ADAM10 Knockout from Human Glioblastoma and Colon Cancer Cells Modulates Diverse Signalling Networks and Inhibits Tumour Growth In Vivo
Abstract
1. Introduction
2. Results
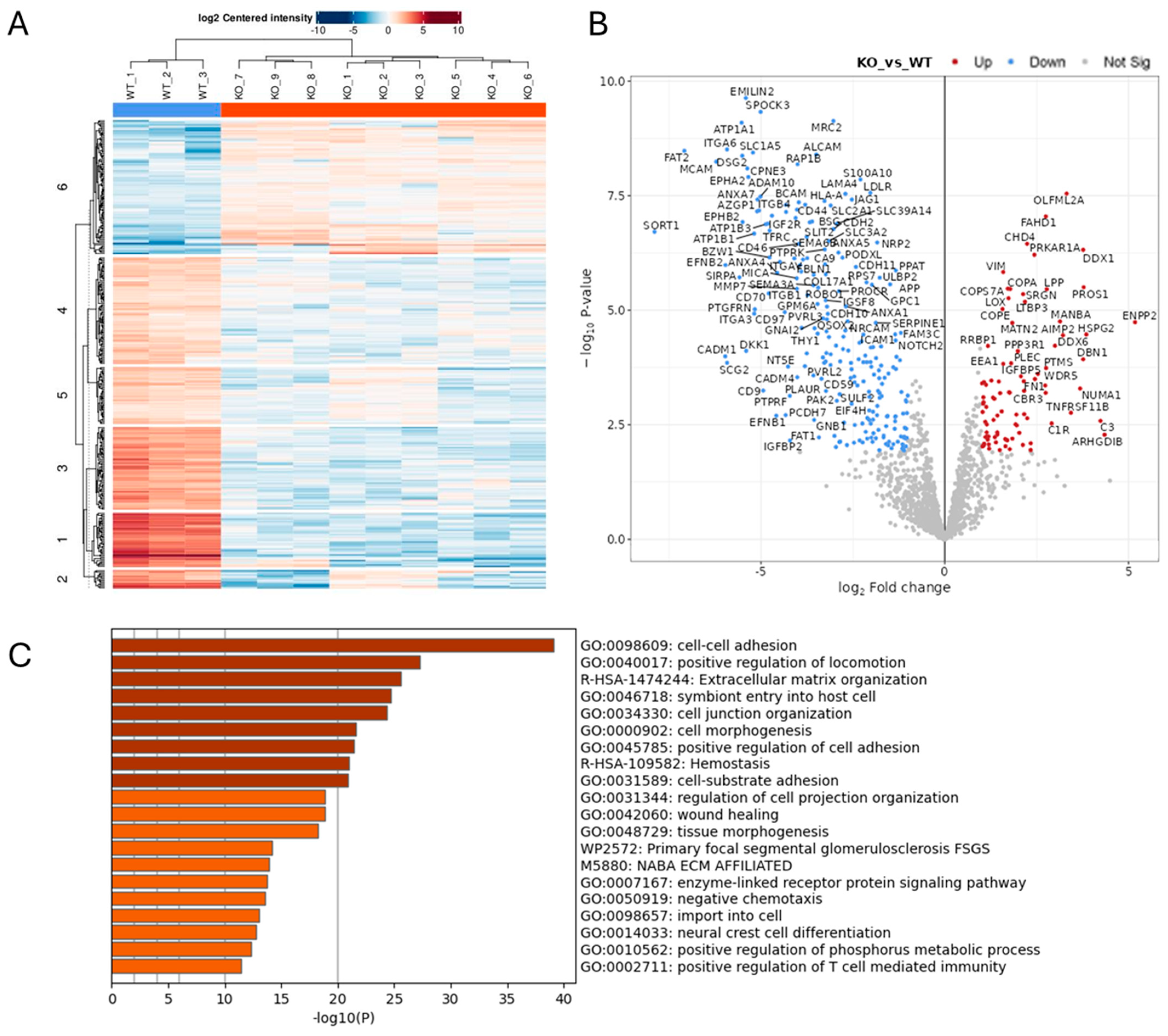
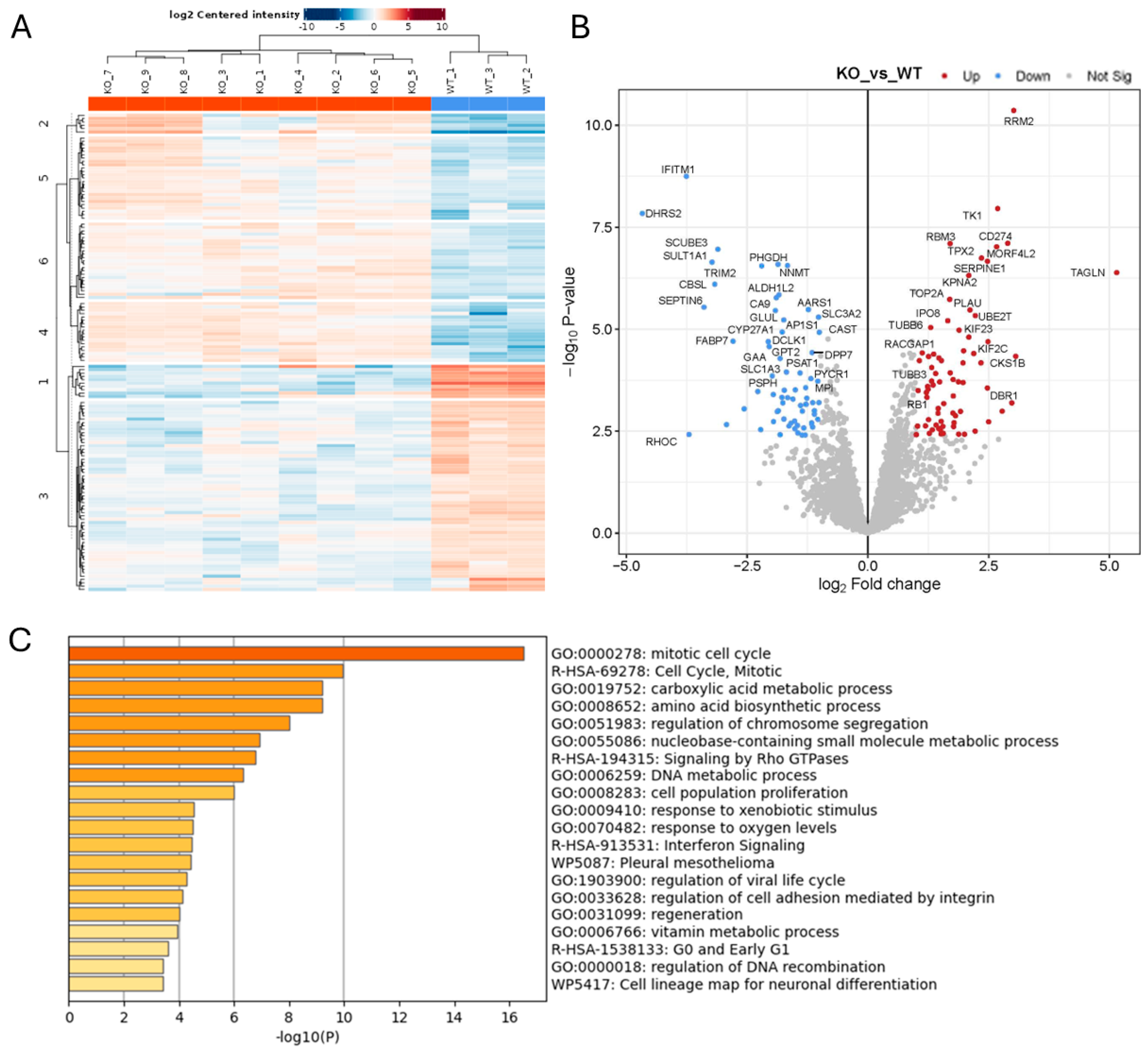
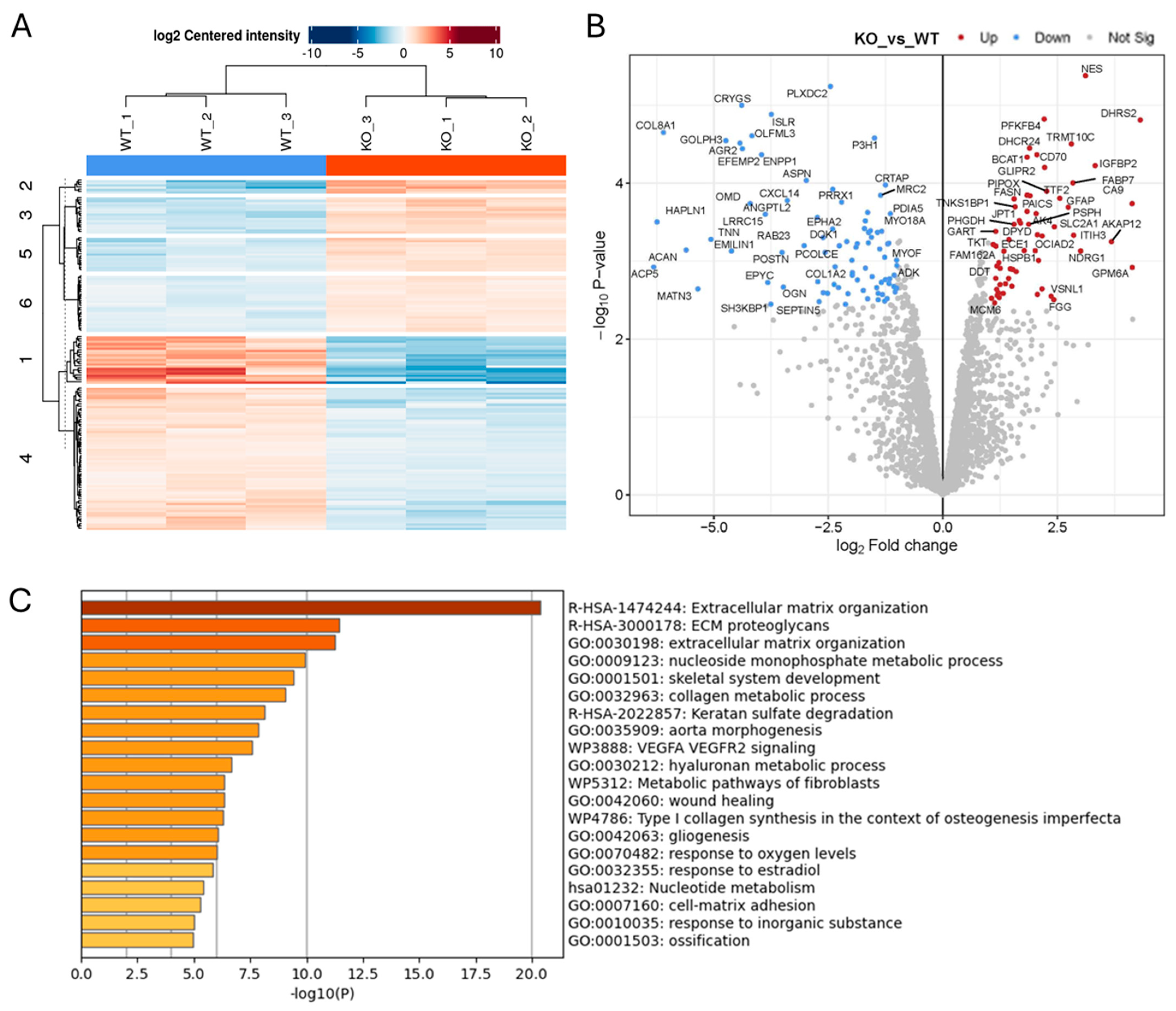
2.1. Effect of ADAM10 KO on the Tumour Cell Secretome
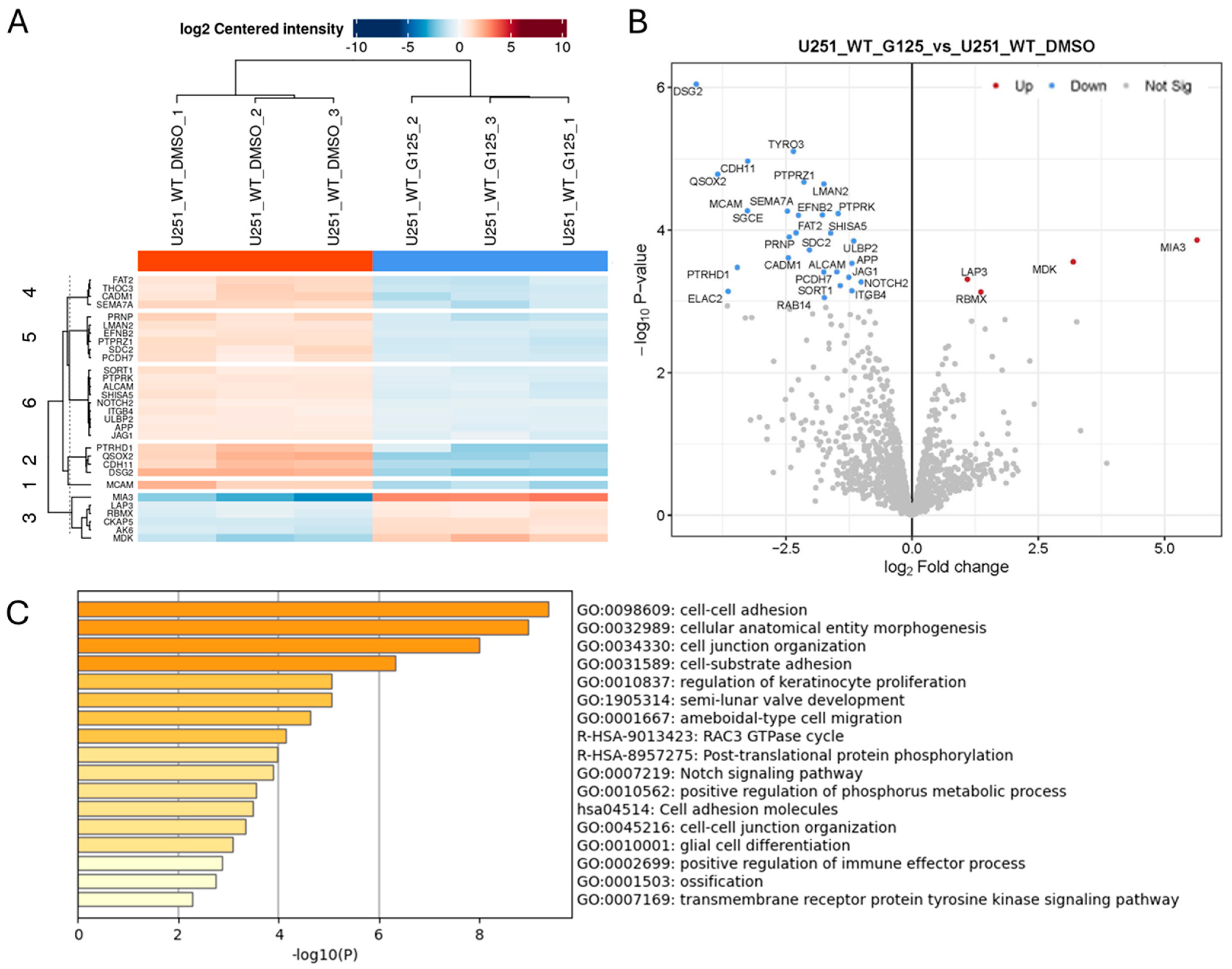
2.2. Effect of ADAM10 Inhibitor Treatment on the Tumour Cell Secretome
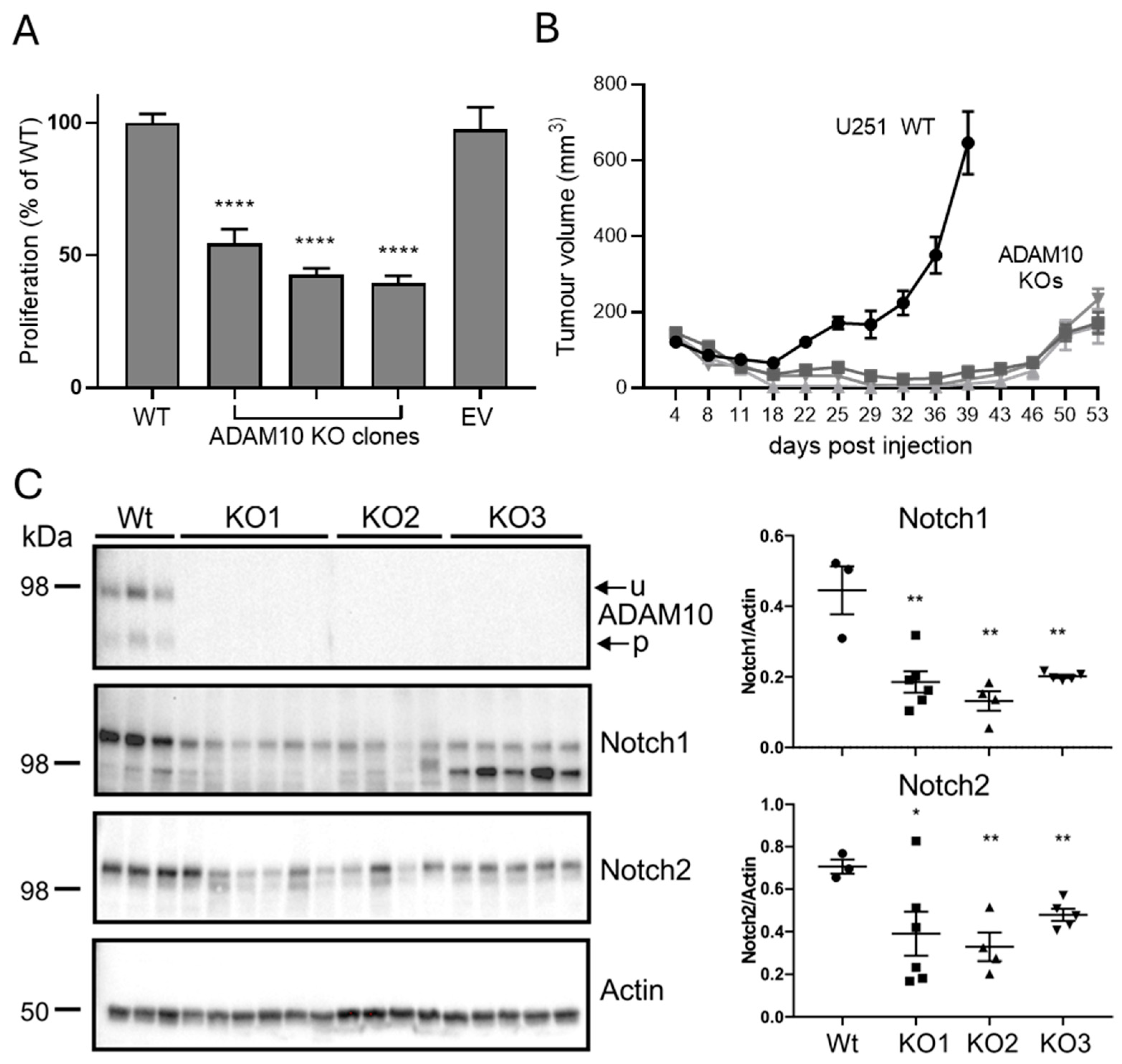
2.3. In Vitro and Functional Analysis of ADAM10 KO Tumour Cells
2.4. Analysis of ADAM10 KO Tumour Xenografts
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Maintenance, and Proliferation Assay
4.2. Generation of ADAM10 Knockout Cell Lines
4.3. Mass Spectrometry Experimental Design and Data Analysis
4.4. Protein Extracts for Immunoprecipitation and Immuno-Blotting
4.5. Immunohistochemistry (IHC) and Microscopy
4.6. Second Harmonic Generation (SHG) Imaging
4.7. MSC Differentiation Assay
4.8. Tumour Xenograft Models
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reiss, K.; Saftig, P. The “A Disintegrin And Metalloprotease” (ADAM) family of sheddases: Physiological and cellular functions. Semin. Cell Dev. Biol. 2009, 20, 126–137. [Google Scholar] [CrossRef]
- Bozkulak, E.C.; Weinmaster, G. Selective Use of ADAM10 and ADAM17 in Activation of Notch1 Signaling. Mol. Cell. Biol. 2009, 29, 5679–5695. [Google Scholar] [CrossRef]
- Hartmann, D.; de Strooper, B.; Serneels, L.; Craessaerts, K.; Herreman, A.; Annaert, W.; Umans, L.; Lübke, T.; Illert, A.L.; von Figura, K.; et al. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum. Mol. Genet. 2002, 11, 2615–2624. [Google Scholar] [CrossRef] [PubMed]
- Alabi, R.O.; Farber, G.; Blobel, C.P. Intriguing Roles for Endothelial ADAM10/Notch Signaling in the Development of Organ-Specific Vascular Beds. Physiol. Rev. 2018, 98, 2025–2061. [Google Scholar] [CrossRef] [PubMed]
- Hattori, M.; Osterfield, M.; Flanagan, J.G. Regulated Cleavage of a Contact-Mediated Axon Repellent. Science 2000, 289, 1360–1365. [Google Scholar] [CrossRef]
- Janes, P.W.; Saha, N.; Barton, W.A.; Kolev, M.V.; Wimmer-Kleikamp, S.H.; Nievergall, E.; Blobel, C.P.; Himanen, J.-P.; Lackmann, M.; Nikolov, D.B. Adam Meets Eph: An ADAM Substrate Recognition Module Acts as a Molecular Switch for Ephrin Cleavage in Trans. Cell 2005, 123, 291–304. [Google Scholar] [CrossRef]
