Genome-Wide Association Study Reveals Novel QTNs and Candidate Genes Implicated in Resistance to Northern Corn Leaf Blight in Maize (Zea mays L.)
Abstract
1. Introduction
2. Results
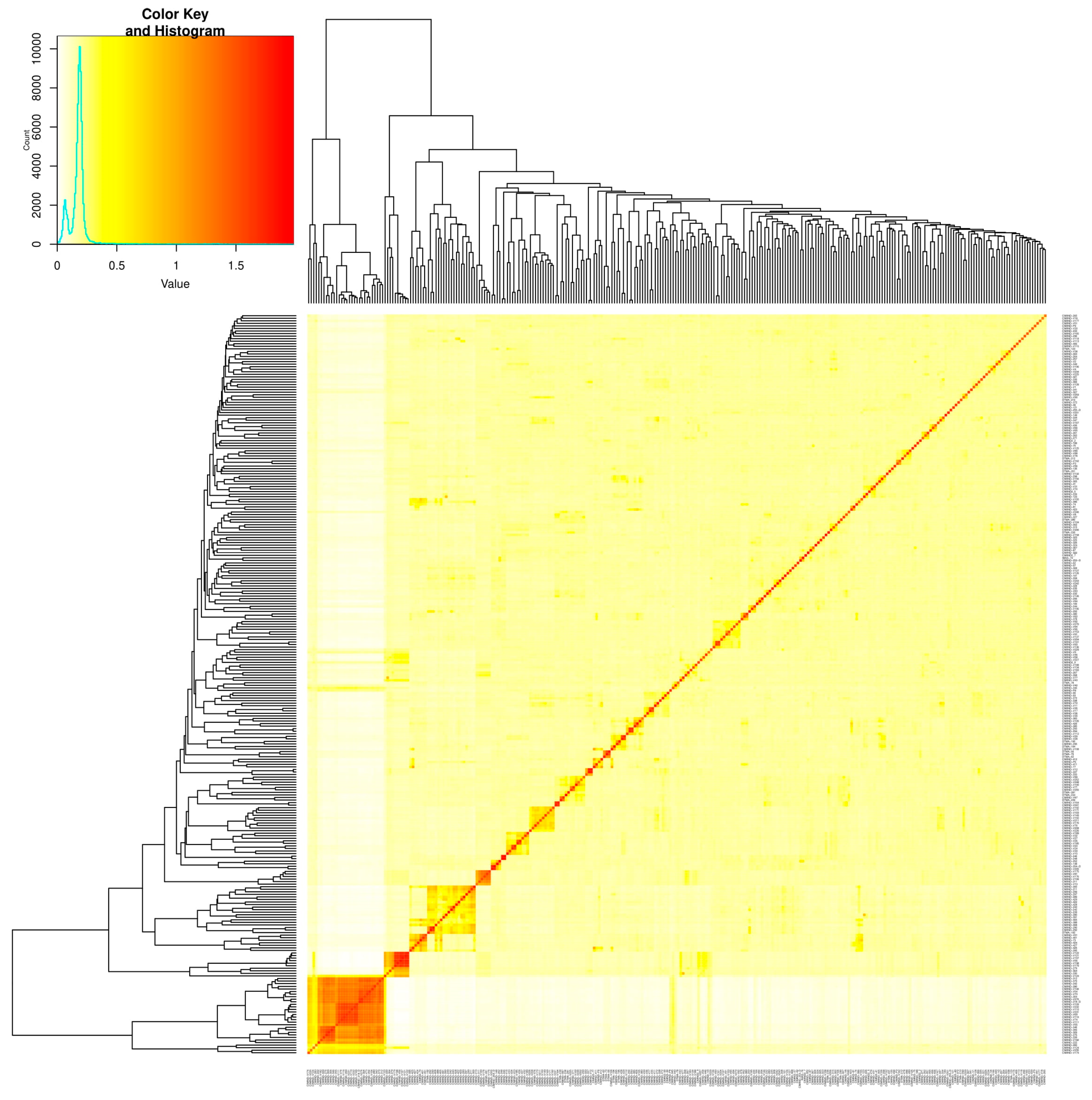
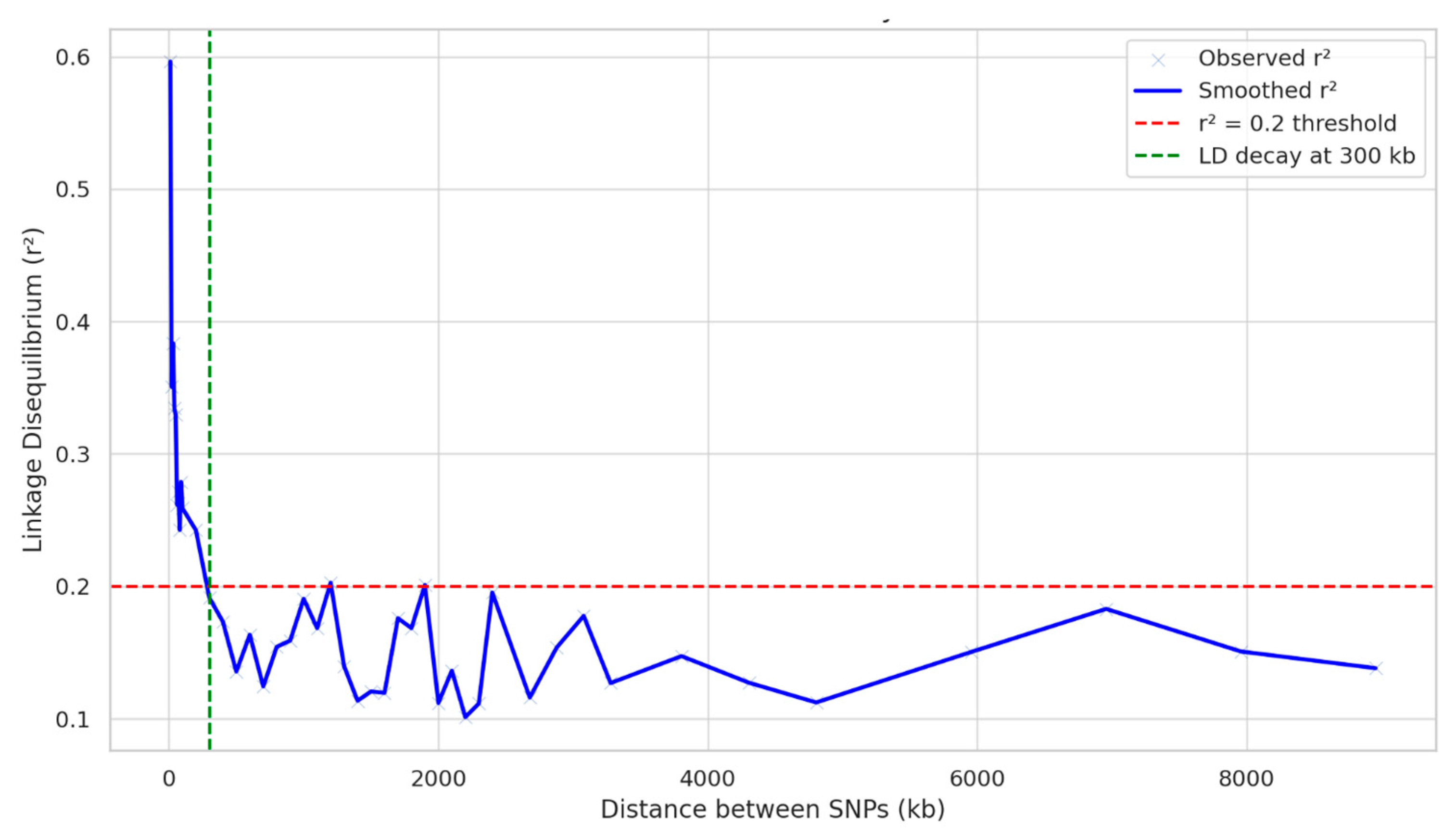
2.1. Population Structure, Kinship, and LD Analysis
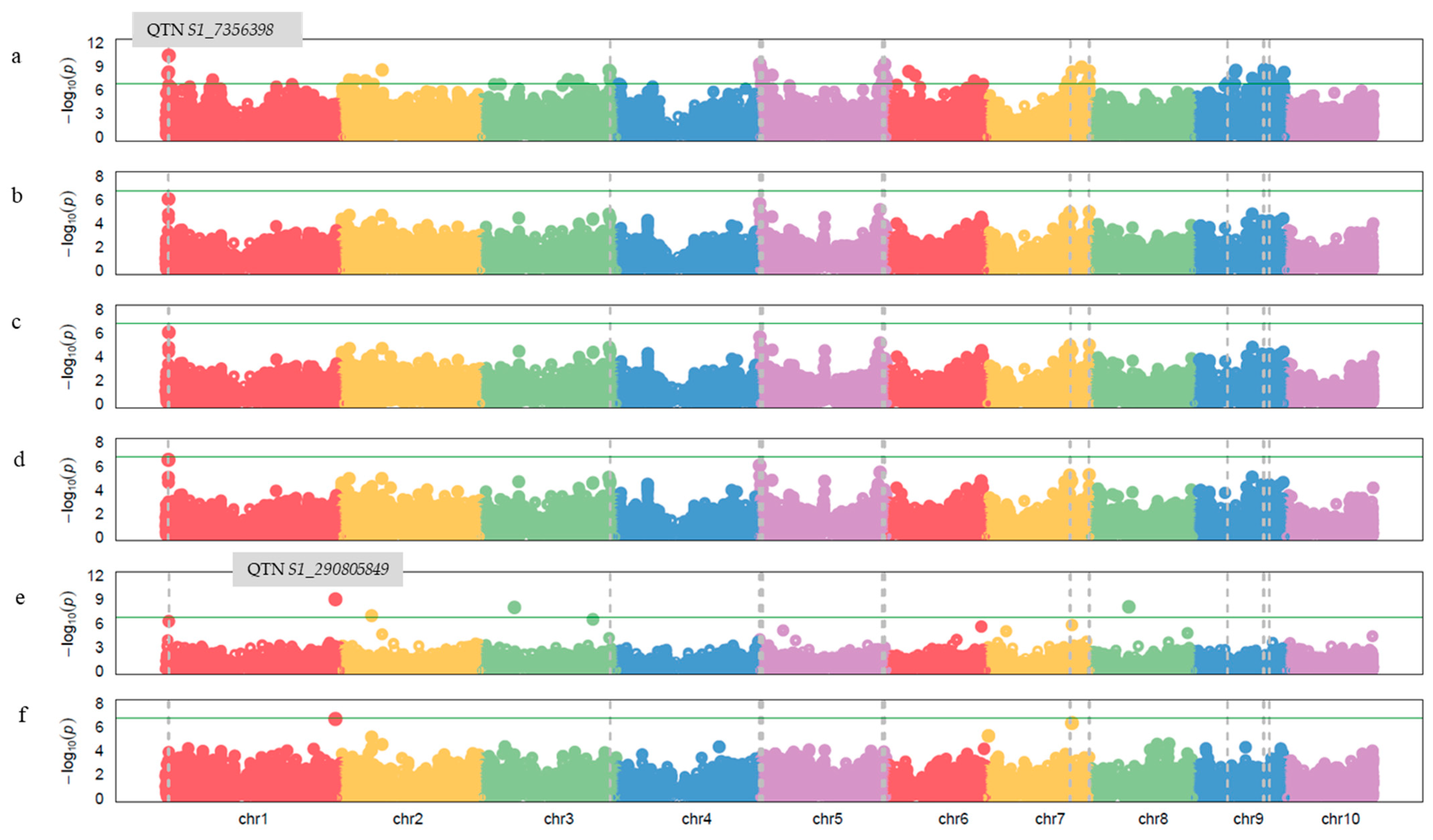
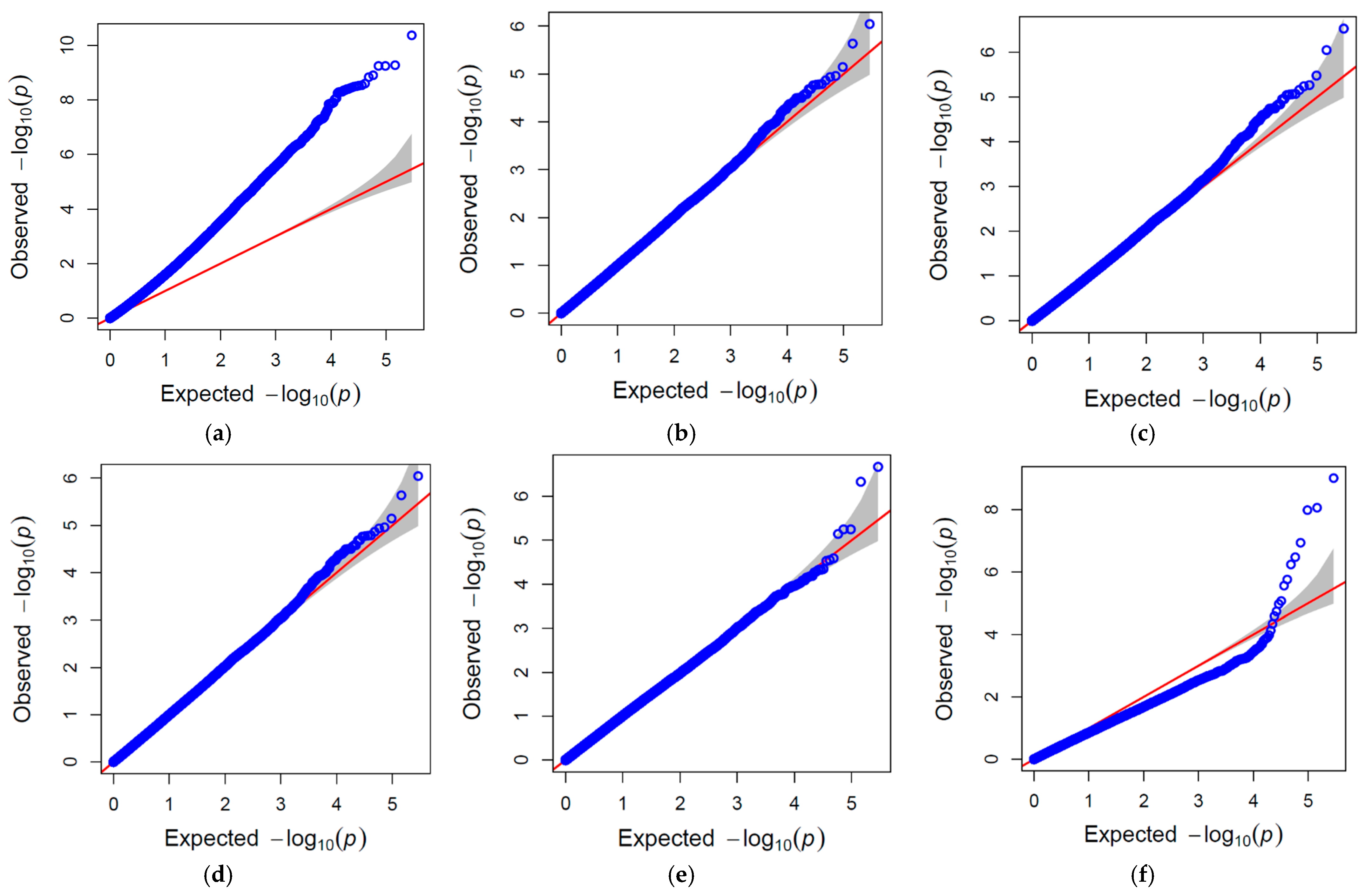
2.2. GWAS Analysis and Candidate Gene Identification
3. Discussion
4. Materials and Methods
4.1. Screening for NCLB
4.2. Statistical Analyses
4.3. Genotyping
4.4. Population Structure and LD Analysis
4.5. GWAS Analysis
4.6. QQ-Plot and Manhattan Plot
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, R.; Mani, V.P.; Koranga, K.S.; Khendelwal, R.S.; Bhandari, P.; Pant, S.K. Identification of additional sources of resistance to Exserohilum turcicum in maize. SABRAO J. Breed. Genet. 2004, 36, 45–47. [Google Scholar]
- De Rossi, R.L.; Guerra, F.A.; Plazas, M.C.; Vuletic, E.E.; Brücher, E.; Guerra, G.D.; Reis, E.M. Crop damage, economic losses, and the economic damage threshold for northern corn leaf blight. Crop Prot. 2022, 154, 105901. [Google Scholar] [CrossRef]
- Galiano-Carneiro, A.L.; Miedaner, T. Genetics of resistance and pathogenicity in the maize/Setosphaeria turcica pathosystem and implications for breeding. Front. Plant Sci. 2017, 8, 1490. [Google Scholar] [CrossRef]
- Rashid, Z.; Sofi, M.; Harlapur, S.I.; Kachapur, R.M.; Dar, Z.A.; Singh, P.K.; Zaidi, P.H.; Vivek, B.S.; Nair, S.K. Genome-wide association studies in tropical maize germplasm reveal novel and known genomic regions for resistance to northern corn leaf blight. Sci. Rep. 2020, 10, 21949. [Google Scholar] [CrossRef]
