.; Posukh, O.L. Different Rates of the SLC26A4-Related Hearing Loss in Two Indigenous Peoples of Southern Siberia (Russia). Diagnostics 2021, 11, 2378. [Google Scholar] [CrossRef]
- Kallel, R.; Niasme-Grare, M.; Belguith-Maalej, S.; Mnif, M.; Abid, M.; Ayadi, H.; Masmoudi, S.; Jonard, L.; Hadj Kacem, H. Screening of SLC26A4 gene in autoimmune thyroid diseases. Int. J. Immunogenet. 2013, 40, 284–291. [Google Scholar] [CrossRef]
- Rajalakshmi, K.; Thirunavukkarasu, J.; Vikraman, M.A.; Maruthy, S.; Sylvester, C.; Kundapur, R. Analysis of SLC26A4 Gene in Individuals with Non Syndromic Hearing Impairment in Relation with GJB2 Associated Mutations. Avicenna J. Med. Biotechnol. 2023, 15, 124–127. [Google Scholar] [CrossRef]
- López-Bigas, N.; Rabionet, R.; de Cid, R.; Govea, N.; Gasparini, P.; Zelante, L.; Arbonés, M.L.; Estivill, X. Splice-site mutation in the PDS gene may result in intrafamilial variability for deafness in Pendred syndrome. Hum. Mutat. 1999, 14, 520–526. [Google Scholar] [CrossRef]
- Massa, G.; Jaenen, N.; de Varebeke, S.J.; Peeters, N.; Wuyts, W. Solitary thyroid nodule as presenting symptom of Pendred syndrome caused by a novel splice-site mutation in intron 8 of the SLC26A4 gene. Eur. J. Pediatr. 2003, 162, 674–677. [Google Scholar] [CrossRef]
- Park, H.J.; Lee, S.J.; Jin, H.S.; Lee, J.O.; Go, S.H.; Jang, H.S.; Moon, S.K.; Lee, S.C.; Chun, Y.M.; Lee, H.K.; et al. Genetic basis of hearing loss associated with enlarged vestibular aqueducts in Koreans. Clin. Genet. 2005, 67, 160–165. [Google Scholar] [CrossRef]
- Yang, J.J.; Tsai, C.C.; Hsu, H.M.; Shiao, J.Y.; Su, C.C.; Li, S.Y. Hearing loss associated with enlarged vestibular aqueduct and Mondini dysplasia is caused by splice-site mutation in the PDS gene. Hear. Res. 2005, 199, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Dossena, S.; Nofziger, C.; Brownstein, Z.; Kanaan, M.; Avraham, K.B.; Paulmichl, M. Functional characterization of pendrin mutations found in the Israeli and Palestinian populations. Cell Physiol. Biochem. 