- Seegar, T.C.M.; Killingsworth, L.B.; Saha, N.; Meyer, P.A.; Patra, D.; Zimmerman, B.; Janes, P.W.; Rubinstein, E.; Nikolov, D.B.; Skiniotis, G.; et al. Structural Basis for Regulated Proteolysis by the α-Secretase ADAM10. Cell 2017, 171, 1638–1648.e7. [Google Scholar] [CrossRef] [PubMed]
- Mullooly, M.; McGowan, P.M.; Crown, J.; Duffy, M.J. The ADAMs family of proteases as targets for the treatment of cancer. Cancer Biol. Ther. 2016, 17, 870–880. [Google Scholar] [CrossRef]
- Arora, S.; Scott, A.M.; Janes, P.W. ADAM Proteases in Cancer: Biological Roles, Therapeutic Challenges, and Emerging Opportunities. Cancers 2025, 17, 1703. [Google Scholar] [CrossRef]
- Smith, T.M., Jr.; Tharakan, A.; Martin, R.K. Targeting ADAM10 in Cancer and Autoimmunity. Front. Immunol. 2020, 11, 499. [Google Scholar] [CrossRef]
- Musumeci, G.; Magro, G.; Cardile, V.; Coco, M.; Marzagalli, R.; Castrogiovanni, P.; Imbesi, R.; Graziano, A.C.E.; Barone, F.; Di Rosa, M.; et al. Characterization of matrix metalloproteinase-2 and -9, ADAM-10 and N-cadherin expression in human glioblastoma multiforme. Cell Tissue Res. 2015, 362, 45–60. [Google Scholar] [CrossRef]
- Qu, M.; Qiu, B.; Xiong, W.; Chen, D.; Wu, A. Expression of a-disintegrin and metalloproteinase 10 correlates with grade of malignancy in human glioma. Oncol. Lett. 2015, 9, 2157–2162. [Google Scholar] [CrossRef]
- Bulstrode, H.; Jones, L.M.; Siney, E.J.; Sampson, J.M.; Ludwig, A.; Gray, W.P.; Willaime-Morawek, S. A-Disintegrin and Metalloprotease (ADAM) 10 and 17 promote self-renewal of brain tumor sphere forming cells. Cancer Lett. 2012, 326, 79–87. [Google Scholar] [CrossRef]
- Siney, E.J.; Holden, A.; Casselden, E.; Bulstrode, H.; Thomas, G.J.; Willaime-Morawek, S. Metalloproteinases ADAM10 and ADAM17 Mediate Migration and Differentiation in Glioblastoma Sphere-Forming Cells. Mol. Neurobiol. 2016, 54, 3893–3905. [Google Scholar] [CrossRef] [PubMed]
- Dang, N.-N.; Li, X.-B.; Zhang, M.; Han, C.; Fan, X.-Y.; Huang, S.-H. NLGN3 Upregulates Expression of ADAM10 to Promote the Cleavage of NLGN3 via Activating the LYN Pathway in Human Gliomas. Front. Cell Dev. Biol. 2021, 9, 662763. [Google Scholar] [CrossRef] [PubMed]
- Kohga, K.; Takehara, T.; Tatsumi, T.; Miyagi, T.; Ishida, H.; Ohkawa, K.; Kanto, T.; Hiramatsu, N.; Hayashi, N. Anticancer chemotherapy inhibits MHC class I-related chain a ectodomain shedding by downregulating ADAM10 expression in hepatocellular carcinoma. Cancer Res. 2009, 69, 8050–8057. [Google Scholar] [CrossRef]
