Functional Characterization of Fp2Cas9, a Cold-Adapted Type II-C CRISPR Nuclease from Flavobacterium psychrophilum
Abstract
1. Introduction
2. Results
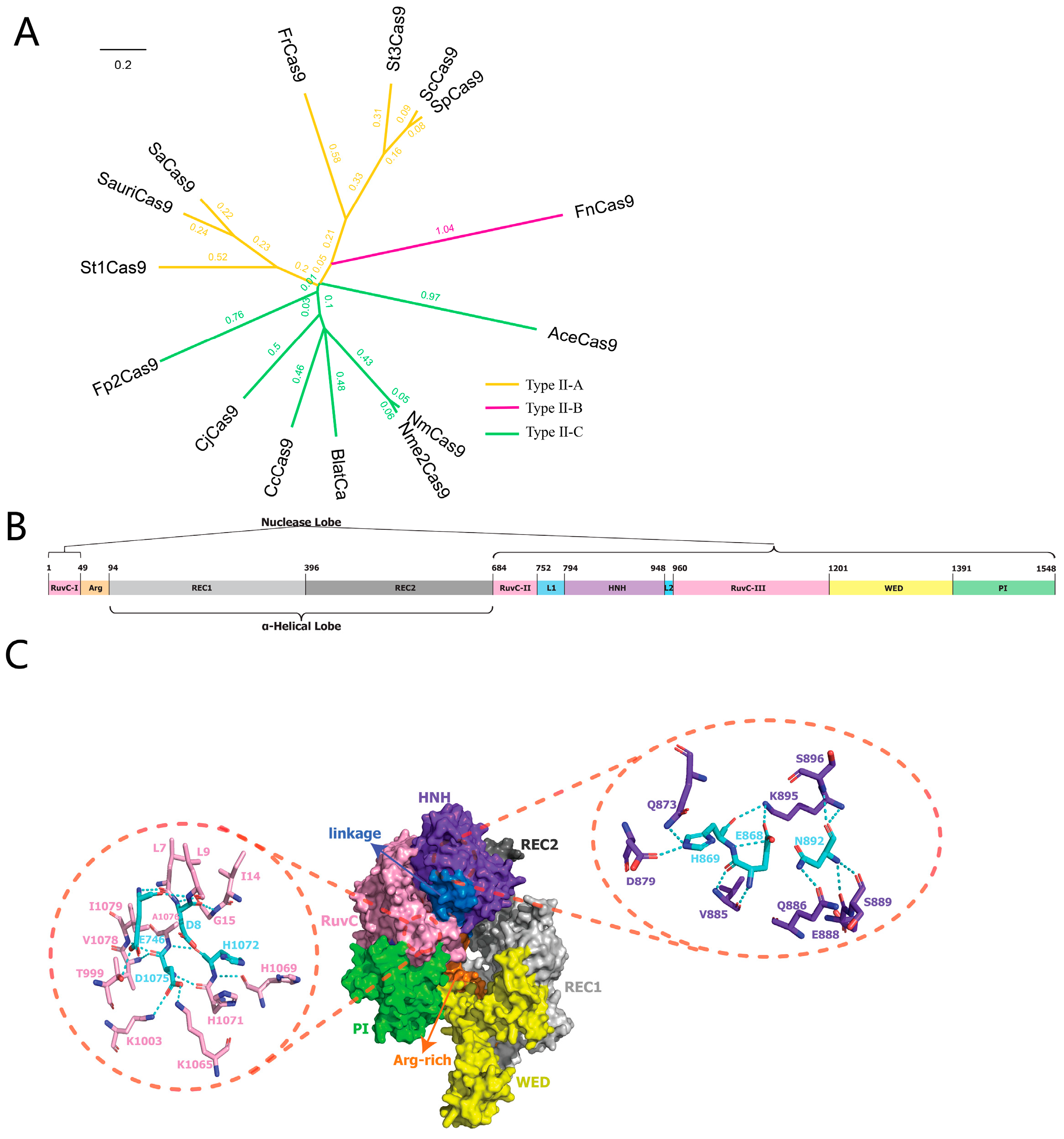
2.1. Bioinformatic Characterization of Fp2Cas9
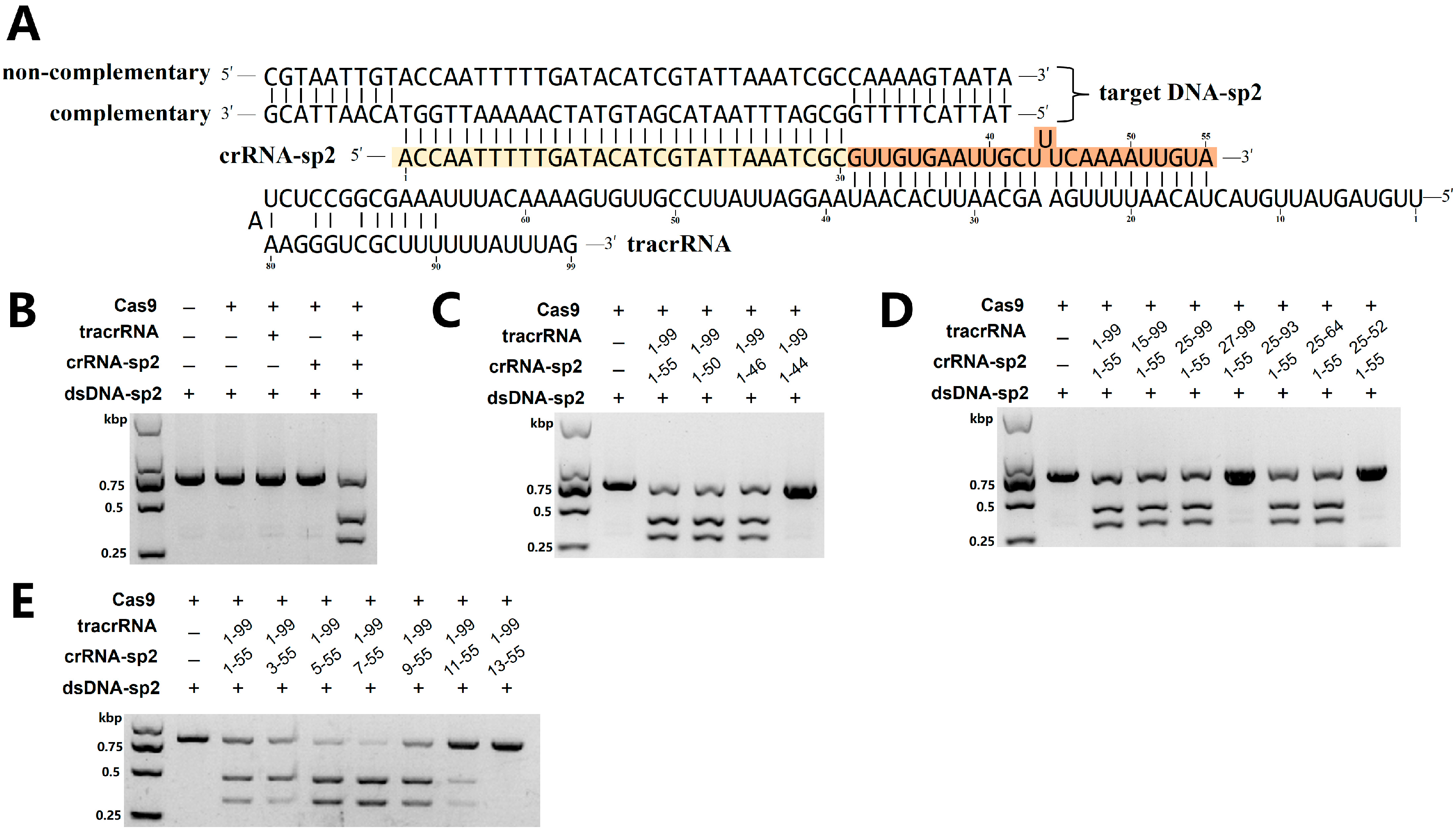
2.2. Characterization of Fp2Cas9 Nuclease
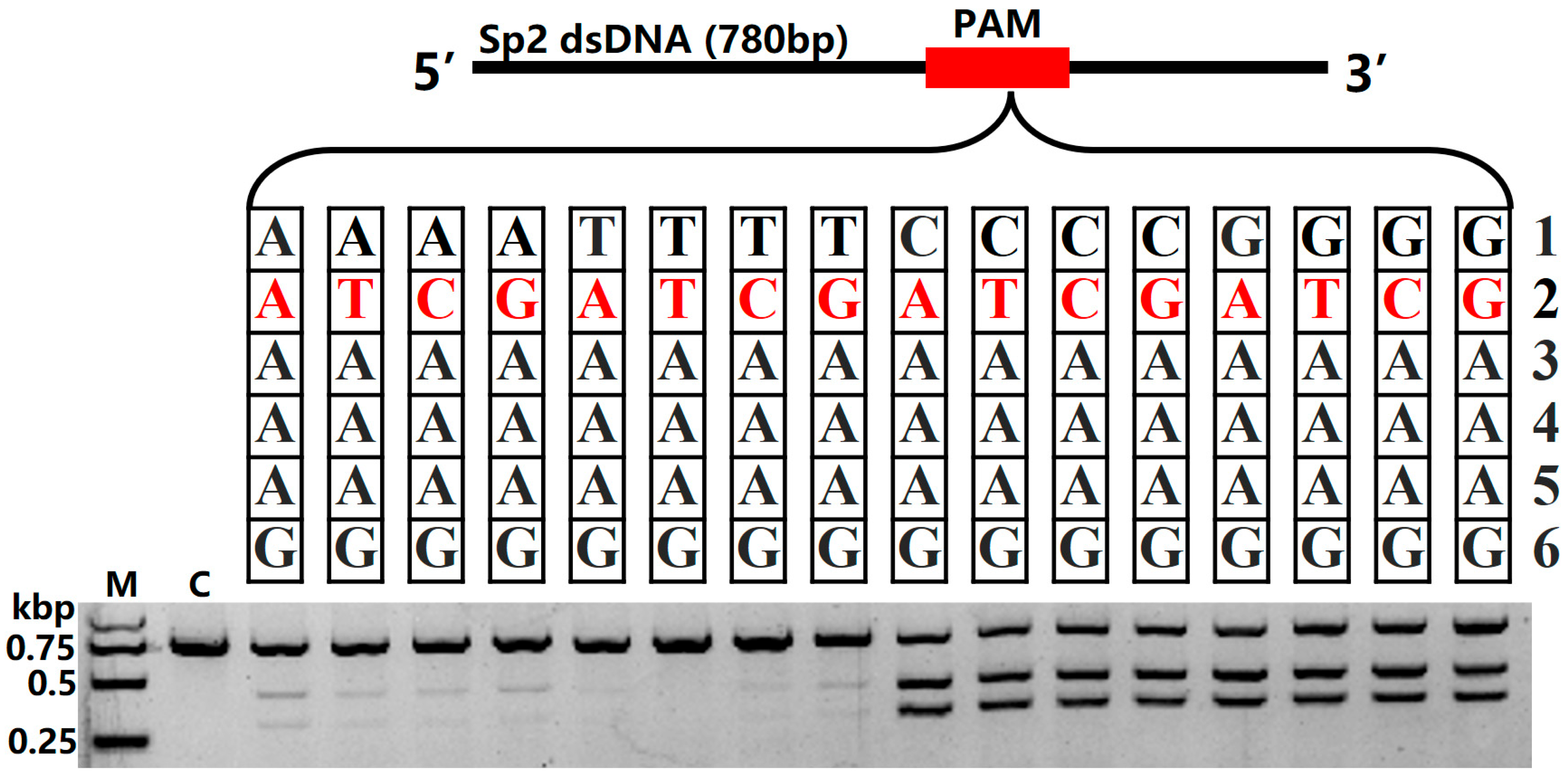
2.3. Fp2Cas9 In Vitro PAM Analysis
2.4. Fp2Cas9 sgRNA Design
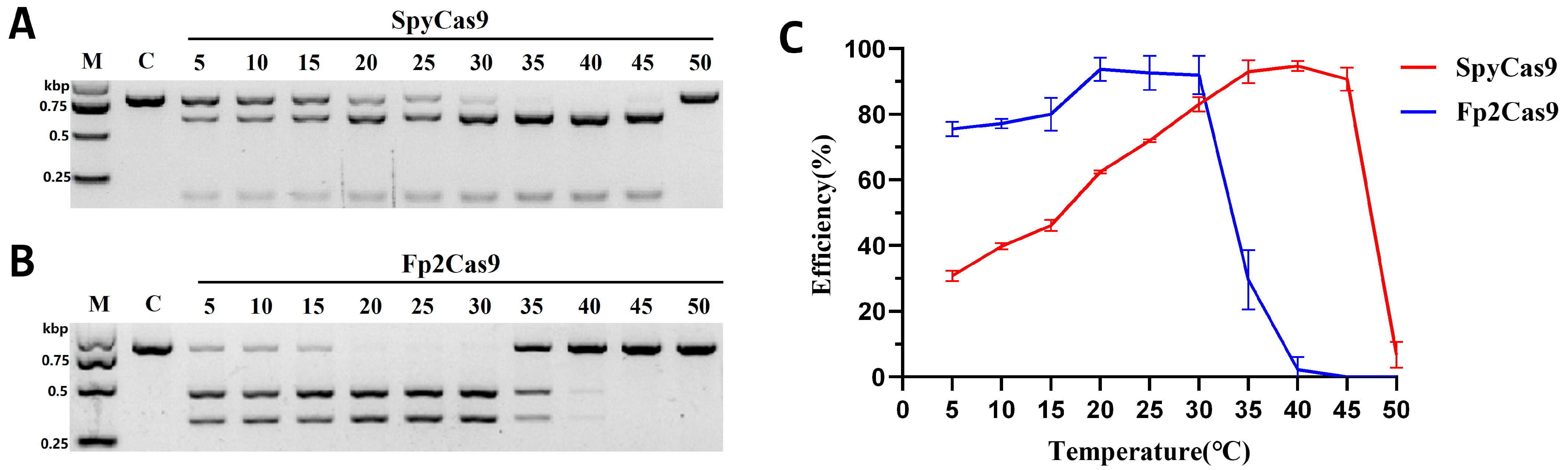
2.5. Fp2Cas9 Temperature Preferences
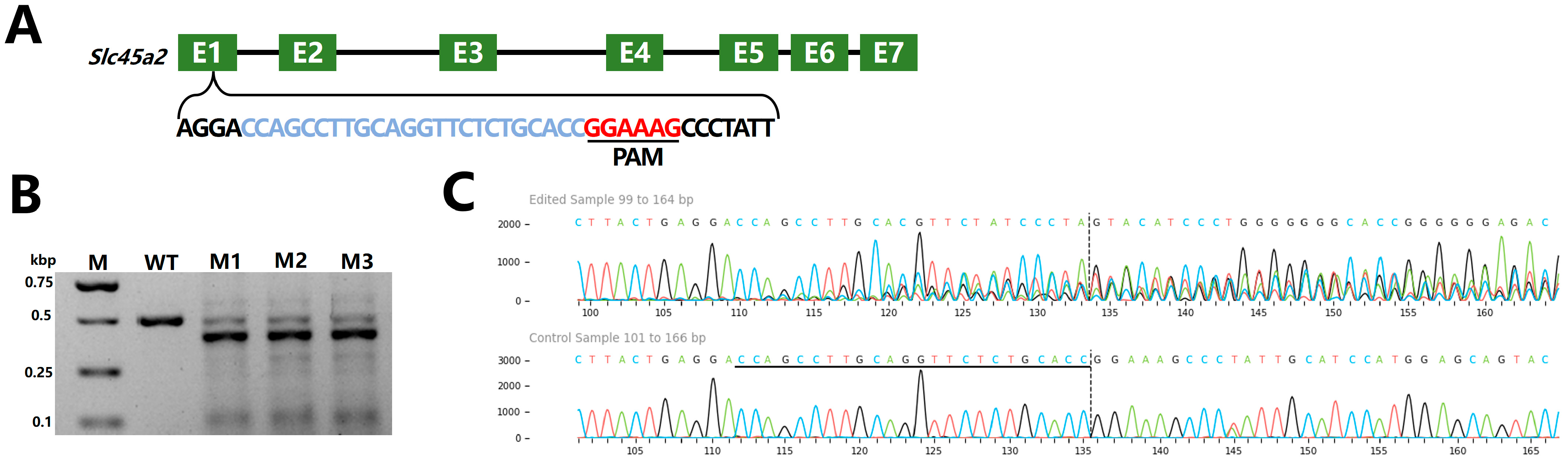
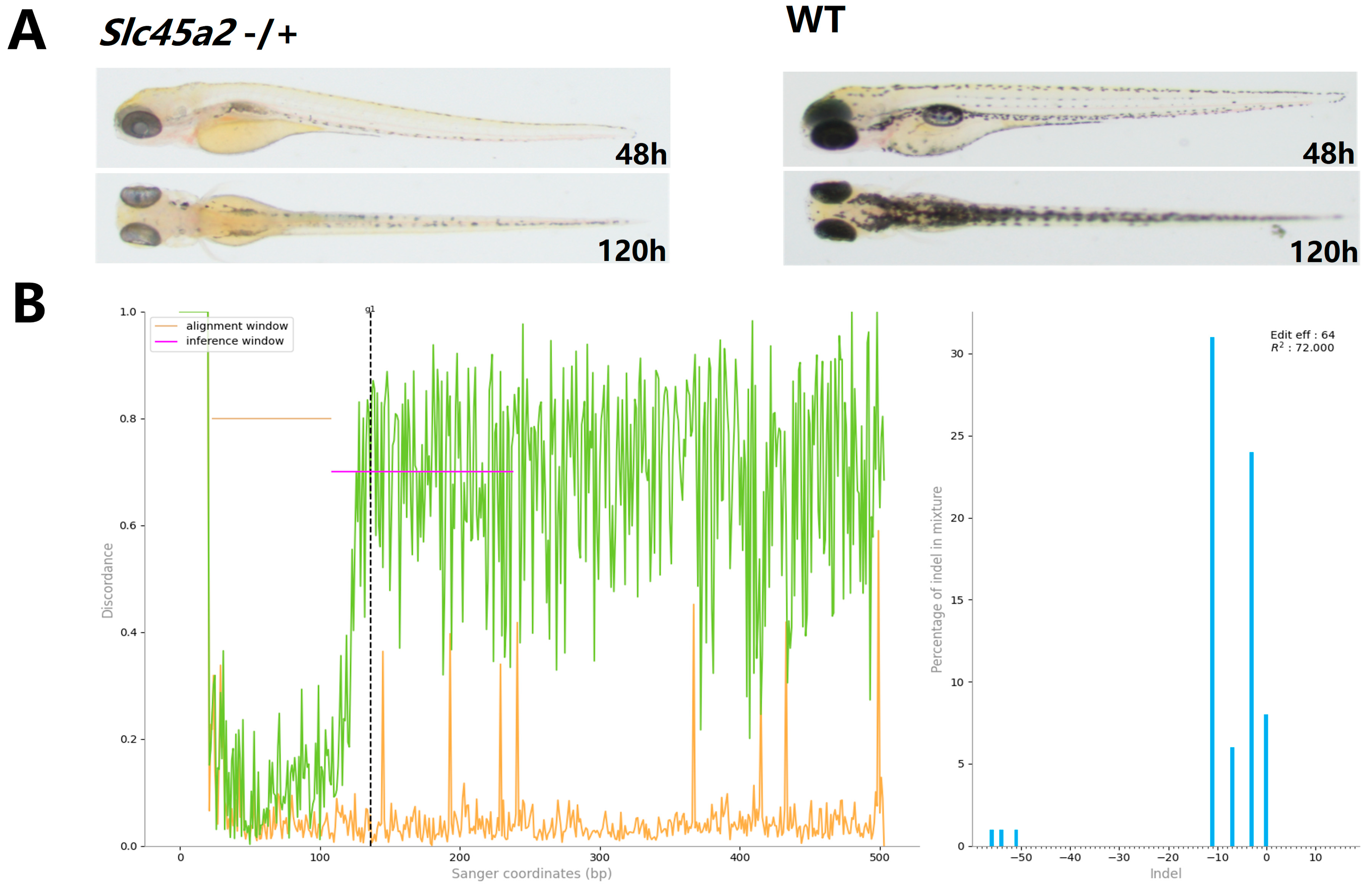
2.6. Nuclease Activity of Fp2Cas9 in Zebrafish Embryos
3. Discussion
4. Materials and Methods
4.1. Bioinformatical Predictions of Fp2Cas9
4.2. Fp2Cas9 Expression and Purification
4.3. In Vitro Transcription and Purification of RNA
4.4. In Vitro DNA Cleavage Assays
4.5. CRISPR/Fp2Cas9 Targeted Editing of Zebrafish slc45a2 and Phenotypic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| PAM | Protospacer Adjacent Motif |
| sgRNA | single-guide RNA |
| RNP | Ribonucleoprotein |
| NLS | Nuclear Localization Signal |
| hpf | hours post-fertilization |
References
- Marraffini, L.A.; Sontheimer, E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 2008, 322, 1843–1845. [Google Scholar] [CrossRef]