- Welz, H.G.; Geiger, H.H. Genes for resistance to northern corn leaf blight in diverse maize populations. Plant Breed. 2000, 119, 1–14. [Google Scholar] [CrossRef]
- Ranganatha, H.M.; Lohithaswa, H.C.; Pandravada, A. Mapping and validation of major quantitative trait loci for resistance to northern corn leaf blight along with the determination of the relationship between resistance to multiple foliar pathogens of maize. Front. Genet. 2021, 11, 548407. [Google Scholar] [CrossRef]
- Lohithaswa, H.C.; Balasundara, D.C.; Mallikarjuna, M.G.; Sowmya, M.S.; Mallikarjuna, N.; Kulkarni, R.S.; Pandravada, A.S.; Bhatia, B.S. Experimental evaluation of effectiveness of genomic selection for resistance to northern corn leaf blight in maize. J. Appl. Genet. 2024, 66, 493–521. [Google Scholar] [CrossRef]
- Young, N.D. QTL mapping and quantitative disease resistance in plants. Annu. Rev. Phytopathol. 1996, 34, 479–501. [Google Scholar] [CrossRef]
- Chen, G.; Wang, X.; Long, S.; Jaqueth, J.; Li, B.; Yan, J.; Ding, J. Mapping of QTL conferring resistance to northern corn leaf blight using high-density SNPs in maize. Mol. Breed. 2016, 36, 4. [Google Scholar] [CrossRef]
- Wang, J.; Xu, Z.; Yang, J.; Lu, X.; Zhou, Z.; Zhang, C.; Zheng, L.; Tian, R.; Hao, Z.; Yong, H.; et al. qNCLB7.02, a novel QTL for resistance to northern corn leaf blight in maize. Mol. Breed. 2018, 38, 54. [Google Scholar] [CrossRef]
