Analysis of MSX1, RYK, NFκB p65, and CCL4 Proteins and MSX2, RYK, and PTX3 Genes in Human Cleft Lip Tissue
Abstract
1. Introduction
2. Results
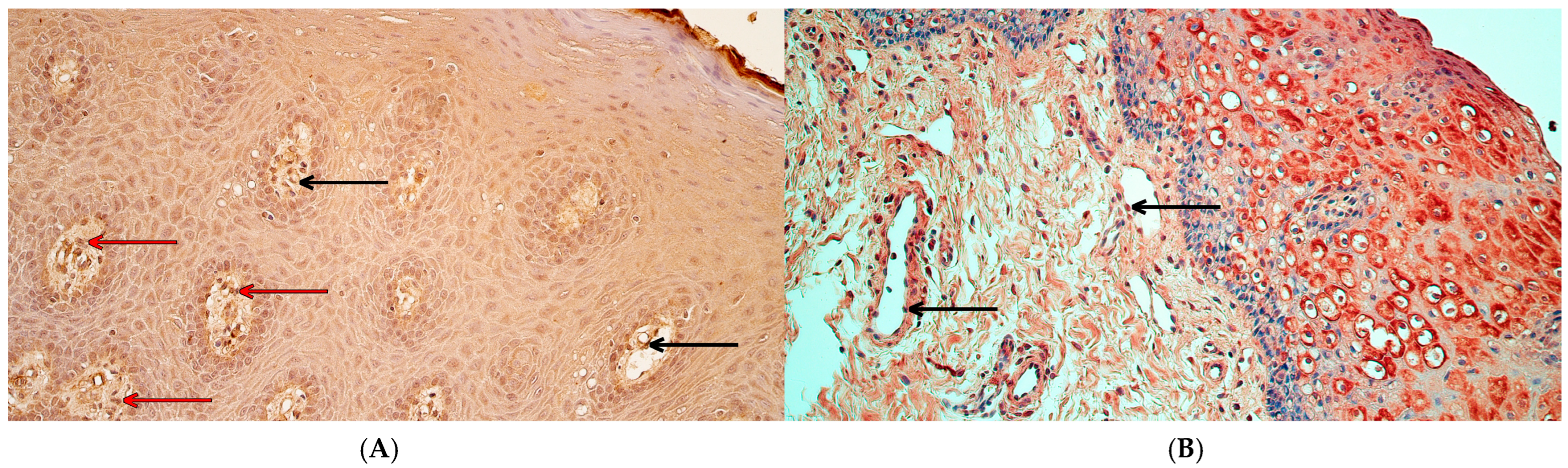
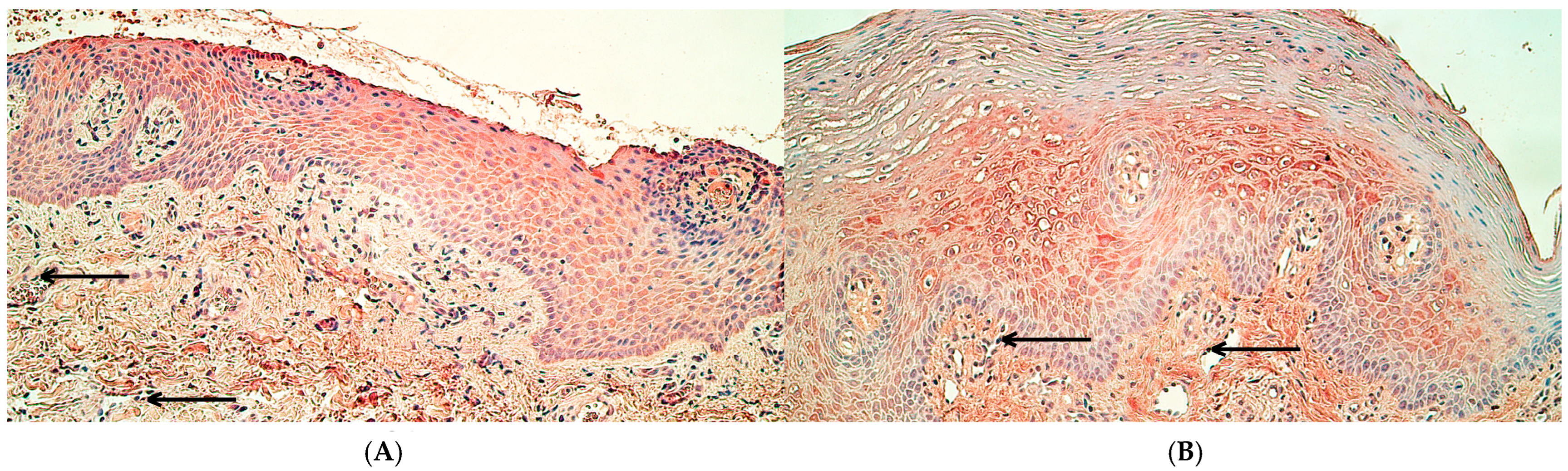
2.1. Immunohistochemistry
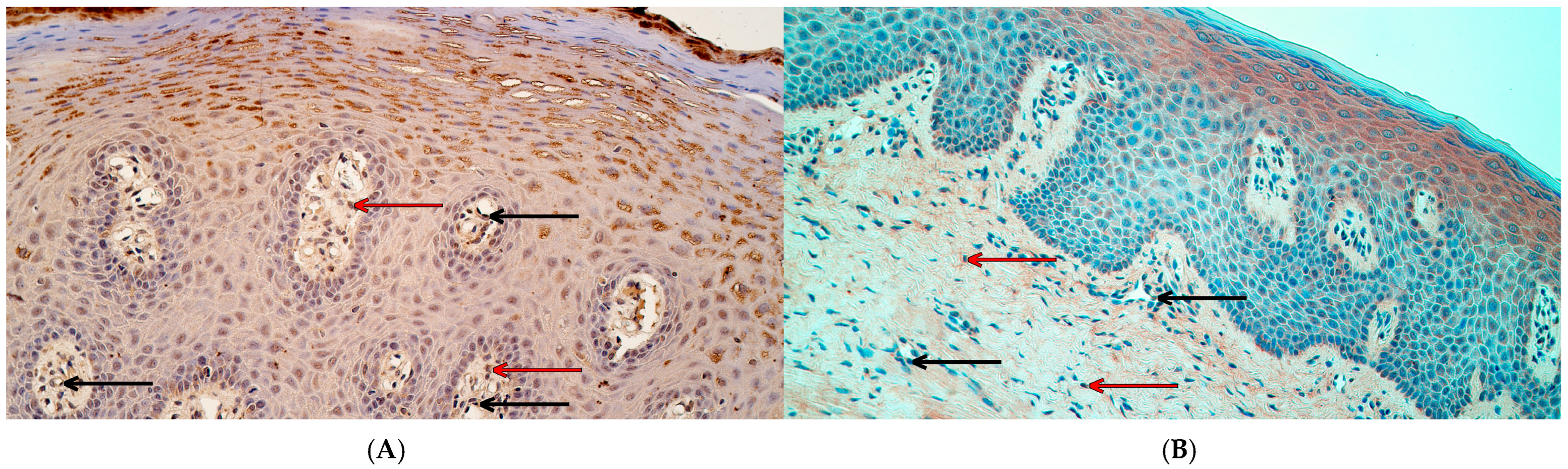
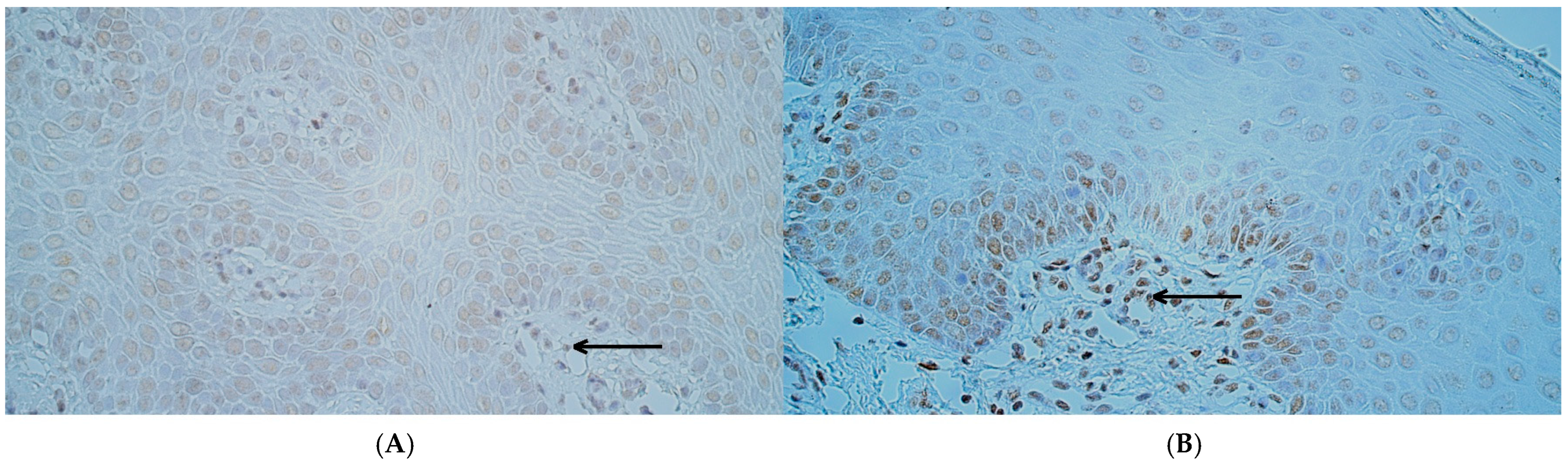
2.2. Chromogenic In Situ Hybridization
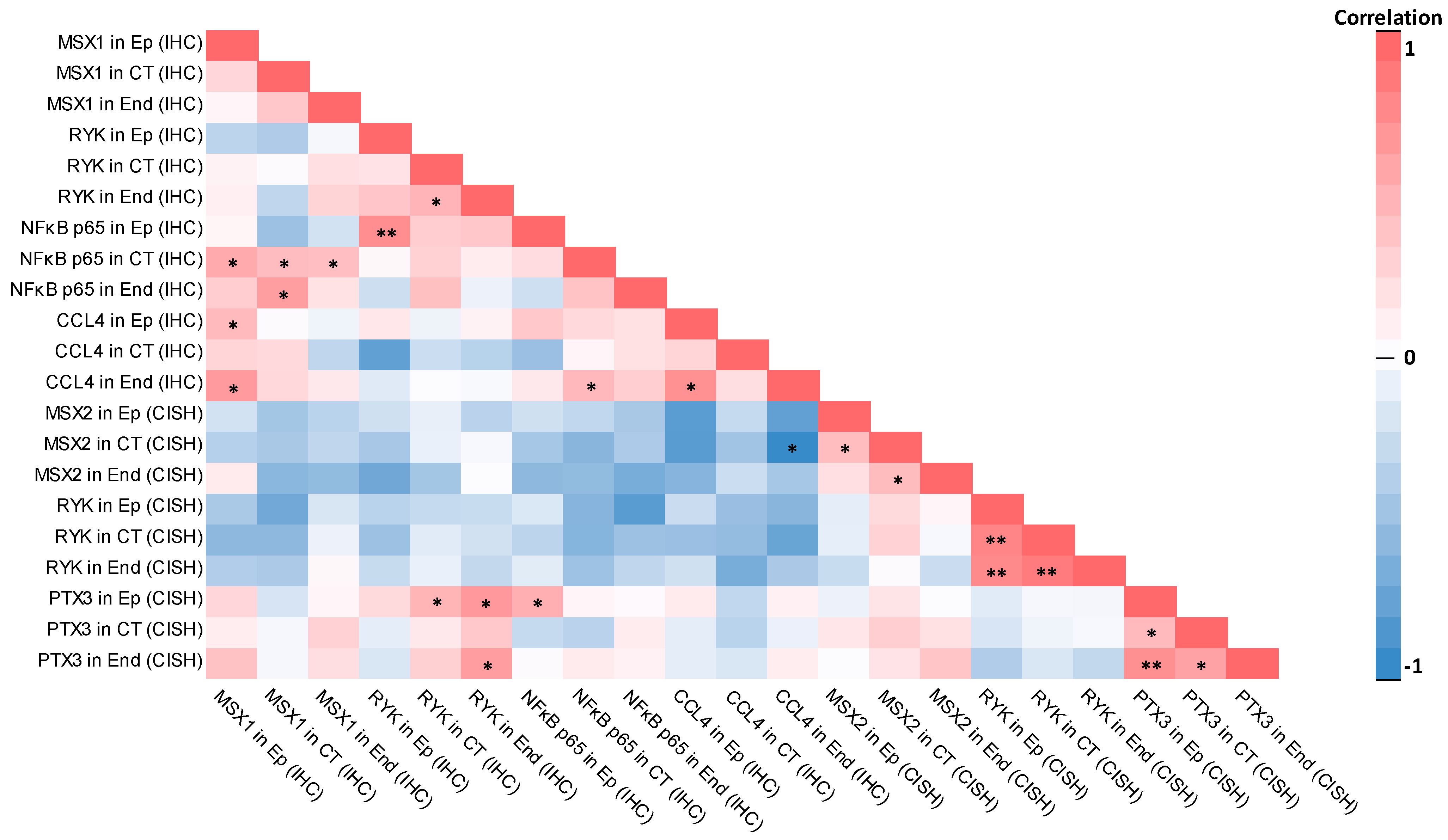
2.3. Correlations
2.3.1. Correlations in Control Group
2.3.2. Correlations in Patient Group
3. Discussion
3.1. Immunoreactivity of MSX1, NFκB p65, and CCL4 Proteins Was Significantly Decreased in Cleft Lip Connective Tissue and Endothelium but Not in Surface Epithelium
3.2. RYK Protein Immunoreactivity Was Significantly Decreased Only in Cleft Lip-Affected Connective Tissue
3.3. The Number of MSX2 and RYK Gene-Signal-Containing Cells Was Significantly Increased in Cleft Lip Tissue While the Number PTX3 Gene-Signal-Containing Cells Did Not Differ from Controls
3.4. Statistically Significant Correlations Between Factors Were Mostly Different Within Control and Patient Groups
3.5. Limitations of This Study
3.6. Clinical Application of Data and Possible Future Research Directions
4. Materials and Methods
4.1. Patient Group and Control Group Characteristics
- Diagnosis of cleft lip;
- Cleft lip surgery performed before/during the age of primary dentition;
- No malignancy, excessive inflammation, fibrosis, or any other pathological change in the soft tissue of the lip present.
- Individuals without cleft lip and palate;
- No cleft lip and palate in family anamnesis;
- Individuals before/during the age of primary dentition (or as close as possible);
- No malignancy, excessive inflammation, fibrosis, or any other pathological change in the soft tissue of the lip present.
4.2. Immunohistochemistry (IHC) and Chromogenic In Situ Hybridization (CISH)
- MSX1 antibodies (LS-C47382/11448, rabbit, polyclonal, dilution 1:200, LifeSpan BioSciences Inc., Seattle, WA, USA);
- RYK antibodies (orb38371, rabbit, polyclonal, dilution 1:100, Biorbyt Ltd., Cambridge, UK);
- NFκB p65 antibodies (orb37069, rabbit, polyclonal, dilution 1:100, Biorbyt Ltd., Cambridge, UK);
- CCL4 antibodies (ab235978, rabbit, polyclonal, dilution 1:100, Abcam Inc., Cambridge, UK).