- Marraffini, L.A. CRISPR-Cas immunity in prokaryotes. Nature 2015, 526, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Sontheimer, E.J.; Barrangou, R. The Bacterial Origins of the CRISPR Genome-Editing Revolution. Hum. Gene Ther. 2015, 26, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Brouns, S.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.; Snijders, A.P.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; van der Oost, J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef]
- Mohanraju, P.; Makarova, K.S.; Zetsche, B.; Zhang, F.; Koonin, E.V.; van der Oost, J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 2016, 353, aad5147. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Chylinski, K.; Makarova, K.S.; Charpentier, E.; Koonin, E.V. Classification and evolution of type II CRISPR-Cas systems. Nucleic. Acids. Res. 2014, 42, 6091–6105. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017, 15, 169–182. [Google Scholar] [CrossRef]
- Wright, A.V.; Nuñez, J.K.; Doudna, J.A. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 2016, 164, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Selkova, P.; Vasileva, A.; Arseniev, A.; Abramova, M.; Musharova, O.; Malysheva, P.; Khodorkovskii, M.; Severinov, K. Characterization of Streptococcus uberis Cas9 (SuCas9)—A Type II-A Ortholog Functional in Human Cells. BioRxiv 2023. BioRxiv:2023.12.11.571105. [Google Scholar] [CrossRef]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.-Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Zhang, Y.; Propson, N.E.; Howden, S.E.; Chu, L.F.; Sontheimer, E.J.; Thomson, J.A. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 2013, 110, 15644–15649. [Google Scholar] [CrossRef] [PubMed]
- Matveeva, A.; Ryabchenko, A.; Petrova, V.; Prokhorova, D.; Zhuravlev, E.; Zakabunin, A.; Tikunov, A.; Stepanov, G. 17Expression and Functional Analysis of the Compact Thermophilic Anoxybacillus flavithermus Cas9 Nuclease. IJMS 2023, 24, 17121. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, H.; Xu, X.; Wang, Y.; Chen, W.; Wang, Y.; Wu, Z.; Tang, N.; Wang, Y.; Zhao, S.; et al. Catalytic-state structure and engineering of Streptococcus thermophilus Cas9. Nat. Catal. 2020, 3, 813–823. [Google Scholar] [CrossRef]
- Mir, A.; Edraki, A.; Lee, J.; Sontheimer, E.J. Type II-C CRISPR-Cas9 Biology, Mechanism, and Application. ACS Chem. Biol. 2018, 13, 357–365. [Google Scholar] [CrossRef]
- Xu, R.; Qin, R.; Xie, H.; Li, J.; Liu, X.; Zhu, M.; Sun, Y.; Yu, Y.; Lu, P.; Wei, P. Genome editing with type II-C CRISPR-Cas9 systems from Neisseria meningitidis in rice. Plant Biotechnol. J. 2022, 20, 350–359. [Google Scholar] [CrossRef]