- Crossa, J.; Perez-Rodriguez, P.; Cuevas, J.; Montesinos-Lopez, O.; Jarquin, D.; de los Campos, G.; Burgueno, J.; Gonzalez-Camacho, J.M.; Perez-Elizalde, S.; Beyene, Y.; et al. Genomic selection in plant breeding: Methods, models, and perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Xue, Y.; Tang, X.; Zhu, X.; Zhang, R.; Yao, Y.; Cao, D.; He, W.; Liu, Q.; Luan, X.; Shu, Y.; et al. Leveraging GWAS-Identified Markers in Combination with Bayesian and Machine Learning Models to Improve Genomic Selection in Soybean. Intl. J. Mol. Sci. 2025, 26, 9586. [Google Scholar] [CrossRef]
- Zheng, Y.; Yuan, F.; Huang, Y.; Zhao, Y.; Jia, X.; Zhu, L.; Guo, J. Genome-wide association studies of grain quality traits in maize. Sci. Rep. 2021, 11, 9797. [Google Scholar] [CrossRef]
- Van Inghelandt, D.; Melchinger, A.E.; Martinant, J.P.; Stich, B. Genome-wide association mapping of flowering time and northern corn leaf blight (Setosphaeria turcica) resistance in a vast commercial maize germplasm set. BMC Plant Biol. 2012, 12, 56. [Google Scholar] [CrossRef]
- Ding, J.; Ali, F.; Chen, G.; Li, H.; Mahuku, G.; Yang, N.; Narro, L.; Magorokosho, C.; Makumbi, D.; Yan, J. Genome-wide QTL mapping for NCLB resistance in tropical maize. BMC Plant Biol. 2015, 15, 206. [Google Scholar] [CrossRef]
- Omondi, D.O.; Dida, M.M.; Berger, D.K.; Beyene, Y.; Nsibo, D.L.; Juma, C.; Mahabaleswara, S.L.; Gowda, M. Combination of linkage and association mapping with genomic prediction to infer QTL regions associated with gray leaf spot and northern corn leaf blight resistance in tropical maize. Front. Genet. 2023, 14, 1282673. [Google Scholar] [CrossRef]