- MSX2 probe (MSX2-20-DIG, Empire Genomics Corp., Williamsville, NY, USA);
- RYK probe (RYK-20-DIG, Empire Genomics Corp., Williamsville, NY, USA);
- PTX3 probe (PTX3-20-DIG, Empire Genomics Corp., Williamsville, NY, USA).
4.3. Semiquantitative Counting Method and Data Analysis
- 0—no protein-containing/gene-signal-containing cells in the visual field (0.0%);
- 0/+—a rare occurrence of protein-containing/gene-signal-containing cells in the visual field (0.0–12.5%);
- +—a few protein-containing/gene-signal-containing cells in the visual field (12.5–25.0%);
- +/++—few to moderate protein-containing/gene-signal-containing cells in the visual field (25.0–37.5%);
- ++—a moderate number of protein-containing/gene-signal-containing cells in the visual field (37.5–50.0%);
- ++/+++—moderate to numerous protein-containing/gene-signal-containing cells in the visual field (50.0–62.5%),
- +++—numerous protein-containing/gene-signal-containing cells in the visual field (62.5–75.0%),
- +++/++++—numerous to abundant protein-containing/gene-signal-containing cells in the visual field (75.0–87.5%);
- ++++—abundant protein-containing/gene-signal-containing cells in the visual field (87.5–100.0%).
5. Conclusions
- 1.
- Immunoreactivity of MSX1, NFκB p65, and CCL4 proteins was significantly decreased in cleft lip connective tissue and endothelium but not in surface epithelium; a decrease in MSX1 has been previously associated with disturbed growth and fusion of the developing upper lip primordia, while the decrease in NFκB p65 and CCL4 proteins could be explained by the characteristics of the patient group and lack of active inflammation within tissue.
- 2.
- RYK protein immunoreactivity was significantly decreased only in cleft lip-affected connective tissue, possibly due to a disrupted WNT signaling pathway in cleft lip tissue.
- 3.
- The number of MSX2 and RYK gene-signal-containing cells was significantly increased in cleft lip tissue, probably due to WNT signaling disturbances and gene activation in cleft-affected tissue.
- 4.
- The number of PTX3 gene-signal-containing cells did not differ from controls, possibly due to characteristics of the patient group and the lack of active inflammation in tissue.
- 5.
- Statistically significant correlations between factors were mostly different within the control and patient groups, with some minor overlap being present; negative correlations predominantly were identified in the control group and not the cleft lip tissue group, indicating a disturbance of factor regulation within cleft lip tissue; positive correlations in the cleft lip group mainly involved the PTX3 gene, NFκB p65, and MSX1 protein, which was not seen in the control group, again possibly due to disturbed signaling pathways in cleft lip tissue.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MSX1 | Muscle segment homeobox 1 |
| MSX2 | Muscle segment homeobox 2 |
| RYK | Receptor-like tyrosine kinase |
| NFκB p65 | Nuclear factor kappa-light-chain-enhancer of activated B cells protein 65 |
| CCL4 | C-C motif chemokine ligand 4 |
| PTX3 | Pentraxin 3 |
| IHC | Immunohistochemistry |
| CISH | Chromogenic in situ hybridization |
| RSU | Rīga Stradiņš University |
| NFκB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| WNT | Wingless-related integration site |
| TNF-α | Tumor necrosis factor alpha |
| DIG | Digoxigenin |
| DNP | Dinitrophenol |
| AP | Alkaline phosphatase |
| HRP | Horseradish peroxidase |
| Ep | Epithelium |
| CT | Connective tissue |
| End | Endothelium |
| SPSS | Statistical Product and Service Solutions |
| U | Mann–Whitney U test value |
| p | p-value |
References
- Sandy, J.; Davies, A.; Humphries, K.; Ireland, T.; Wren, Y. Cleft Lip and Palate: Care Configuration, National Registration, and Research Strategies. J. World Fed. Orthod. 2020, 9, S40–S44. [Google Scholar] [CrossRef]
- Smarius, B.; Loozen, C.; Manten, W.; Bekker, M.; Pistorius, L.; Breugem, C. Accurate Diagnosis of Prenatal Cleft Lip/Palate by Understanding the Embryology. World J. Methodol. 2017, 7, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Fell, M.; Bradley, D.; Chadha, A.; Butterworth, S.; Davies, A.; Russell, C.; Richard, B.; Wren, Y.; Lewis, S.; Chong, D. Sidedness in Unilateral Orofacial Clefts: A Systematic Scoping Review. Cleft Palate Craniofac. J. 2025, 62, 730–743. [Google Scholar] [CrossRef] [PubMed]
- Shkoukani, M.A.; Chen, M.; Vong, A. Cleft Lip—A Comprehensive Review. Front. Pediatr. 2013, 1, 53. [Google Scholar] [CrossRef]
- Babai, A.; Irving, M. Orofacial Clefts: Genetics of Cleft Lip and Palate. Genes 2023, 14, 1603. [Google Scholar] [CrossRef] [PubMed]
- Leslie, E.J.; Marazita, M.L. Genetics of Cleft Lip and Cleft Palate. Am. J. Med. Genet. C Semin. Med. Genet. 2013, 163C, 246–258. [Google Scholar] [CrossRef]
- Goida, J.; Pilmane, M. The Presence and Distribution of Various Genes in Postnatal CLP-Affected Palatine Tissue. Maxillofac. Plast. Reconstr. Surg. 2024, 46, 1. [Google Scholar] [CrossRef]
- Sakuma, C.; Imura, H.; Yamada, T.; Hirata, A.; Ikeda, Y.; Ito, M.; Natsume, N. Histological and Immunohistochemical Studies to Determine the Mechanism of Cleft Palate Induction after Palatal Fusion in Mice Exposed to TCDD. Int. J. Mol. Sci. 2022, 23, 2069. [Google Scholar] [CrossRef]