2011, 28, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, H.R.; Kim, J.; Shin, J.W.; Park, H.J.; Choi, J.Y.; Kim, U.K.; Lee, K.A. A novel synonymous mutation causing complete skipping of exon 16 in the SLC26A4 gene in a Korean family with hearing loss. Biochem. Biophys. Res. Commun. 2013, 430, 1147–1150. [Google Scholar] [CrossRef] [PubMed]
- Sauna, Z.E.; Kimchi-Sarfaty, C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011, 12, 683–691. [Google Scholar] [CrossRef]
- Hunt, R.C.; Simhadri, V.L.; Iandoli, M.; Sauna, Z.E.; Kimchi-Sarfaty, C. Exposing synonymous mutations. Trends Genet. 2014, 30, 308–321. [Google Scholar] [CrossRef]
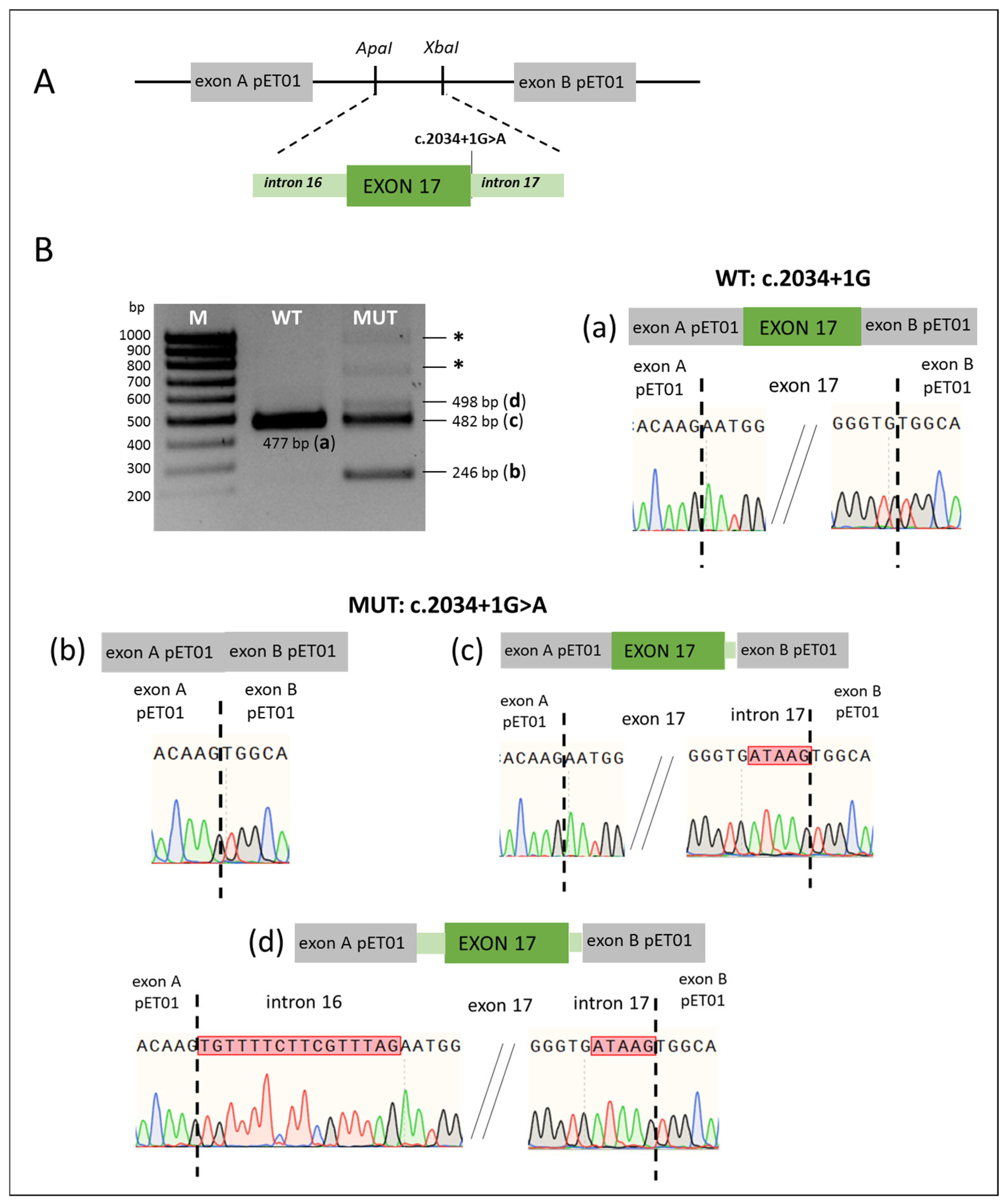
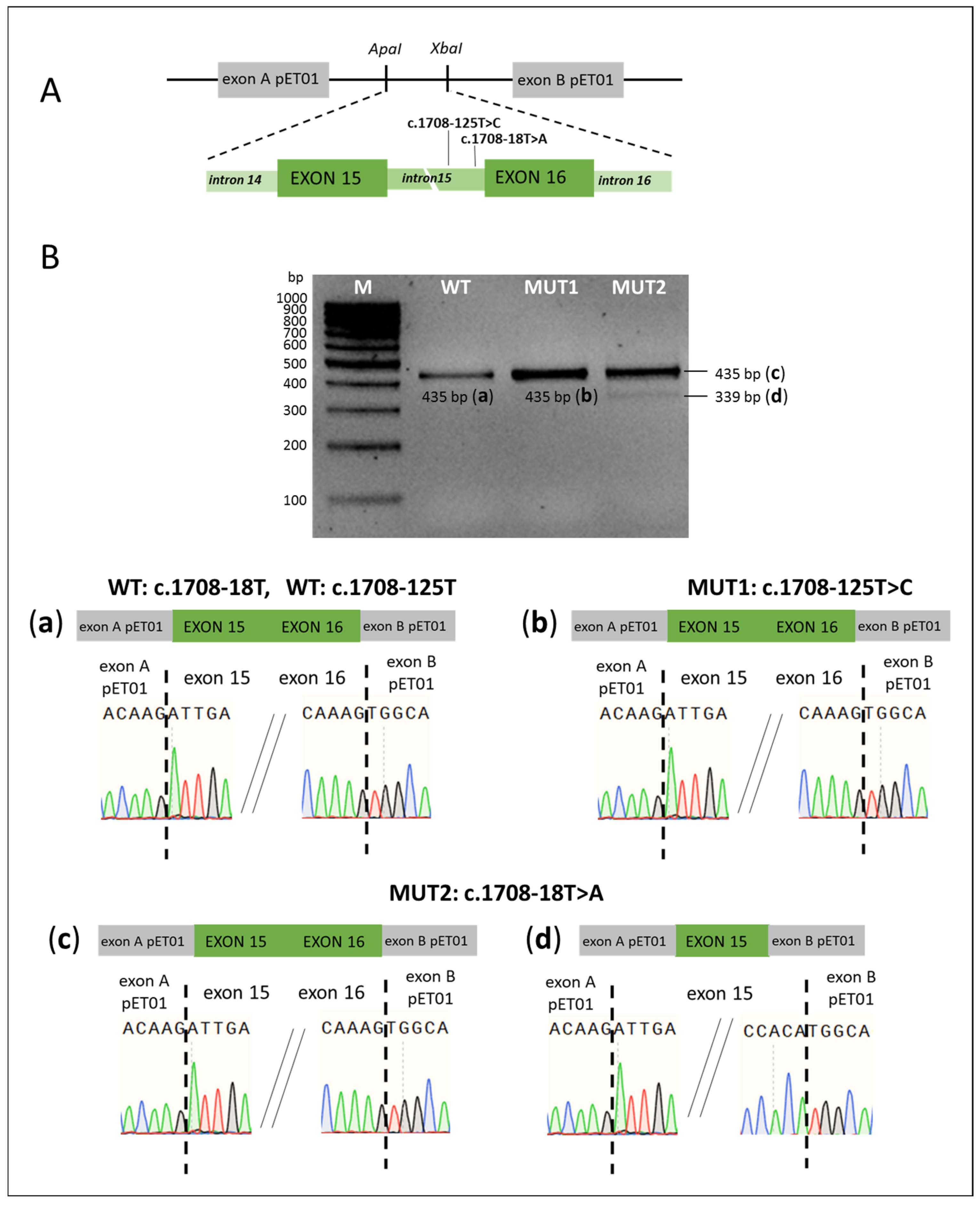
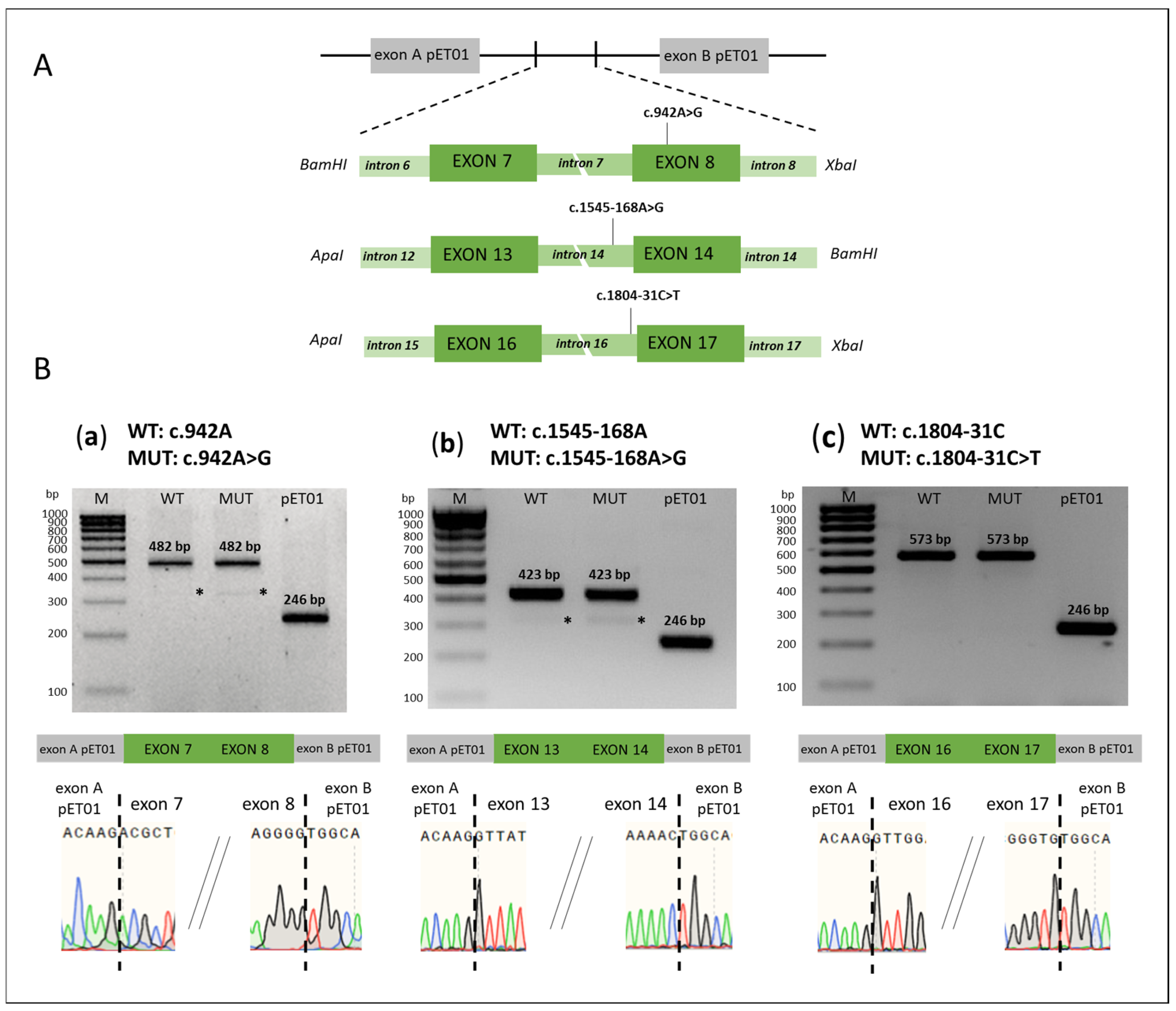
| Intronic Variants | ||||||
| Variant (Location in the SLC26A4 Gene) | Position GRCh38.p14: chr 7, NC_000007.14: | Human Splicing Finder (HSF) | SpliceAI | CADD | FATHMM-MKL Non-Coding Score | FATHM-XF Non-Coding Score |
| c.2034+1G>A rs759683649 (intron 17) | g.107702058G>A | New Donor splice site: Activation of a cryptic Donor site. Potential alteration of splicing (HSF) Broken WT Donor Site: Alteration of the WT Donor site, most probably affecting splicing (HSF) Broken WT Donor Site: Alteration of the WT Donor site, most probably affecting splicing (MaxEnt) | Donor Loss 1.0 Donor Gain 0.81 | 35 | 0.99333 pathogenic | 0.991095 pathogenic |
| c.1545-168A>G * rs1791783004 (intron 13) | g.107697874A>G | Alteration of auxiliary sequences: Significant alteration of ESE/ESS motifs ratio (5) | 0.0 | 18.2 | 0.93758 pathogenic | 0.373789 benign |
| c.1708-125T>C (intron 15) | g.107700976T>C | Alteration of auxiliary sequences: Significant alteration of ESE/ESS motifs ratio (6) | 0.0 | 3.529 | 0.17111 benign | 0.048611 benign |