- Chitadze, G.; Lettau, M.; Luecke, S.; Wang, T.; Janssen, O.; Fürst, D.; Mytilineos, J.; Wesch, D.; Oberg, H.-H.; Held-Feindt, J.; et al. NKG2D- and T-cell receptor-dependent lysis of malignant glioma cell lines by human γδ T cells: Modulation by temozolomide and A disintegrin and metalloproteases 10 and 17 inhibitors. OncoImmunology 2016, 5, e1093276. [Google Scholar] [CrossRef] [PubMed]
- Meurette, O.; Mehlen, P. Notch Signaling in the Tumor Microenvironment. Cancer Cell 2018, 34, 536–548. [Google Scholar] [CrossRef]
- Venkatesh, H.S.; Tam, L.T.; Woo, P.J.; Lennon, J.; Nagaraja, S.; Gillespie, S.M.; Ni, J.; Duveau, D.Y.; Morris, P.J.; Zhao, J.J.; et al. Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature 2017, 549, 533–537. [Google Scholar] [CrossRef]
- Bazzoni, R.; Bentivegna, A. Role of Notch Signaling Pathway in Glioblastoma Pathogenesis. Cancers 2019, 11, 292. [Google Scholar] [CrossRef]
- Wang, S.; Gu, S.; Chen, J.; Yuan, Z.; Liang, P.; Cui, H. Mechanism of Notch Signaling Pathway in Malignant Progression of Glioblastoma and Targeted Therapy. Biomolecules 2024, 14, 480. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, N.; Rose-John, S.; Schmidt-Arras, D. ADAM-Mediated Signalling Pathways in Gastrointestinal Cancer Formation. Int. J. Mol. Sci. 2020, 21, 5133. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, P.J. Role of ADAM10 in intestinal crypt homeostasis and tumorigenesis. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2017, 1864, 2228–2239. [Google Scholar] [CrossRef]
- Yuan, S.; Yin, Y.; Wang, K.; Zhou, H.; Qian, C. Tetraspanin-29 activates Notch signaling by interacting with ADAM10 to enhance its activity in colorectal cancer. Biochem. Cell Biol. 2022, 100, 292–300. [Google Scholar] [CrossRef]
- Atapattu, L.; Saha, N.; Chheang, C.; Eissman, M.F.; Xu, K.; Vail, M.E.; Hii, L.; Llerena, C.; Liu, Z.; Horvay, K.; et al. An activated form of ADAM10 is tumor selective and regulates cancer stem-like cells and tumor growth. J. Exp. Med. 2016, 213, 1741–1757. [Google Scholar] [CrossRef] [PubMed]
- Saha, N.; Baek, D.-S.; Mendoza, R.P.; Robev, D.; Xu, Y.; Goldgur, Y.; De La Cruz, M.J.; de Stanchina, E.; Janes, P.W.; Xu, K.; et al. Fully human monoclonal antibody targeting activated ADAM10 on colorectal cancer cells. Biomed. Pharmacother. 2023, 161, 114494. [Google Scholar] [CrossRef]