- Lin, L.; Petersen, T.S.; Jensen, K.T.; Bolund, L.; Kühn, R.; Luo, Y. Fusion of SpCas9 to E. coli Rec A protein enhances CRISPR-Cas9 mediated gene knockout in mammalian cells. J. Biotechnol. 2017, 247, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Bauer, D.E.; Canver, M.C.; Orkin, S.H. Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. J. Vis. Exp. 2015, 83, e52118. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Kurokawa, S.; Rahman, H.; Yamanaka, N.; Ishizaki, C.; Islam, S.; Aiso, T.; Hirata, S.; Yamamoto, M.; Kobayashi, K.; Kaya, H. A Simple Heat Treatment Increases SpCas9-Mediated Mutation Efficiency in Arabidopsis. Plant Cell Physiol. 2021, 62, 1676–1686. [Google Scholar] [CrossRef]
- Harrington, L.B.; Paez-Espino, D.; Staahl, B.T.; Chen, J.S.; Ma, E.; Kyrpides, N.C.; Doudna, J.A. A thermostable Cas9 with increased lifetime in human plasma. Nat. Commun. 2017, 8, 1424. [Google Scholar] [CrossRef] [PubMed]
- Mougiakos, I.; Mohanraju, P.; Bosma, E.F.; Vrouwe, V.; Finger Bou, M.; Naduthodi, M.I.S.; Gussak, A.; Brinkman, R.B.L.; van Kranenburg, R.; van der Oost, J. Characterizing a thermostable Cas9 for bacterial genome editing and silencing. Nat. Commun. 2017, 8, 1647. [Google Scholar] [CrossRef]
- Handeland, S.O.; Björnsson; Arnesen, A.M.; Stefansson, S.O. Seawater adaptation and growth of post-smolt Atlantic salmon (Salmo salar) of wild and farmed strains. Aquaculture 2003, 220, 367–384. [Google Scholar] [CrossRef]