- Bi, Y.; Jiang, X.; Yin, X.; Shaw, R.K.; Guo, R.; Wang, J.; Fan, X. Identification of candidate gene associated with maize northern leaf blight resistance in a multi-parent population. Plant Cell Rep. 2024, 43, 189. [Google Scholar] [CrossRef]
- Poland, J.A.; Bradbury, P.J.; Buckler, E.S.; Nelson, R.J. Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc. Natl. Acad. Sci. USA 2011, 108, 6893–6898. [Google Scholar] [CrossRef]
- Li, C.; Ling, F.; Su, G.; Sun, W.; Liu, H.; Su, Y.; Qi, X. Location and mapping of the NCLB resistance genes in maize by bulked segregant analysis using whole genome re-sequencing. Mol. Breed. 2020, 40, 92. [Google Scholar] [CrossRef]
- Zhai, R.; Huang, A.; Mo, R.; Zou, C.; Wei, X.; Yang, M.; Tan, H.; Huang, K.; Qin, J. SNP-based bulk segregant analysis revealed disease resistance QTLs associated with northern corn leaf blight in maize. Front. Genet. 2022, 13, 1038948. [Google Scholar] [CrossRef]
- Cazares-Alvarez, J.E.; Baez-Astorga, P.A.; Arroyo-Becerra, A.; Maldonado-Mendoza, E.I. Genome-wide identification of a maize chitinase gene family and the induction of its expression by Fusarium verticillioides infection. Genes 2024, 15, 1087. [Google Scholar] [CrossRef] [PubMed]
- Hughes, G.R.; Hooker, A.L. Gene action conditioning resistance to northern corn leaf blight in maize. Crop Sci. 1971, 11, 180–183. [Google Scholar] [CrossRef]