- Siewert, A.; Reiz, B.; Krug, C.; Heggemann, J.; Mangold, E.; Dickten, H.; Ludwig, K.U. Analysis of Candidate Genes for Cleft Lip ± Cleft Palate Using Murine Single-Cell Expression Data. Front. Cell Dev. Biol. 2023, 11, 1091666. [Google Scholar] [CrossRef]
- Babajko, S.; de La Dure-Molla, M.; Jedeon, K.; Berdal, A. MSX2 in Ameloblast Cell Fate and Activity. Front. Physiol. 2014, 5, 510. [Google Scholar] [CrossRef]
- Han, J.; Ishii, M.; Bringas, P., Jr.; Maas, R.L.; Maxson, R.E., Jr.; Chai, Y. Concerted Action of Msx1 and Msx2 in Regulating Cranial Neural Crest Cell Differentiation during Frontal Bone Development. Mech. Dev. 2007, 124, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Pengelly, R.J.; Arias, L.; Martínez, J.; Upstill-Goddard, R.; Seaby, E.G.; Gibson, J.; Ennis, S.; Collins, A.; Briceño, I. Deleterious Coding Variants in Multi-Case Families with Non-Syndromic Cleft Lip and/or Palate Phenotypes. Sci. Rep. 2016, 6, 30457. [Google Scholar] [CrossRef][Green Version]
- Yu, Y.; Zuo, X.; He, M.; Gao, J.; Fu, Y.; Qin, C.; Meng, L.; Wang, W.; Song, Y.; Cheng, Y.; et al. Genome-Wide Analyses of Non-Syndromic Cleft Lip with Palate Identify 14 Novel Loci and Genetic Heterogeneity. Nat. Commun. 2017, 8, 14364. [Google Scholar] [CrossRef]
- Gajera, M.; Desai, N.; Suzuki, A.; Li, A.; Zhang, M.; Jun, G.; Jia, P.; Zhao, Z.; Iwata, J. MicroRNA-655-3p and microRNA-497-5p Inhibit Cell Proliferation in Cultured Human Lip Cells through the Regulation of Genes Related to Human Cleft Lip. BMC Med. Genomics. 2019, 12, 70. [Google Scholar] [CrossRef]
- Khadka, D.; Luo, T.; Sargent, T.D. Msx1 and Msx2 Have Shared Essential Functions in Neural Crest but May Be Dispensable in Epidermis and Axis Formation in Xenopus. Int. J. Dev. Biol. 2006, 50, 499–502. [Google Scholar] [CrossRef]
- Catron, K.M.; Wang, H.; Hu, G.; Shen, M.M.; Abate-Shen, C. Comparison of MSX-1 and MSX-2 Suggests a Molecular Basis for Functional Redundancy. Mech. Dev. 1996, 55, 185–199. [Google Scholar] [CrossRef]
- Green, J.; Nusse, R.; van Amerongen, R. The Role of Ryk and Ror Receptor Tyrosine Kinases in Wnt Signal Transduction. Cold Spring Harb. Perspect. Biol. 2014, 6, a009175. [Google Scholar] [CrossRef]
- Zapata-García, J.A.; Jave-Suárez, L.F.; Aguilar-Lemarroy, A. Delving into the Role of Receptor-like Tyrosine Kinase (RYK) in Cancer: In Silico Insights into Its Diagnostic and Prognostic Utility. J. Mol. Pathol. 2024, 5, 66–80. [Google Scholar] [CrossRef]
- Kim, H.-T.; Panza, P.; Kikhi, K.; Nakamichi, Y.; Atzberger, A.; Guenther, S.; Ruppert, C.; Guenther, A.; Stainier, D.Y.R. WNT/RYK Signaling Functions as an Antiinflammatory Modulator in the Lung Mesenchyme. Proc. Natl. Acad. Sci. USA 2022, 119, e2201707119. [Google Scholar] [CrossRef]
- Zhong, J.; Kim, H.-T.; Lyu, J.; Yoshikawa, K.; Nakafuku, M.; Lu, W. The Wnt Receptor Ryk Controls Specification of GABAergic Neurons versus Oligodendrocytes during Telencephalon Development. Development 2011, 138, 409–419. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lanoue, V.; Langford, M.; White, A.; Sempert, K.; Fogg, L.; Cooper, H.M. The Wnt Receptor Ryk Is a Negative Regulator of Mammalian Dendrite Morphogenesis. Sci. Rep. 2017, 7, 5965. [Google Scholar] [CrossRef]
- Watanabe, A.; Akita, S.; Tin, N.T.D.; Natsume, N.; Nakano, Y.; Niikawa, N.; Uchiyama, T.; Yoshiura, K.-I. A Mutation in RYK Is a Genetic Factor for Nonsyndromic Cleft Lip and Palate. Cleft Palate Craniofac. J. 2006, 43, 310–316. [Google Scholar] [CrossRef]
- Giridharan, S.; Srinivasan, M. Mechanisms of NF-κB P65 and Strategies for Therapeutic Manipulation. J. Inflamm. Res. 2018, 11, 407–419. [Google Scholar] [CrossRef]
- Du, Y.; Yang, Y.; Zhang, W.; Yang, C.; Xu, P. Human β-Defensin-3 and Nuclear Factor-Kappa B P65 Synergistically Promote the Cell Proliferation and Invasion of Oral Squamous Cell Carcinoma. Transl. Oncol. 2023, 27, 101582. [Google Scholar] [CrossRef]
- Kamperos, G.; Nikitakis, N.; Sfakianou, A.; Avgoustidis, D.; Sklavounou-Andrikopoulou, A. Expression of NF-κB and IL-6 in Oral Precancerous and Cancerous Lesions: An Immunohistochemical Study. Med. Oral Patol. Oral Cir. Bucal 2016, 21, e6–e13. [Google Scholar] [CrossRef]
- Li, J.-H.; Yu, J.-P.; Yu, H.-G.; Xu, X.-M.; Yu, L.-L.; Liu, S.-Q. Expression and Significance of Nuclear Factor kappaB P65 in Colon Tissues of Rats with TNBS-Induced Colitis. World J. Gastroenterol. 2005, 11, 1759–1763. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.-L.; Yu, H.-G.; Yu, J.-P.; Luo, H.-S.; Xu, X.-M.; Li, J.-H. Nuclear Factor-kappaB P65 (RelA) Transcription Factor Is Constitutively Activated in Human Colorectal Carcinoma Tissue. World J. Gastroenterol. 2004, 10, 3255–3260. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-I.; Kim, Y.-B.; Koh, K.-M.; Youn, Y.-K.; Suh, G.-J.; Cho, E.-S.; Leem, D.-H.; Baek, J.-A.; Shin, H.-K.; Ko, S.-O. Activation of NF-κB Pathway in Oral Buccal Mucosa during Small Intestinal Ischemia-Reperfusion Injury. J. Surg. Res. 2013, 179, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.H.; Kobayashi, Y.; Van Bui, D.; Yun, Y.; Nguyen, L.M.; Mitani, A.; Suzuki, K.; Asako, M.; Kanda, A.; Iwai, H. CCL4 Regulates Eosinophil Activation in Eosinophilic Airway Inflammation. Int. J. Mol. Sci. 2022, 23, 16149. [Google Scholar] [CrossRef]
- Mukaida, N.; Sasaki, S.-I.; Baba, T. CCL4 Signaling in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1231, 23–32. [Google Scholar] [CrossRef]