- Jiang, X.; Dong, S.; Liu, R.; Huang, M.; Dong, K.; Ge, J.; Gao, Q.; Zhou, Y. Effects of temperature, dissolved oxygen, and their interaction on the growth performance and condition of rainbow trout (Oncorhynchus mykiss). J. Therm. Biol. 2021, 98, 102928. [Google Scholar] [CrossRef]
- Xiang, G.; Zhang, X.; An, C.; Cheng, C.; Wang, H. Temperature effect on CRISPR-Cas9 mediated genome editing. J. Genet. Genom. 2017, 44, 199–205. [Google Scholar] [CrossRef]
- Chen, F.; Wang, D.; Lu, T.; Li, S. Identification of a novel type II-C Cas9 from the fish pathogen Flavobacterium psychrophilum. Front. Microbiol. 2023, 14, 1181303. [Google Scholar] [CrossRef]
- Briner, A.E.; Donohoue, P.D.; Gomaa, A.A.; Selle, K.; Slorach, E.M.; Nye, C.H.; Haurwitz, R.E.; Beisel, C.L.; May, A.P.; Barrangou, R. Guide RNA Functional Modules Direct Cas9 Activity and Orthogonality. Mol. Cell 2014, 56, 333–339. [Google Scholar] [CrossRef]
- Amrani, N.; Gao, X.D.; Liu, P.; Edraki, A.; Mir, A.; Ibraheim, R.; Gupta, A.; Sasaki, K.E.; Wu, T.; Donohoue, P.D.; et al. NmeCas9 is an intrinsically high-fidelity genome-editing platform. Genome Biol. 2018, 19, 214. [Google Scholar] [CrossRef]
- Mengstie, M.A.; Azezew, M.T.; Dejenie, T.A.; Teshome, A.A.; Admasu, F.T.; Teklemariam, A.B.; Mulu, A.T.; Agidew, M.M.; Adugna, D.G.; Geremew, H.; et al. Recent Advancements in Reducing the Off-Target Effect of CRISPR-Cas9 Genome