- Zhou, G.; Hao, D.; Xue, L.; Chen, G.; Lu, H.; Zhang, Z.; Shi, M.; Huang, X.; Mao, Y. Genome-wide association study of kernel moisture content at harvest stage in maize. Breed. Sci. 2018, 68, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Cebeci, Z.; Bayraktar, M.; Gokce, G. Comparison of the statistical methods for genome-wide association studies on simulated quantitative traits of domesticated goats (Capra hircus L.). Small Rumin. Res. 2023, 227, 107053. [Google Scholar] [CrossRef]
- He, L.; Xiao, J.; Rashid, K.Y.; Yao, Z.; Li, P.; Jia, G.; Wang, X.; Cloutier, S.; You, F.M. Genome-wide association studies for pasmo resistance in flax (Linum usitatissimum L.). Front. Plant Sci. 2019, 9, 1982. [Google Scholar] [CrossRef]
- Yin, B.; Xu, L.; Li, J.; Zheng, Y.; Song, W.; Hou, P.; Zhu, L.; Jia, X.; Zhao, Y.; Song, W.; et al. Integrated transcriptome and GWAS analysis to identify candidate genes for Ustilago maydis resistance in maize. Agriculture 2024, 14, 958. [Google Scholar] [CrossRef]
- Qian, F.; Jing, J.; Zhang, Z.; Chen, S.; Sang, Z.; Li, W. GWAS and meta-QTL analysis of yield-related ear traits in maize. Plants 2023, 12, 3806. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767, Erratum in PLoS Genet. 2016, 12, e1005957. [Google Scholar] [CrossRef]
- Manghwar, H.; Hussain, A.; Ali, Q.; Liu, F. Brassinosteroids (BRs) role in plant development and coping with different stresses. Int. J. Mol. Sci. 2022, 23, 1012. [Google Scholar] [CrossRef]
- Jia, H.; Li, M.; Li, W.; Liu, L.; Jian, Y.; Yang, Z.; Shen, X.; Ning, Q.; Du, Y.; Zhao, R.; et al. A serine/threonine protein kinase encoding gene KERNEL NUMBER PER ROW6 regulates maize grain yield. Nat. Commun. 2020, 11, 988. [Google Scholar] [CrossRef]