- Yan, H.; Vail, M.E.; Hii, L.; Guo, N.; McMurrick, P.J.; Oliva, K.; Wilkins, S.; Saha, N.; Nikolov, D.B.; Lee, F.-T.; et al. Preferential Antibody and Drug Conjugate Targeting of the ADAM10 Metalloprotease in Tumours. Cancers 2022, 14, 3171. [Google Scholar] [CrossRef]
- Groth, E.; Pruessmeyer, J.; Babendreyer, A.; Schumacher, J.; Pasqualon, T.; Dreymueller, D.; Higashiyama, S.; Lorenzen, I.; Grötzinger, J.; Cataldo, D.; et al. Stimulated release and functional activity of surface expressed metalloproteinase ADAM17 in exosomes. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2016, 1863, 2795–2808. [Google Scholar] [CrossRef]
- Klessner, J.L.; Desai, B.V.; Amargo, E.V.; Getsios, S.; Green, K.J. EGFR and ADAMs Cooperate to Regulate Shedding and Endocytic Trafficking of the Desmosomal Cadherin Desmoglein 2. Mol. Biol. Cell 2009, 20, 328–337. [Google Scholar] [CrossRef]
- Kamekura, R.; Nava, P.; Feng, M.; Quiros, M.; Nishio, H.; Weber, D.A.; Parkos, C.A.; Nusrat, A. Inflammation-induced desmoglein-2 ectodomain shedding compromises the mucosal barrier. Mol. Biol. Cell 2015, 26, 3165–3177. [Google Scholar] [CrossRef]
- Wojtalewicz, N.; Sadeqzadeh, E.; Weiß, J.V.; Tehrani, M.M.; Klein-Scory, S.; Hahn, S.; Schmiegel, W.; Warnken, U.; Schnölzer, M.; De Bock, C.E.; et al. A Soluble Form of the Giant Cadherin Fat1 Is Released from Pancreatic Cancer Cells by ADAM10 Mediated Ectodomain Shedding. PLoS ONE 2014, 9, e90461. [Google Scholar] [CrossRef]
- Miller, M.A.; Meyer, A.S.; Beste, M.T.; Lasisi, Z.; Reddy, S.; Jeng, K.W.; Chen, C.-H.; Han, J.; Isaacson, K.; Griffith, L.G.; et al. ADAM-10 and -17 regulate endometriotic cell migration via concerted ligand and receptor shedding feedback on kinase signaling. Proc. Natl. Acad. Sci. USA 2013, 110, E2074–E2083. [Google Scholar] [CrossRef]
- Shitomi, Y.; Thøgersen, I.B.; Ito, N.; Leitinger, B.; Enghild, J.J.; Itoh, Y. ADAM10 controls collagen signaling and cell migration on collagen by shedding the ectodomain of discoidin domain receptor 1 (DDR1). Mol. Biol. Cell 2015, 26, 659–673. [Google Scholar] [CrossRef]
- Zingoni, A.; Vulpis, E.; Loconte, L.; Santoni, A. NKG2D Ligand Shedding in Response to Stress: Role of ADAM10. Front. Immunol. 2020, 11, 447. [Google Scholar] [CrossRef]
- Orme, J.J.; Jazieh, K.A.; Xie, T.; Harrington, S.; Liu, X.; Ball, M.; Madden, B.; Charlesworth, M.C.; Azam, T.U.; Lucien, F.; et al. ADAM10 and ADAM17 cleave PD-L1 to mediate PD-(L)1 inhibitor resistance. Oncoimmunology 2020, 9, 1744980. [Google Scholar] [CrossRef]