- e la Fuente López, M.; Landskron, G.; Parada, D.; Dubois-Camacho, K.; Simian, D.; Martinez, M.; Romero, D.; Roa, J.C.; Chahuán, I.; Gutiérrez, R.; et al. The Relationship between Chemokines CCL2, CCL3, and CCL4 with the Tumor Microenvironment and Tumor-Associated Macrophage Markers in Colorectal Cancer. Tumour Biol. 2018, 40, 101042831881005. [Google Scholar] [CrossRef]
- Koch, A.E.; Kunkel, S.L.; Shah, M.R.; Fu, R.; Mazarakis, D.D.; Haines, G.K.; Burdick, M.D.; Pope, R.M.; Strieter, R.M. Macrophage Inflammatory Protein-1 Beta: A C-C Chemokine in Osteoarthritis. Clin. Immunol. Immunopathol. 1995, 77, 307–314. [Google Scholar] [CrossRef]
- O’Grady, N.P.; Tropea, M.; Preas, H.L., 2nd; Reda, D.; Vandivier, R.W.; Banks, S.M.; Suffredini, A.F. Detection of Macrophage Inflammatory Protein (MIP)-1alpha and MIP-1beta during Experimental Endotoxemia and Human Sepsis. J. Infect. Dis. 1999, 179, 136–141. [Google Scholar] [CrossRef]
- Ciechanowska, A.; Popiolek-Barczyk, K.; Pawlik, K.; Ciapała, K.; Oggioni, M.; Mercurio, D.; De Simoni, M.-G.; Mika, J. Changes in Macrophage Inflammatory Protein-1 (MIP-1) Family Members Expression Induced by Traumatic Brain Injury in Mice. Immunobiology 2020, 225, 151911. [Google Scholar] [CrossRef] [PubMed]
- Zehnder, M.; Greenspan, J.S.; Greenspan, D.; Bickel, M. Chemokine Gene Expression in Human Oral Mucosa. Eur. J. Oral Sci. 1999, 107, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-C.; Tsai, H.-C.; Yang, D.-Y.; Wang, S.-W.; Tsai, M.-H.; Hua, C.-H.; Chen, K.-J.; Chen, M.Y.-C.; Lien, M.-Y.; Tang, C.-H. The Chemokine CCL4 Stimulates Angiopoietin-2 Expression and Angiogenesis via the MEK/ERK/STAT3 Pathway in Oral Squamous Cell Carcinoma. Biomedicines 2022, 10, 1612. [Google Scholar] [CrossRef]
- Lien, M.-Y.; Tsai, H.-C.; Chang, A.-C.; Tsai, M.-H.; Hua, C.-H.; Wang, S.-W.; Tang, C.-H. Chemokine CCL4 Induces Vascular Endothelial Growth Factor C Expression and Lymphangiogenesis by miR-195-3p in Oral Squamous Cell Carcinoma. Front. Immunol. 2018, 9, 412. [Google Scholar] [CrossRef] [PubMed]
- Massimino, A.M.; Colella, F.E.; Bottazzi, B.; Inforzato, A. Structural Insights into the Biological Functions of the Long Pentraxin PTX3. Front. Immunol. 2023, 14, 1274634. [Google Scholar] [CrossRef]
- Woo, J.M.; Kwon, M.-Y.; Shin, D.-Y.; Kang, Y.-H.; Hwang, N.; Chung, S.W. Human Retinal Pigment Epithelial Cells Express the Long Pentraxin PTX3. Mol. Vis. 2013, 19, 303–310. [Google Scholar]
- Kim, Y.; Park, J.-S.; Park, H.-J.; Kim, M.-K.; Kim, Y.-I.; Bae, S.-K.; Kim, H.J.; Jeong, C.-H.; Bae, M.-K. Pentraxin 3 Modulates the Inflammatory Response in Human Dental Pulp Cells. J. Endod. 2018, 44, 1826–1831. [Google Scholar] [CrossRef]
- Brunetta, E.; Folci, M.; Bottazzi, B.; De Santis, M.; Gritti, G.; Protti, A.; Mapelli, S.N.; Bonovas, S.; Piovani, D.; Leone, R.; et al. Macrophage Expression and Prognostic Significance of the Long Pentraxin PTX3 in COVID-19. Nat. Immunol. 2021, 22, 19–24. [Google Scholar] [CrossRef]
- Jain, N.; Pilmane, M. Evaluating the Expression of Candidate Homeobox Genes and Their Role in Local-Site Inflammation in Mucosal Tissue Obtained from Children with Non-Syndromic Cleft Lip and Palate. J. Pers. Med. 2021, 11, 1135. [Google Scholar] [CrossRef]
- Blackburn, J.; Kawasaki, K.; Porntaveetus, T.; Kawasaki, M.; Otsuka-Tanaka, Y.; Miake, Y.; Ota, M.S.; Watanabe, M.; Hishinuma, M.; Nomoto, T.; et al. Excess NF-κB Induces Ectopic Odontogenesis in Embryonic Incisor Epithelium. J. Dent. Res. 2015, 94, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Geng, R.; Wang, Z.; Liu, H.; Wang, W. Anatomical Structure, and Expression of CCL4 and CCL13-like during the Development of Maxillary Barbel in Paramisgurnus Dabryanus. Organogenesis 2019, 15, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Park, J.-Y.; Park, H.-J.; Kim, M.-K.; Kim, Y.-I.; Kim, H.J.; Bae, S.-K.; Bae, M.-K. Pentraxin-3 Modulates Osteogenic/Odontogenic Differentiation and Migration of Human Dental Pulp Stem Cells. Int. J. Mol. Sci. 2019, 20, 5778. [Google Scholar] [CrossRef]
- Liang, J.; Von den Hoff, J.; Lange, J.; Ren, Y.; Bian, Z.; Carels, C.E.L. MSX1 Mutations and Associated Disease Phenotypes: Genotype-Phenotype Relations. Eur. J. Hum. Genet. 2016, 24, 1663–1670. [Google Scholar] [CrossRef]
- Ding, T.; Liu, H.; Yu, G. Novel MSX1 Gene Variants in Chinese Children with Non-Syndromic Tooth Agenesis: A Clinical and Genetic Analysis. Children 2024, 11, 1418. [Google Scholar] [CrossRef]
- Modesto, A.; Moreno, L.M.; Krahn, K.; King, S.; Lidral, A.C. MSX1 and Orofacial Clefting with and without Tooth Agenesis. J. Dent. Res. 2006, 85, 542–546. [Google Scholar] [CrossRef]
- Nakatomi, M.; Wang, X.-P.; Key, D.; Lund, J.J.; Turbe-Doan, A.; Kist, R.; Aw, A.; Chen, Y.; Maas, R.L.; Peters, H. Genetic Interactions between Pax9 and Msx1 Regulate Lip Development and Several Stages of Tooth Morphogenesis. Dev. Biol. 2010, 340, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Nakatomi, M.; Ludwig, K.U.; Knapp, M.; Kist, R.; Lisgo, S.; Ohshima, H.; Mangold, E.; Peters, H. Msx1 Deficiency Interacts with Hypoxia and Induces a Morphogenetic Regulation during Mouse Lip Development. Development 2020, 147, dev189175. [Google Scholar] [CrossRef]