- Poloni, A.; Garde, R.; Dittiger, L.D.; Heidrich, T.; Muller, C.; Drechsler, F.; Zhao, Y.; Mazumdar, T.; Schirawski, J. Transcriptome analysis reveals contrasting plant responses of Sorghum bicolor upon colonization by two formae speciales of Sporisorium reilianum. Int. J. Mol. Sci. 2022, 23, 8864. [Google Scholar] [CrossRef]
- Chung, C.; Longfellow, J.M.; Walsh, E.K.; Kerdieh, Z.; Van Esbroeck, G.; Balint-Kurti, P.; Nelson, R.J. Resistance loci affecting distinct stages of fungal pathogenesis: Use of introgression lines for QTL mapping and characterization in the maize-Setosphaeria turcica pathosystem. BMC Plant Biol. 2010, 10, 103. [Google Scholar] [CrossRef]
- Mitiku, M.; Eshte, Y.; Shiferaw, W. Evaluation of maize variety for northern leaf blight (Trichometasphaeria turcica) in south Omo zone. World J. Agric. Res. 2014, 2, 237–239. [Google Scholar] [CrossRef]
- Robinson, H.F.; Comstock, R.E.; Harvey, P.H. Estimates of heritability and the degree of dominance in corn. Agron. J. 1949, 41, 353–359. [Google Scholar] [CrossRef]
- Johnson, H.W.; Robinson, H.F.; Comstock, R.E. Estimates of genetic and environmental variability in soybeans. Agron. J. 1955, 47, 314–318. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Yu, J.M.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, B.J.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Zhang, Z.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Segura, V.; Vilhjálmsson, B.J.; Platt, A.; Korte, A.; Seren, Ü.; Long, Q.; Nordborg, M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 2012, 44, 825–830. [Google Scholar] [CrossRef]