Editing. Biol. Targets Ther. 2024, 18, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Shinkado, S.; Saito, H.; Yamazaki, M.; Kotera, S.; Arazoe, T.; Arie, T.; Kamakura, T. Genome editing using a versatile vector-based CRISPR/Cas9 system in Fusarium species. Sci. Rep. 2022, 12, 16243. [Google Scholar] [CrossRef]
- Hu, P.; Zhao, X.; Zhang, Q.; Li, W.; Zu, Y. Comparison of Various Nuclear Localization Signal-Fused Cas9 Proteins and Cas9 mRNA for Genome Editing in Zebrafish. G3 Genes|Genomes|Genet. 2018, 8, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Belato, H.B.; Knight, A.L.; Alexandra, M.D.; Pindi, C.; Fan, Z.; Luo, J.; Palermo, G.; Jogl, G.; Lisi, G.P. Structural and dynamic impacts of single-atom disruptions to guide RNA interactions within the recognition lobe of Geobacillus stearothermophilus Cas9. eLife 2025, 13, RP99275. [Google Scholar] [CrossRef]
- Montecillo, J.A.V.; Chu, L.L.; Bae, H. CRISPR-Cas9 System for Plant Genome Editing: Current Approaches and Emerging Developments. Agronomy 2020, 10, 1033. [Google Scholar] [CrossRef]
- Li, S.; Chai, J.; Knupp, C.; Nicolas, P.; Wang, D.; Cao, Y.; Deng, F.; Chen, F.; Lu, T.; Loch, T.P. Phenotypic and Genetic Characterization of Flavobacterium psychrophilum Recovered from Diseased Salmonids in China. Microbiol. Spectr. 2021, 9, e00330-21. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Siddiqui, K.S.; Andrews, D.; Sowers, K.R. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 2002, 13, 253–261. [Google Scholar] [CrossRef]