- Vandersmissen, I.; Craps, S.; Depypere, M.; Coppiello, G.; van Gastel, N.; Maes, F.; Carmeliet, G.; Schrooten, J.; Jones, E.A.V.; Umans, L.; et al. Endothelial Msx1 Transduces Hemodynamic Changes into an Arteriogenic Remodeling Response. J. Cell Biol. 2015, 210, 1239–1256. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.; Goupille, O.; Saint Cloment, C.; Lallemand, Y.; Cumano, A.; Robert, B. Msx Genes Define a Population of Mural Cell Precursors Required for Head Blood Vessel Maturation. Development 2011, 138, 3055–3066. [Google Scholar] [CrossRef] [PubMed]
- Seidel, C.L.; Percivalle, E.; Tschaftari, M.; Weider, M.; Strobel, K.; Willershausen, I.; Unertl, C.; Schmetzer, H.M.; Weber, M.; Schneider, M.; et al. Orofacial Clefts Lead to Increased Pro-Inflammatory Cytokine Levels on Neonatal Oral Mucosa. Front. Immunol. 2022, 13, 1044249. [Google Scholar] [CrossRef]
- Mussbacher, M.; Salzmann, M.; Brostjan, C.; Hoesel, B.; Schoergenhofer, C.; Datler, H.; Hohensinner, P.; Basílio, J.; Petzelbauer, P.; Assinger, A.; et al. Cell Type-Specific Roles of NF-κB Linking Inflammation and Thrombosis. Front. Immunol. 2019, 10, 85. [Google Scholar] [CrossRef]
- Tatara, Y.; Ohishi, M.; Yamamoto, K.; Shiota, A.; Hayashi, N.; Iwamoto, Y.; Takeda, M.; Takagi, T.; Katsuya, T.; Ogihara, T.; et al. Macrophage Inflammatory Protein-1beta Induced Cell Adhesion with Increased Intracellular Reactive Oxygen Species. J. Mol. Cell. Cardiol. 2009, 47, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Quandt, J.; Dorovini-Zis, K. The Beta Chemokines CCL4 and CCL5 Enhance Adhesion of Specific CD4+ T Cell Subsets to Human Brain Endothelial Cells. J. Neuropathol. Exp. Neurol. 2004, 63, 350–362. [Google Scholar] [CrossRef]
- Menten, P.; Wuyts, A.; Van Damme, J. Macrophage Inflammatory Protein-1. Cytokine Growth Factor Rev. 2002, 13, 455–481. [Google Scholar] [CrossRef]
- Jeong, S.-Y.; Lyu, J.; Kim, J.-A.; Oh, I.-H. Ryk Modulates the Niche Activity of Mesenchymal Stromal Cells by Fine-Tuning Canonical Wnt Signaling. Exp. Mol. Med. 2020, 52, 1140–1151. [Google Scholar] [CrossRef]
- Reynolds, K.; Kumari, P.; Sepulveda Rincon, L.; Gu, R.; Ji, Y.; Kumar, S.; Zhou, C.J. Wnt Signaling in Orofacial Clefts: Crosstalk, Pathogenesis and Models. Dis. Model. Mech. 2019, 12, dmm037051. [Google Scholar] [CrossRef]
- Song, L.; Li, Y.; Wang, K.; Wang, Y.-Z.; Molotkov, A.; Gao, L.; Zhao, T.; Yamagami, T.; Wang, Y.; Gan, Q.; et al. Lrp6-Mediated Canonical Wnt Signaling Is Required for Lip Formation and Fusion. Development 2009, 136, 3161–3171. [Google Scholar] [CrossRef]
- Fujii, S.; Fujimoto, K.; Goto, N.; Kanawa, M.; Kawamoto, T.; Pan, H.; Srivatanakul, P.; Rakdang, W.; Pornprasitwech, J.; Saskianti, T.; et al. Characteristic Expression of MSX1, MSX2, TBX2 and ENTPD1 in Dental Pulp Cells. Biomed. Rep. 2015, 3, 566–572. [Google Scholar] [CrossRef][Green Version]
- Vaivads, M.; Akota, I.; Pilmane, M. Cleft Candidate Genes and Their Products in Human Unilateral Cleft Lip Tissue. Diseases 2021, 9, 26. [Google Scholar] [CrossRef]
- Macheda, M.L.; Sun, W.W.; Kugathasan, K.; Hogan, B.M.; Bower, N.I.; Halford, M.M.; Zhang, Y.F.; Jacques, B.E.; Lieschke, G.J.; Dabdoub, A.; et al. The Wnt Receptor Ryk Plays a Role in Mammalian Planar Cell Polarity Signaling. J. Biol. Chem. 2012, 287, 29312–29323. [Google Scholar] [CrossRef]
- Famili, F.; Perez, L.G.; Naber, B.A.; Noordermeer, J.N.; Fradkin, L.G.; Staal, F.J. The Non-Canonical Wnt Receptor Ryk Regulates Hematopoietic Stem Cell Repopulation in Part by Controlling Proliferation and Apoptosis. Cell Death Dis. 2016, 7, e2479. [Google Scholar] [CrossRef]
- Kamitori, K.; Machide, M.; Osumi, N.; Kohsaka, S. Expression of Receptor Tyrosine Kinase RYK in Developing Rat Central Nervous System. Brain Res. Dev. Brain Res. 1999, 114, 149–160. [Google Scholar] [CrossRef]
- Simoneaux, D.K.; Fletcher, F.A.; Jurecic, R.; Shilling, H.G.; Van, N.T.; Patel, P.; Belmont, J.W. The Receptor Tyrosine Kinase-Related Gene (Ryk) Demonstrates Lineage and Stage-Specific Expression in Hematopoietic Cells. J. Immunol. 1995, 154, 1157–1166. [Google Scholar] [CrossRef] [PubMed]
- Dolcemascolo, R.; Goiriz, L.; Montagud-Martínez, R.; Rodrigo, G. Gene Regulation by a Protein Translation Factor at the Single-Cell Level. PLoS Comput. Biol. 2022, 18, e1010087. [Google Scholar] [CrossRef]
- Cenik, C.; Cenik, E.S.; Byeon, G.W.; Grubert, F.; Candille, S.I.; Spacek, D.; Alsallakh, B.; Tilgner, H.; Araya, C.L.; Tang, H.; et al. Integrative Analysis of RNA, Translation, and Protein Levels Reveals Distinct Regulatory Variation across Humans. Genome Res. 2015, 25, 1610–1621. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Kodama, T.; Daida, H. Pentraxin 3: A Novel Biomarker for Inflammatory Cardiovascular Disease. Int. J. Vasc. Med. 2012, 2012, 657025. [Google Scholar] [CrossRef]
- Magrini, E.; Mantovani, A.; Garlanda, C. The Dual Complexity of PTX3 in Health and Disease: A Balancing Act? Trends Mol. Med. 2016, 22, 497–510. [Google Scholar] [CrossRef]
- Zhai, Y.; Iura, A.; Yeasmin, S.; Wiese, A.B.; Wu, R.; Feng, Y.; Fearon, E.R.; Cho, K.R. MSX2 Is an Oncogenic Downstream Target of Activated WNT Signaling in Ovarian Endometrioid Adenocarcinoma. Oncogene 2011, 30, 4152–4162. [Google Scholar] [CrossRef]
- Srivastava, A.; Rikhari, D.; Srivastava, S. RSPO2 as Wnt Signaling Enabler: Important Roles in Cancer Development and Therapeutic Opportunities. Genes Dis. 2024, 11, 788–806. [Google Scholar] [CrossRef]