| Source of Variation | Degrees of Freedom | Mean Sum of Squares | |
|---|---|---|---|
| Season 1 | Season 2 | ||
| Entry | 335 | 2.06 *** | 2.42 *** |
| Replication | 1 | 1.74 | 1.90 * |
| Replication: Block | 22 | 1.66 | 1.71 |
| Residuals | 313 | 0.24 | 0.77 |
| Source of Variation | Degrees of Freedom | Mean Sum of Squares |
|---|---|---|
| Genotype | 335 | 117.70 *** |
| Year | 1 | 7.70 * |
| Replication | 1 | 6.46 |
| Genotype: Year | 335 | 48.59 ** |
| Residuals | 1339 | 1.40 |
| QTNs | Chromosome | Candidate Gene | Gene Name | Predicted Function | Reference |
|---|---|---|---|---|---|
| S1_7356398 (downstream) | 1 | GRMZM2G099598 | Chitinase | Defence response to fungus Polysaccharide catabolic process Cell wall macromolecule catabolic process Chitinase activity Chitin catabolic process | [21] |
| 1 | GRMZM2G099598 | Putative serine/threonine protein kinase | Protein phosphorylation Brassinosteroid mediated signalling pathway Protein kinase activity ATP binding Salt and drought tolerance | - | |
| S1_290805849 (upstream) | 1 | GRMZM2G038964 | Aldehyde oxygenase (deformylating) | Fatty acid biosynthetic process Iron ion binding Fatty acid alpha-hydroxylase activity Aldehyde oxygenase (deformylating) activity Fatty acid metabolic process Octadecanal decarbonylase activity | - |