| c.1708-18T>A rs55701254 (intron 15) | g.10770108 3T>A | New Acceptor splice site: Activation of a cryptic Acceptor site. Potential alteration of splicing (HSF) Broken WT Acceptor Site: Alteration of the WT Acceptor site, most probably affecting splicing (MaxEnt) | Acceptor Loss 0.01 | 21.2 | 0.98823 pathogenic | 0.565967 pathogenic |
| c.1804-31C>T (intron 16) | g.107701796C>T | Broken WT Branch Point: Alteration of the WT Branch Point, may affect splicing | Acceptor Gain 0.04 | 4.452 | 0.19947 benign | 0.073999 benign |
| Exonic Variant | ||||||
| Variant (Location in the SLC26A4 gene) | Position GRCh38.p14: chr 7, NC_000007.14: | Human Splicing Finder (HSF) | SpliceAI | RBPmap | SpliceAid | EX-SKIP |
| c.942A>G * (p.Ser314=) rs2535310634 (exon 8) | g.107683478A>G | New Donor splice site: Activation of a cryptic Donor site. Potential alteration of splicing (HSF) | Donor Loss 0.02 | Loss or disruption of binding sites of RNA-binding proteins | Loss or disruption of binding sites of RNA-binding proteins | Allele c.942A>G has a higher chance of exon skipping than allele c.942A |
| SLC26A4 Variant | Location | In Silico Predictions for Splicing Defect | In Vitro Analysis | Reference |
|---|---|---|---|---|
| Canonical Splice Acceptor and Donor Sites (Positions −1, −2, +1, +2) | ||||
| c.601-1G>A | CSAS (intron 5) | Pathogenic | A partial loss of exon 5 | [31] |
| c.919-2A>G | CSAS (intron 7) | Pathogenic | Splicing errors: (1) exon 8 skipping [45]; (2) exons 7 and 8 skipping (60% transcripts), exon 8 skipping (40% transcripts) [31] | [31,45] |
| c.1002-1G>T | CSAS (intron 8) | Pathogenic | Exon 9 skipping | [33] |
| c.1149+1G>A | CSDS (intron 9) | Pathogenic | Exon 9 skipping | [37] |
| c.1545-2A>G | CSAS (intron 13) | Pathogenic | Splicing errors: (1) exon 14 skipping (~80%); (2) 9 bp (~10%) and 24 bp (~10%) deletions of exon 14, respectively, due to activation of cryptic splice acceptor site | [33] |