- Wu, G.; Zhang, S.; Zhang, H.; Zhang, S.; Liu, Z. A novel esterase from a psychrotrophic bacterium Psychrobacter celer 3Pb1 showed cold-adaptation and salt-tolerance. J. Mol. Catal. B Enzym. 2013, 98, 119–126. [Google Scholar] [CrossRef]
- Fu, J.; Leiros, H.-K.S.; de Pascale, D.; Johnson, K.A.; Blencke, H.-M.; Landfald, B. Functional and structural studies of a novel cold-adapted esterase from an Arctic intertidal metagenomic library. Appl. Microbiol. Biotechnol. 2013, 97, 3965–3978. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Wu, G.; Zhan, T.; Shao, Z.; Liu, Z. Characterization of a cold-adapted and salt-tolerant esterase from a psychrotrophic bacterium Psychrobacter pacificensis. Extremophiles 2013, 17, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S.; et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science 2014, 343, 1247997. [Google Scholar] [CrossRef]
- Sun, W.; Yang, J.; Cheng, Z.; Amrani, N.; Liu, C.; Wang, K.; Ibraheim, R.; Edraki, A.; Huang, X.; Wang, M.; et al. Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States. Mol. Cell 2019, 76, 938–952.e5. [Google Scholar] [CrossRef]