- Morinaga, J.; Kadomatsu, T.; Miyata, K.; Endo, M.; Terada, K.; Tian, Z.; Sugizaki, T.; Tanigawa, H.; Zhao, J.; Zhu, S.; et al. Angiopoietin-like protein 2 increases renal fibrosis by accelerating transforming growth factor-β signaling in chronic kidney disease. Kidney Int. 2016, 89, 327–341. [Google Scholar] [CrossRef]
- Kim, J.-S.; Kim, T.H.; Kang, D.L.; Baek, S.Y.; Lee, Y.; Koh, Y.-G.; Kim, Y.I. Chondrogenic differentiation of human ASCs by stiffness control in 3D fibrin hydrogel. Biochem. Biophys. Res. Commun. 2020, 522, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Qiang, L.; Yang, Y.; Ma, Y.-J.; Chen, F.-H.; Zhang, L.-B.; Liu, W.; Qi, Q.; Lu, N.; Tao, L.; Wang, X.-T.; et al. Isolation and characterization of cancer stem like cells in human glioblastoma cell lines. Cancer Lett. 2009, 279, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, H.; Yuasa, T.; Kubota, Y.; Seita, M.; Sasamoto, H.; Shahid, J.M.; Hayashi, T.; Nakahara, H.; Hassan, R.; Iwamuro, M.; et al. Characteristics of CD133+ human colon cancer SW620 cells. Cell Transplant. 2010, 19, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, R.; Xi, B.; Nie, D.; Xu, H.; Liu, A. Mechanisms of Senescence-Related NKG2D Ligands Release and Immune Escape Induced by Chemotherapy in Neuroblastoma Cells. Front. Cell Dev. Biol. 2022, 10, 829404. [Google Scholar] [CrossRef]
- Ludwig, A.; Hundhausen, C.; Lambert, M.H.; Broadway, N.; Andrews, R.C.; Bickett, D.M.; Leesnitzer, M.A.; Becherer, J.D. Metalloproteinase Inhibitors for the Disintegrin-Like Metalloproteinases ADAM10 and ADAM17 that Differentially Block Constitutive and Phorbol Ester-Inducible Shedding of Cell Surface Molecules. Comb. Chem. High Throughput Screen. 2005, 8, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Witters, L.; Scherle, P.; Friedman, S.; Fridman, J.; Caulder, E.; Newton, R.; Lipton, A. Synergistic Inhibition with a Dual Epidermal Growth Factor Receptor/HER-2/neu Tyrosine Kinase Inhibitor and a Disintegrin and Metalloprotease Inhibitor. Cancer Res. 2008, 68, 7083–7089. [Google Scholar] [CrossRef] [PubMed]
- Saha, N.; Robev, D.; Himanen, J.P.; Nikolov, D.B. ADAM proteases: Emerging role and targeting of the non-catalytic domains. Cancer Lett. 2019, 467, 50–57. [Google Scholar] [CrossRef]