| c.1614+1G>C | CSDS (intron 14) | Pathogenic | Exon 14 skipping | [33] |
| c.2034+1G>A | CSDS (intron 17) | Pathogenic | Splicing errors: (1) exon 17 skipping; (2) 5 bp retention of intron 17 due to activation of a cryptic donor site in intron 17; (3) 16 bp retention of intron 16 and 5 bp retention of intron 17 due to activation of a cryptic donor and acceptor sites | This study |
| Intronic variants outside canonical splice sites (within 50 nucleotides from intron–exon boundaries) | ||||
| c.165-5G>A a | NSAS (intron 2) | Pathogenic | No splicing error | [33] |
| c.165-13T>G a | NSAS (intron 2) | Pathogenic | No splicing error | [33] |
| c.415+4A>G a | NSDS (intron 4) | Pathogenic | No splicing error [30]; exon 4 skipping [33] | [30,33] |
| c.415+7A>G | NSDS (intron 4) | Pathogenic | Activation of a cryptic splice donor site in intron 4 leading to 6 bp retention of intron 4 | [33,42] |
| c.765+3A>C | NSDS (intron 6) | Pathogenic | Exon 6 skipping | [33] |
| c.765+3A>T | NSDS (intron 6) | Pathogenic | Exon 6 skipping | [30,33] |
| c.765+4A>C a | NSDS (intron 6) | Pathogenic | No splicing error | [33] |
| c.765+4A>G | NSDS (intron 6) | Pathogenic | Exon 6 skipping | [36] |
| c.1001+4A>G | NSDS (intron 8) | Pathogenic | Activation of a cryptic splice donor site in intron 8 leading to 40 bp retention of intron 8 | [30,33] |
| c.1001+5G>C | NSDS (intron 8) | Pathogenic | Activation of a cryptic splice donor site in intron 8 leading to 40 bp retention of intron 8 | [30,33] |
| c.1001+5G>T | NSDS (intron 8) | Pathogenic | Activation of a cryptic splice donor site in intron 8 leading to 40 bp retention of intron 8 | [30,33] |
| c.1001+30A>G a | NSDS (intron 8) | Ambiguous | No splicing error | [33] |
| c.1002-4C>G | NSAS (intron 8) | Pathogenic | Exon 9 skipping | [33,43] |
| c.1002-8C>G | NSAS (intron 8) | Pathogenic | Exon 9 skipping | [33] |
| c.1002-9A>C a | NSAS (intron 8) | Pathogenic | No splicing error | [29,33] |