- Tolmachev, D.; Mamistvalov, G.; Lukasheva, N.; Larin, S.; Karttunen, M. Effects of Amino Acid Side-Chain Length and Chemical Structure on Anionic Polyglutamic and Polyaspartic Acid Cellulose-Based Polyelectrolyte Brushes. Polymers 2021, 13, 1789. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, R.; Zhu, J.; Wang, J.; Wang, D.; Liu, X.; Han, L.; Li, S. Functional Characterization of Fp2Cas9, a Cold-Adapted Type II-C CRISPR Nuclease from Flavobacterium psychrophilum. Int. J. Mol. Sci. 2025, 26, 10681. https://doi.org/10.3390/ijms262110681
Zhao R, Zhu J, Wang J, Wang D, Liu X, Han L, Li S. Functional Characterization of Fp2Cas9, a Cold-Adapted Type II-C CRISPR Nuclease from Flavobacterium psychrophilum. International Journal of Molecular Sciences. 2025; 26(21):10681. https://doi.org/10.3390/ijms262110681
Chicago/Turabian StyleZhao, Ran, Jianqiang Zhu, Jing Wang, Di Wang, Xinting Liu, Lanlan Han, and Shaowu Li. 2025. "Functional Characterization of Fp2Cas9, a Cold-Adapted Type II-C CRISPR Nuclease from Flavobacterium psychrophilum" International Journal of Molecular Sciences 26, no. 21: 10681. https://doi.org/10.3390/ijms262110681
APA StyleZhao, R., Zhu, J., Wang, J., Wang, D., Liu, X., Han, L., & Li, S. (2025). Functional Characterization of Fp2Cas9, a Cold-Adapted Type II-C CRISPR Nuclease from Flavobacterium psychrophilum. International Journal of Molecular Sciences, 26(21), 10681. https://doi.org/10.3390/ijms262110681