- Kuhn, P.-H.; Colombo, A.V.; Schusser, B.; Dreymueller, D.; Wetzel, S.; Schepers, U.; Herber, J.; Ludwig, A.; Kremmer, E.; Montag, D.; et al. Systematic substrate identification indicates a central role for the metalloprotease ADAM10 in axon targeting and synapse function. eLife 2016, 5, e12748. [Google Scholar] [CrossRef]
- Wang, Y.; Herrera, A.H.; Li, Y.; Belani, K.K.; Walcheck, B. Regulation of Mature ADAM17 by Redox Agents for L-Selectin Shedding. J. Immunol. 2009, 182, 2449–2457. [Google Scholar] [CrossRef]
- Willems, S.H.; Tape, C.J.; Stanley, P.L.; Taylor, N.A.; Mills, I.G.; Neal, D.E.; McCafferty, J.; Murphy, G. Thiol isomerases negatively regulate the cellular shedding activity of ADAM17. Biochem. J. 2010, 428, 439–450. [Google Scholar] [CrossRef]
- Düsterhöft, S.; Jung, S.; Hung, C.-W.; Tholey, A.; Sönnichsen, F.D.; Grötzinger, J.; Lorenzen, I. Membrane-Proximal Domain of a Disintegrin and Metalloprotease-17 Represents the Putative Molecular Switch of Its Shedding Activity Operated by Protein-disulfide Isomerase. J. Am. Chem. Soc. 2013, 135, 5776–5781. [Google Scholar] [CrossRef]
- Quéré, M.; Alberto, J.-M.; Broly, F.; Hergalant, S.; Christov, C.; Gauchotte, G.; Guéant, J.-L.; Namour, F.; Battaglia-Hsu, S.-F. ALDH1L2 Knockout in U251 Glioblastoma Cells Reduces Tumor Sphere Formation by Increasing Oxidative Stress and Suppressing Methionine Dependency. Nutrients 2022, 14, 1887. [Google Scholar] [CrossRef]
- Thomas, P.; Pranatharthi, A.; Ross, C.; Srivastava, S. RhoC: A fascinating journey from a cytoskeletal organizer to a Cancer stem cell therapeutic target. J. Exp. Clin. Cancer Res. 2019, 38, 328. [Google Scholar] [CrossRef]
- Zatovicova, M.; Kajanova, I.; Takacova, M.; Jelenska, L.; Sedlakova, O.; Labudova, M.; Pastorekova, S. ADAM10 mediates shedding of carbonic anhydrase IX ectodomain non-redundantly to ADAM17. Oncol. Rep. 2022, 49, 27. [Google Scholar] [CrossRef] [PubMed]
- Mueller, A.C.; Piper, M.; Goodspeed, A.; Bhuvane, S.; Williams, J.S.; Bhatia, S.; Phan, A.V.; Van Court, B.; Zolman, K.L.; Peña, B.; et al. Induction of ADAM10 by Radiation Therapy Drives Fibrosis, Resistance, and Epithelial-to-Mesenchyal Transition in Pancreatic Cancer. Cancer Res. 2021, 81, 3255–3269. [Google Scholar] [CrossRef] [PubMed]
- Zbodakova, O.; Chalupsky, K.; Sarnova, L.; Kasparek, P.; Jirouskova, M.; Gregor, M.; Sedlacek, R. ADAM10 and ADAM17 regulate EGFR, c-Met and TNF RI signalling in liver regeneration and fibrosis. Sci. Rep. 2021, 11, 11414. [Google Scholar] [CrossRef]