| c.1002-9A>G | NSAS (intron 8) | Pathogenic | Exon 9 skipping | [33] |
| c.1149+3A>G | NSDS (intron 9) | Pathogenic | Exon 9 skipping | [30,44] |
| c.1341+3A>C | NSDS (intron 11) | Pathogenic | Exon 11 skipping | [30] |
| c.1544+5G>A | NSDS (intron 13) | Pathogenic | Exon 13 skipping | [30,33] |
| c.1544+9C>G a | NSDS (intron 13) | Ambiguous | Activation of a cryptic splice donor site in intron 13, leading to 9 bp retention of intron 13 | [33] |
| c.1544+9C>T a | NSDS (intron 13) | Ambiguous | No splicing error | [29,33] |
| c.1545-5T>G a | NSAS (intron 13) | Pathogenic | No splicing error | [29,33] |
| c.1614+5G>A | NSDS (intron 14) | Pathogenic | Exon 14 skipping | [32] |
| c.1614+7A>G a | NSDS (intron 14) | Ambiguous | No splicing error | [33] |
| c.1707+5G>A | NSDS (intron 15) | Pathogenic | A complete loss of exon 15 | [30,31] |
| c.1708-18T>A a | NSAS (intron 15) | Pathogenic | (1) no splicing error; (2) exon 16 skipping in a small proportion of transcripts | This study |
| c.1804-31C>T a | NSAS (intron 16) | Ambiguous | No splicing error | This study |
| c.2089+3A>T | NSDS (intron 18) | Pathogenic | Exon 18 skipping | [37] |
| Deep intronic variants | ||||
| c.304+941C>T a | DIV (intron 3) | Ambiguous | Activation of a cryptic donor and acceptor sites leading to 126 bp retention of intron 3 | [35] |
| c.1545-168A>G a | DIV (intron 13) | Ambiguous | No splicing error | This study |
| c.1707+94C>T a | DIV (intron 15) | Benign | Skipping of exons 15 and 16 | [33] |
| c.1708-125T>C a | DIV (intron 15) | Ambiguous | No splicing error | This study |
| Exonic variants | ||||
| c.162C>T (p.Cys54=) | Exon 2 | Pathogenic | Splicing errors: (1) exon 2 skipping; (2) 4 bp deletion of exon 2 due to activation of cryptic splice donor site | [34] |
| c.304G>A (p.Gly102Arg) | Last nucleotide of exon 3 | Pathogenic | Exon 3 skipping | [30] |