- Ma, B.; Hottiger, M.O. Crosstalk between Wnt/β-Catenin and NF-κB Signaling Pathway during Inflammation. Front. Immunol. 2016, 7, 378. [Google Scholar] [CrossRef]
- Ahmad, R.; Kochumon, S.; Chandy, B.; Shenouda, S.; Koshy, M.; Hasan, A.; Arefanian, H.; Al-Mulla, F.; Sindhu, S. TNF-α Drives the CCL4 Expression in Human Monocytic Cells: Involvement of the SAPK/JNK and NF-κB Signaling Pathways. Cell. Physiol. Biochem. 2019, 52, 908–921. [Google Scholar] [CrossRef]
- Rathore, M.; Girard, C.; Ohanna, M.; Tichet, M.; Ben Jouira, R.; Garcia, E.; Larbret, F.; Gesson, M.; Audebert, S.; Lacour, J.-P.; et al. Cancer Cell-Derived Long Pentraxin 3 (PTX3) Promotes Melanoma Migration through a Toll-like Receptor 4 (TLR4)/NF-κB Signaling Pathway. Oncogene 2019, 38, 5873–5889. [Google Scholar] [CrossRef]
- Alappat, S.; Zhang, Z.Y.; Chen, Y.P. Msx Homeobox Gene Family and Craniofacial Development. Cell Res. 2003, 13, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Mulliken, J.B.; Martínez-Pérez, D. The Principle of Rotation Advancement for Repair of Unilateral Complete Cleft Lip and Nasal Deformity: Technical Variations and Analysis of Results. Plast. Reconstr. Surg. 1999, 104, 1247–1260. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.-Y.; Timbang, M.R.; Tollefson, T.T. Nuance in Bilateral Cleft Lip Repair. Oper. Tech. Otolayngol. Head Neck Surg. 2020, 31, 62–70. [Google Scholar] [CrossRef]
- Hsu, S.M.; Raine, L.; Fanger, H. The Use of Antiavidin Antibody and Avidin-Biotin-Peroxidase Complex in Immunoperoxidase Technics. Am. J. Clin. Pathol. 1981, 75, 816–821. [Google Scholar] [CrossRef]
- Tanner, M.; Gancberg, D.; Di Leo, A.; Larsimont, D.; Rouas, G.; Piccart, M.J.; Isola, J. Chromogenic in Situ Hybridization: A Practical Alternative for Fluorescence in Situ Hybridization to Detect HER-2/Neu Oncogene Amplification in Archival Breast Cancer Samples. Am. J. Pathol. 2000, 157, 1467–1472. [Google Scholar] [CrossRef]
- Pilmane, M.; Shine, J.; Iismaa, T.P. Distribution of Galanin Immunoreactivity in the Bronchi of Humans with Tuberculosis. Ann. N. Y. Acad. Sci. 1998, 863, 445–449. [Google Scholar] [CrossRef] [PubMed]


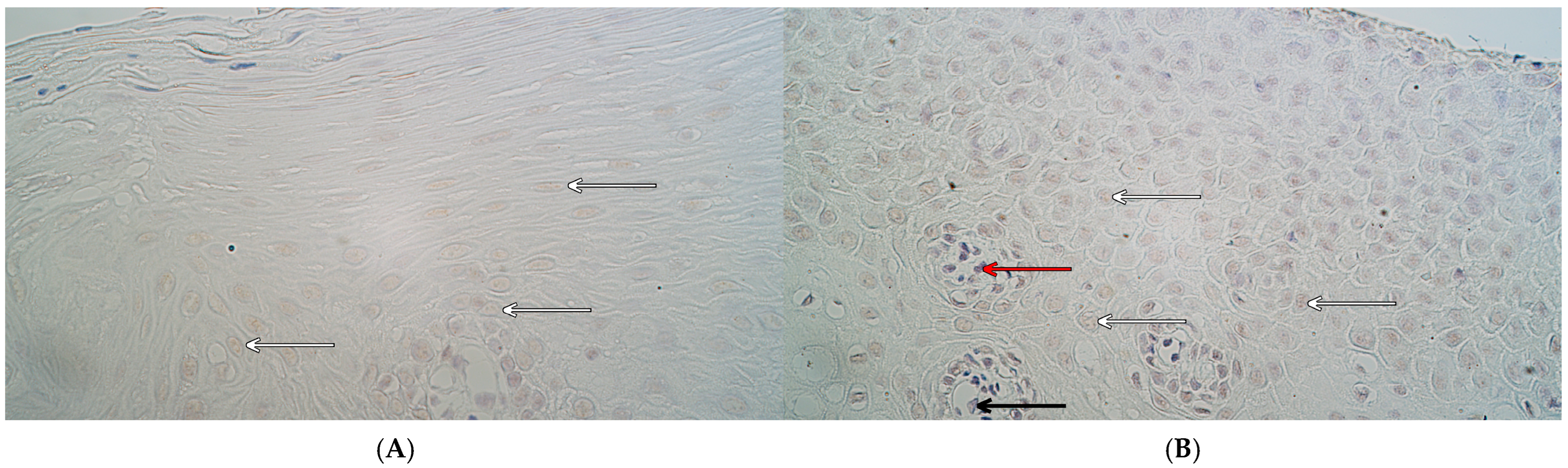

| Group | Number | MSX1 (IHC) | RYK (IHC) | NFκB p65 (IHC) | CCL4 (IHC) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ep | CT | End | Ep | CT | End | Ep | CT | End | Ep | CT | End | ||
| Controls | 1 | ++ | ++ | ++ | +++ | +++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| 2 | +++ | ++/+++ | ++ | +++/++++ | +++ | +++/++++ | ++ | ++ | ++ | +/++ | ++ | +/++ | |
| 3 | +++/++++ | ++/+++ | +++ | +++ | +++ | +++ | ++/+++ | +/++ | +/++ | +/++ | ++ | + | |
| 4 | +++ | ++ | +/++ | ++/+++ | ++/+++ | +++ | ++/+++ | ++ | ++ | + | ++ | +/++ | |
| 5 | ++ | ++ | +/++ | +++/++++ | +++/++++ | +++ | +++ | +++ | ++/+++ | ++/+++ | ++/+++ | ++ | |
| 6 | +/++ | +/++ | ++ | +++ | ++/+++ | +++ | ++/+++ | + | +/++ | ++ | + | + | |
| Median | ++/+++ | ++ | ++ | +++ | +++ | +++ | ++/+++ | ++ | ++ | +/++–++ | ++ | +/++ | |
| Patients | 1 | ++/+++ | 0/+ | + | +++ | ++/+++ | +++ | ++/+++ | + | + | ++ | 0/+ | + |
| 2 | ++ | ++ | +/++ | ++ | +++ | ++/+++ | +/++ | + | +/++ | + | + | 0/+ | |
| 3 | +/++ | 0/+ | 0/+ | +++/++++ | ++/+++ | +++ | +++ | 0/+ | 0/+ | +/++ | 0/+ | 0/+ | |
| 4 | ++ | + | + | +++ | ++ | ++ | ++/+++ | + | 0 | + | 0/+ | 0/+ | |
| 5 | ++/+++ | +/++ | + | +++ | ++ | +++ | ++ | + | + | +/++ | + | 0/+ | |
| 6 | ++/+++ | + | + | ++/+++ | ++ | +++ | ++/+++ | + | 0/+ | ++/+++ | +/++ | + | |
| 7 | ++ | +/++ | + | +++ | +/++ | ++ | ++ | 0/+ | + | ++/+++ | + | + | |
| 8 | ++ | + | + | +++/++++ | ++/+++ | ++/+++ | +++ | + | + | ++ | 0 | 0/+ | |
| 9 | +/++ | 0/+ | + | +++ | ++/+++ | +++ | ++ | 0/+ | 0/+ | +/++ | + | 0 | |
| 10 | ++/+++ | +/++ | + | +++/++++ | +++ | +++ | +++ | +/++ | + | ++/+++ | + | + | |
| 11 | ++/+++ | +/++ | 0/+ | ++ | ++/+++ | ++ | ++ | + | +/++ | ++ | ++ | + | |
| 12 | +/++ | + | 0/+ | ++/+++ | + | ++ | +/++ | 0/+ | 0/+ | + | 0/+ | 0 | |
| 13 | ++ | +/++ | +/++ | +++ | ++ | ++/+++ | ++ | +/++ | + | ++ | + | + | |
| 14 | ++ | 0/+ | 0/+ | ++ | + | + | +/++ | 0/+ | 0 | +/++ | + | 0/+ | |
| 15 | ++ | + | 0/+ | +++ | ++ | ++ | ++/+++ | + | + | ++ | ++ | 0/+ | |
| Median | ++ | + | + | +++ | ++ | ++/+++ | ++ | + | + | ++ | + | 0/+ | |
| U | 30.5 | 5.0 | 2.0 | 33.5 | 12.0 | 23.5 | 35.5 | 7.0 | 2.0 | 44.0 | 10.5 | 6.0 | |
| p | 0.267 | 0.001 | <0.001 | 0.381 | 0.008 | 0.095 | 0.470 | 0.002 | <0.001 | 0.970 | 0.005 | 0.001 | |
| Group | Number | MSX2 (CISH) | RYK (CISH) | PTX3 (CISH) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ep | CT | End | Ep | CT | End | Ep | CT | End | ||
| Controls | 1 | 0 | 0 | 0 | 0/+ | 0 | + | ++ | +/++ | 0 |
| 2 | 0 | 0 | 0 | 0/+ | 0 | 0 | 0/+ | 0 | 0 | |
| 3 | 0/+ | 0/+ | + | + | + | + | +/++ | +/++ | 0/+ | |
| 4 | + | 0/+ | + | 0/+ | 0/+ | 0/+ | 0/+ | 0 | + | |
| 5 | 0 | 0 | 0 | 0 | 0 | + | 0/+ | 0/+ | 0 | |
| 6 | + | + | +/++ | + | 0/+ | 0/+ | 0/+ | 0/+ | + | |
| Median | 0/+ | 0–0/+ | 0–0/+ | 0/+ | 0/+ | 0–0/+ | 0/+–+ | 0/+ | 0/+ | |
| Patients | 1 | + | +/++ | ++ | ++ | ++ | + | + | + | + |
| 2 | ++ | + | ++ | ++/+++ | ++ | + | +/++ | + | ++ | |
| 3 | +/++ | + | + | + | 0/+ | +/++ | 0/+ | 0/+ | +/++ | |
| 4 | + | + | ++/+++ | ++ | ++ | 0/+ | 0 | 0 | + | |
| 5 | ++ | ++ | 0/+ | 0/+ | 0 | + | +/++ | + | ++ | |
| 6 | +/++ | +/++ | +++ | ++/+++ | ++ | ++ | 0/+ | + | +/++ | |