- Dominguez, C.X.; Müller, S.; Keerthivasan, S.; Koeppen, H.; Hung, J.; Gierke, S.; Breart, B.; Foreman, O.; Bainbridge, T.W.; Castiglioni, A.; et al. Single-Cell RNA Sequencing Reveals Stromal Evolution into LRRC15+ Myofibroblasts as a Determinant of Patient Response to Cancer Immunotherapy. Cancer Discov. 2020, 10, 232–253. [Google Scholar] [CrossRef]
- Duan, J.-L.; Ruan, B.; Yan, X.-C.; Liang, L.; Song, P.; Yang, Z.-Y.; Liu, Y.; Dou, K.-F.; Han, H.; Wang, L. Endothelial Notch activation reshapes the angiocrine of sinusoidal endothelia to aggravate liver fibrosis and blunt regeneration in mice. Hepatology 2018, 68, 677–690. [Google Scholar] [CrossRef]
- Kepes, J.J.; Rubinstein, L.J.; Chiang, H. The role of astrocytes in the formation of cartilage in gliomas. An immunohistochemical study of four cases. Am. J. Pathol. 1984, 117, 471–483. [Google Scholar] [PubMed]
- Ricci-Vitiani, L.; Pallini, R.; Larocca, L.M.; Lombardi, D.G.; Signore, M.; Pierconti, F.; Petrucci, G.; Montano, N.; Maira, G.; De Maria, R. Mesenchymal differentiation of glioblastoma stem cells. Cell Death Differ. 2008, 15, 1491–1498. [Google Scholar] [CrossRef]
- Mead, T.J.; Yutzey, K.E. Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proc. Natl. Acad. Sci. USA 2009, 106, 14420–14425. [Google Scholar] [CrossRef] [PubMed]
- HaileMariam, M.; Eguez, R.V.; Singh, H.; Bekele, S.; Ameni, G.; Pieper, R.; Yu, Y. S-Trap, an Ultrafast Sample-Preparation Approach for Shotgun Proteomics. J. Proteome Res. 2018, 17, 2917–2924. [Google Scholar] [CrossRef]
- Humphrey, S.J.; Karayel, O.; James, D.E.; Mann, M. High-throughput and high-sensitivity phosphoproteomics with the EasyPhos platform. Nat. Protoc. 2018, 13, 1897–1916. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Kong, A.T.; LePrevost, F.V.; Avtonomov, D.M.; Mellacheruvu, D.; Nesvizhskii, A.I. MSFragger: Ultrafast and comprehensive peptide identification in mass spectrometry–based proteomics. Nat. Methods 2017, 14, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Teo, G.C.; Polasky, D.A.; Yu, F.; Nesvizhskii, A.I. Fast Deisotoping Algorithm and Its Implementation in the MSFragger Search Engine. J. Proteome Res. 2020, 20, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Haynes, S.E.; Nesvizhskii, A.I. IonQuant Enables Accurate and Sensitive Label-Free Quantification with FDR-Controlled Match-Between-Runs. Mol. Cell. Proteom. 2021, 20, 100077. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.D.; Goode, R.J.A.; Huang, C.; Powell, D.R.; Schittenhelm, R.B. LFQ-Analyst: An Easy-To-Use Interactive Web Platform To Analyze and Visualize Label-Free Proteomics Data Preprocessed with MaxQuant. J. Proteome Res. 2019, 19, 204–211. [Google Scholar] [CrossRef]