| c.392G>T a (p.Gly131Val) | Exon 4 | Pathogenic | (1) exon 4 skipping; (2) no splicing error | [31] |
| c.918G>A a (p.Val306=) | Last nucleotide of exon 7 | Ambiguous | Exon 7 skipping | [33] |
| c.942A>G a (p.Ser314=) | Exon 8 | Pathogenic | No splicing error | This study |
| c.1001G>T (p.Gly334Val) | Last nucleotide of exon 8 | Pathogenic | Activation of a cryptic splice donor site in intron 8 leading to 40 bp retention of intron 8 | [30,31,33,46] |
| c.1261C>A a (p.Gln421Lys) | Exon 10 | Pathogenic | No splicing error | [30] |
| c.1262A>C a (p.Gln421Pro) | Exon 10 | Pathogenic | No splicing error | [30] |
| c.1262A>G a (p.Gln421Arg) | Exon 10 | Pathogenic | No splicing error | [30] |
| c.1262A>T a (p.Gln421Leu) | Exon 10 | Pathogenic | No splicing error | [30] |
| c.1544T>G a (p.Phe515Cys) | Last nucleotide of exon 13 | Pathogenic | No splicing error | [30] |
| c.1545T>G a (p.Phe515Leu) | First nucleotide of exon 14 | Ambiguous | No splicing error | [38] |
| c.1614C>T a (p.Asn538=) | Last nucleotide of exon 14 | Ambiguous | No splicing error | [33] |
| c.1803G>A (p.Lys601=) | Last nucleotide of exon 16 | Pathogenic | Exon 16 skipping | [30,47] |
| Fragments | Primer Sequences | Product Size, bp | Restriction Site | Reference |
|---|---|---|---|---|
| Primers used to amplify of the SLC26A4 gene fragments | ||||
| Exons 7–8 | F: GAATGGGATCCTCACCCAGTTTTTCCTTTCC | 867 | BamHI | This study |
| R: AAGTTTCTAGAAGGACTCTGGTGTTAACCGTA | XbaI | |||
| Exons 13–14 | F: TATCATGGGCCCACTGCACTCC | 2516 | ApaI | [38] |
| R: GCCGGATCCGTCAGAAGGTGCACT | BamHI | |||
| Exons 15–16 | F: TACTGGGCCCTGCGCAACAGAGTGAAA | 1486 | ApaI | This study |
| R:ATTTTCTAGACTTCCACTCCCGCTTGCCTAT | XbaI | |||
| Exons 16–17 | F: TTCTGGGCCCGAGTAGGGTAGCCTGGG | 1322 | ApaI | This study |
| R:CTCCTCTAGATAGTCTGGCTCCAGAGCCT | XbaI | |||
| Exon 17 | F: GTTTGGGCCCTAGACAACATCAAAGTTT | 587 | ApaI | This study |
| R: CTCCTCTAGATAGTCTGGCTCCAGAGCCT | XbaI | |||