| 7 | 0/+ | 0/+ | ++ | +/++ | ++ | 0/+ | + | 0 | 0/+ | |
| 8 | ++ | 0/+ | +++ | ++/+++ | ++/+++ | + | 0/+ | 0 | ++ | |
| 9 | +/++ | + | +++ | ++/+++ | ++ | 0/+ | 0/+ | 0 | +/++ | |
| 10 | + | + | ++ | + | + | + | 0/+ | 0/+ | + | |
| 11 | + | + | +/++ | + | + | 0/+ | 0 | 0 | + | |
| 12 | +/++ | +/++ | ++/+++ | ++/+++ | ++ | 0 | 0 | 0 | +/++ | |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 14 | ++ | ++ | ++/+++ | +/++ | + | 0 | 0/+ | 0 | ++ | |
| 15 | + | 0/+ | +/++ | +/++ | + | 0/+ | 0 | 0 | + | |
| Median | +/++ | +/++ | + | ++ | +/++ | ++ | 0/+ | 0/+ | 0 | |
| U | 11.5 | 12.0 | 13.5 | 10.5 | 12.0 | 13.0 | 44.0 | 29.0 | 31.0 | |
| p | 0.006 | 0.008 | 0.011 | 0.005 | 0.008 | 0.011 | 0.970 | 0.235 | 0.302 | |
| Strength of Correlation | Correlation Between Factors | rs | p |
|---|---|---|---|
| Very strong positive (0.8–1.0) | RYK in Ep (CISH) and MSX2 in End (CISH) | 1.000 | <0.001 |
| NFκB p65 in End (IHC) and NFκB p65 in CT (IHC) | 0.984 | <0.001 | |
| MSX2 in End (CISH) and MSX2 in CT (CISH) | 0.950 | 0.004 | |
| RYK in Ep (CISH) and MSX2 in CT (CISH) | 0.950 | 0.004 | |
| CCL4 in End (IHC) and NFκB p65 in End (IHC) | 0.904 | 0.013 | |
| CCL4 in CT (IHC) and NFκB p65 in CT (IHC) | 0.898 | 0.015 | |
| MSX2 in CT (CISH) and MSX2 in Ep (CISH) | 0.890 | 0.018 | |
| CCL4 in End (IHC) and NFκB p65 in CT (IHC) | 0.889 | 0.018 | |
| MSX1 in CT (IHC) and MSX1 in Ep (IHC) | 0.874 | 0.023 | |
| MSX2 in End (CISH) and MSX2 in Ep (CISH) | 0.874 | 0.023 | |
| RYK in Ep (CISH) and MSX2 in Ep (CISH) | 0.874 | 0.023 | |
| RYK in End (CISH) and MSX2 in End (CISH) | 0.850 | 0.032 | |
| RYK in End (CISH) and RYK in Ep (CISH) | 0.850 | 0.032 | |
| PTX3 in End (CISH) and PTX3 in Ep (CISH) | 0.849 | 0.033 | |
| PTX3 in End (CISH) and PTX3 in CT (CISH) | 0.839 | 0.037 | |
| CCL4 in CT (IHC) and NFκB p65 in End (IHC) | 0.822 | 0.045 | |
| CCL4 in CT (IHC) and RYK in CT (IHC) | 0.822 | 0.045 | |
| Very strong negative (−1.0…−0.8) | MSX2 in End (CISH) and NFκB p65 in CT (IHC) | −0.820 | 0.046 |
| RYK in Ep (CISH) and NFκB p65 in CT (IHC) | −0.820 | 0.046 | |
| RYK in CT (CISH) and CCL4 in CT (IHC) | −0.822 | 0.045 | |
| MSX2 in Ep (CISH) and RYK in Ep (IHC) | −0.826 | 0.043 | |
| MSX2 in End (CISH) and CCL4 in End (IHC) | −0.839 | 0.037 | |
| RYK in Ep (CISH) and CCL4 in End (IHC) | −0.839 | 0.037 | |
| MSX2 in CT (CISH) and RYK in CT (IHC) | −0.867 | 0.025 | |
| MSX2 in Ep (CISH) and RYK in CT (IHC) | −0.890 | 0.018 | |
| RYK in CT (CISH) and CCL4 in End (IHC) | −0.904 | 0.013 | |
| RYK in CT (CISH) and NFκB p65 in CT (IHC) | −0.984 | <0.001 | |
| RYK in CT (CISH) and NFκB p65 in End (IHC) | −1.000 | <0.001 |
| Strength of Correlation | Correlation Between Factors | rs | p |
|---|---|---|---|
| Very strong positive (0.8–1.0) | RYK in End (CISH) and RYK in CT (CISH) | 0.901 | <0.001 |
| RYK in CT (CISH) and RYK in Ep (CISH) | 0.852 | <0.001 | |
| RYK in End (CISH) and RYK in Ep (CISH) | 0.822 | <0.001 | |
| NFκB p65 in Ep (IHC) and RYK in Ep (IHC) | 0.802 | <0.001 | |
| Strong positive (0.6–0.8) | PTX3 in End (CISH) and PTX3 in Ep (CISH) | 0.789 | <0.001 |
| CCL4 in End (IHC) and CCL4 in Ep (IHC) | 0.786 | 0.001 | |
| PTX3 in Ep (CISH) and RYK in End (IHC) | 0.740 | 0.002 | |
| CCL4 in End (IHC) and MSX1 in Ep (IHC) | 0.722 | 0.002 | |
| PTX3 in End (CISH) and RYK in End (IHC) | 0.709 | 0.003 | |
| NFκB p65 in End (IHC) and MSX1 in CT (IHC) | 0.704 | 0.003 | |
| PTX3 in End (CISH) and PTX3 in CT (CISH) | 0.682 | 0.005 | |
| NFκB p65 in CT (IHC) and MSX1 in Ep (IHC) | 0.642 | 0.010 | |
| PTX3 in Ep (CISH) and NFκB p65 in Ep (IHC) | 0.613 | 0.015 | |
| Moderate positive (0.4–0.6) | PTX3 in Ep (CISH) and RYK in CT (IHC) | 0.584 | 0.022 |
| RYK in End (IHC) and RYK in CT (IHC) | 0.578 | 0.024 | |
| CCL4 in End (IHC) and NFκB p65 in CT (IHC) | 0.547 | 0.035 | |
| PTX3 in CT (CISH) and PTX3 in Ep (CISH) | 0.546 | 0.035 | |
| CCL4 in Ep (IHC) and MSX1 in Ep (IHC) | 0.534 | 0.036 | |
| MSX2 in End (CISH) and MSX2 in CT (CISH) | 0.534 | 0.040 | |
| NFκB p65 in CT (IHC) and MSX1 in CT (IHC) | 0.530 | 0.042 | |
| MSX2 in CT (CISH) and MSX2 in Ep (CISH) | 0.527 | 0.044 | |
| NFκB p65 in CT (IHC) and MSX1 in End (IHC) | 0.519 | 0.048 | |
| Moderate negative (−0.6…−0.4) | MSX2 in CT (CISH) and CCL4 in End (IHC) | −0.595 | 0.019 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vaivads, M.; Rone, A.E.; Pilmane, M. Analysis of MSX1, RYK, NFκB p65, and CCL4 Proteins and MSX2, RYK, and PTX3 Genes in Human Cleft Lip Tissue. Int. J. Mol. Sci. 2025, 26, 10599. https://doi.org/10.3390/ijms262110599
Vaivads M, Rone AE, Pilmane M. Analysis of MSX1, RYK, NFκB p65, and CCL4 Proteins and MSX2, RYK, and PTX3 Genes in Human Cleft Lip Tissue. International Journal of Molecular Sciences. 2025; 26(21):10599. https://doi.org/10.3390/ijms262110599
Chicago/Turabian StyleVaivads, Mārtiņš, Alise Elizabete Rone, and Māra Pilmane. 2025. "Analysis of MSX1, RYK, NFκB p65, and CCL4 Proteins and MSX2, RYK, and PTX3 Genes in Human Cleft Lip Tissue" International Journal of Molecular Sciences 26, no. 21: 10599. https://doi.org/10.3390/ijms262110599
APA StyleVaivads, M., Rone, A. E., & Pilmane, M. (2025). Analysis of MSX1, RYK, NFκB p65, and CCL4 Proteins and MSX2, RYK, and PTX3 Genes in Human Cleft Lip Tissue. International Journal of Molecular Sciences, 26(21), 10599. https://doi.org/10.3390/ijms262110599