- Zhang, H.; Steele, J.R.; Kahrood, H.V.; Lucas, D.D.; Shah, A.D.; Schittenhelm, R.B. Phospho-Analyst: An Interactive, Easy-to-Use Web Platform To Analyze Quantitative Phosphoproteomics Data. J Proteome Res. 2023, 22, 2890–2899. [Google Scholar] [CrossRef]
- To, C.; Farnsworth, R.H.; Vail, M.E.; Chheang, C.; Gargett, C.E.; Murone, C.; Llerena, C.; Major, A.T.; Scott, A.M.; Janes, P.W.; et al. Hypoxia-Controlled EphA3 Marks a Human Endometrium-Derived Multipotent Mesenchymal Stromal Cell that Supports Vascular Growth. PLoS ONE 2014, 9, e112106. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, H.; Arora, S.; Hii, L.; Llerena, C.; Vail, M.E.; Allam, A.; Conway, J.R.W.; Steele, J.R.; Lee, H.-C.; Schittenhelm, R.B.; et al. ADAM10 Knockout from Human Glioblastoma and Colon Cancer Cells Modulates Diverse Signalling Networks and Inhibits Tumour Growth In Vivo. Int. J. Mol. Sci. 2025, 26, 10684. https://doi.org/10.3390/ijms262110684
Yan H, Arora S, Hii L, Llerena C, Vail ME, Allam A, Conway JRW, Steele JR, Lee H-C, Schittenhelm RB, et al. ADAM10 Knockout from Human Glioblastoma and Colon Cancer Cells Modulates Diverse Signalling Networks and Inhibits Tumour Growth In Vivo. International Journal of Molecular Sciences. 2025; 26(21):10684. https://doi.org/10.3390/ijms262110684
Chicago/Turabian StyleYan, Hengkang, Sakshi Arora, Linda Hii, Carmen Llerena, Mary E. Vail, Amr Allam, James R. W. Conway, Joel R. Steele, Han-Chung Lee, Ralf B. Schittenhelm, and et al. 2025. "ADAM10 Knockout from Human Glioblastoma and Colon Cancer Cells Modulates Diverse Signalling Networks and Inhibits Tumour Growth In Vivo" International Journal of Molecular Sciences 26, no. 21: 10684. https://doi.org/10.3390/ijms262110684
APA StyleYan, H., Arora, S., Hii, L., Llerena, C., Vail, M. E., Allam, A., Conway, J. R. W., Steele, J. R., Lee, H.-C., Schittenhelm, R. B., Scott, A. M., & Janes, P. W. (2025). ADAM10 Knockout from Human Glioblastoma and Colon Cancer Cells Modulates Diverse Signalling Networks and Inhibits Tumour Growth In Vivo. International Journal of Molecular Sciences, 26(21), 10684. https://doi.org/10.3390/ijms262110684