| Primers used for site-directed mutagenesis of the SLC26A4 variants | ||||
| c.1804-31C>T | F: TTTGATAATTAAGTTGACAGTGTTTTC | - | - | This study |
| R: GATTTGAAATCTTTCAACCATTCATA | ||||
| c.942A>G | F: CATTTCGTATGGAGCCAACCTGG | - | - | This study |
| R: GCAGTAGCAATTATCGTCTG | ||||
| Primers specific to the plasmid region of the pET01 vector | ||||
| cDNA | cDNA primer 1: GATCCACGATGC | - | - | https://www.mobitec.com/ |
| pET01exons | PCR primer 02: GATGGATCCGCTTCCTGCCCC | 246 | BamHI | https://www.mobitec.com/ |
| PCR primer 03: CTCCCGGGCCACCTCCAGTGCC | SmaI | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danilchenko, V.Y.; Panina, E.A.; Zytzar, M.V.; Orishchenko, K.E.; Posukh, O.L. Assessing the Functional Significance of Novel and Rare Variants of the SLC26A4 Gene Found in Patients with Hearing Loss by Minigene Assay. Int. J. Mol. Sci. 2025, 26, 10732. https://doi.org/10.3390/ijms262110732
Danilchenko VY, Panina EA, Zytzar MV, Orishchenko KE, Posukh OL. Assessing the Functional Significance of Novel and Rare Variants of the SLC26A4 Gene Found in Patients with Hearing Loss by Minigene Assay. International Journal of Molecular Sciences. 2025; 26(21):10732. https://doi.org/10.3390/ijms262110732
Chicago/Turabian StyleDanilchenko, Valeriia Yu., Ekaterina A. Panina, Marina V. Zytzar, Konstantin E. Orishchenko, and Olga L. Posukh. 2025. "Assessing the Functional Significance of Novel and Rare Variants of the SLC26A4 Gene Found in Patients with Hearing Loss by Minigene Assay" International Journal of Molecular Sciences 26, no. 21: 10732. https://doi.org/10.3390/ijms262110732
APA StyleDanilchenko, V. Y., Panina, E. A., Zytzar, M. V., Orishchenko, K. E., & Posukh, O. L. (2025). Assessing the Functional Significance of Novel and Rare Variants of the SLC26A4 Gene Found in Patients with Hearing Loss by Minigene Assay. International Journal of Molecular Sciences, 26(21), 10732. https://doi.org/10.3390/ijms262110732