| S1_290805849 (downstream) | 1 | GRMZM2G038494 | Obg-like ATPase 1; OBG-type G domain-containing protein | ATP hydrolysis activity Ribosomal large subunit binding ATP binding GTP binding | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shetty, U.; Sowmya, M.S.; Lohithaswa, H.C.; Mallikarjuna, M.G.; Jadesha, G.; Nayaka, S.C. Genome-Wide Association Study Reveals Novel QTNs and Candidate Genes Implicated in Resistance to Northern Corn Leaf Blight in Maize (Zea mays L.). Int. J. Mol. Sci. 2025, 26, 10677. https://doi.org/10.3390/ijms262110677
Shetty U, Sowmya MS, Lohithaswa HC, Mallikarjuna MG, Jadesha G, Nayaka SC. Genome-Wide Association Study Reveals Novel QTNs and Candidate Genes Implicated in Resistance to Northern Corn Leaf Blight in Maize (Zea mays L.). International Journal of Molecular Sciences. 2025; 26(21):10677. https://doi.org/10.3390/ijms262110677
Chicago/Turabian StyleShetty, Udaya, Muntagodu Shreekanth Sowmya, Hirenallur Chandappa Lohithaswa, Mallana Goudra Mallikarjuna, Ganiga Jadesha, and Siddaiah Chandra Nayaka. 2025. "Genome-Wide Association Study Reveals Novel QTNs and Candidate Genes Implicated in Resistance to Northern Corn Leaf Blight in Maize (Zea mays L.)" International Journal of Molecular Sciences 26, no. 21: 10677. https://doi.org/10.3390/ijms262110677
APA StyleShetty, U., Sowmya, M. S., Lohithaswa, H. C., Mallikarjuna, M. G., Jadesha, G., & Nayaka, S. C. (2025). Genome-Wide Association Study Reveals Novel QTNs and Candidate Genes Implicated in Resistance to Northern Corn Leaf Blight in Maize (Zea mays L.). International Journal of Molecular Sciences, 26(21), 10677. https://doi.org/10.3390/ijms262110677






