Comparative Transcriptomic and Proteomic Analyses Identify Byssogenesis-Associated Genes in the Mediterranean Mussel Mytilus galloprovincialis Lamarck, 1819
Abstract
1. Introduction
2. Results
2.1. Statistics of Transcriptome and Proteome Sequencing
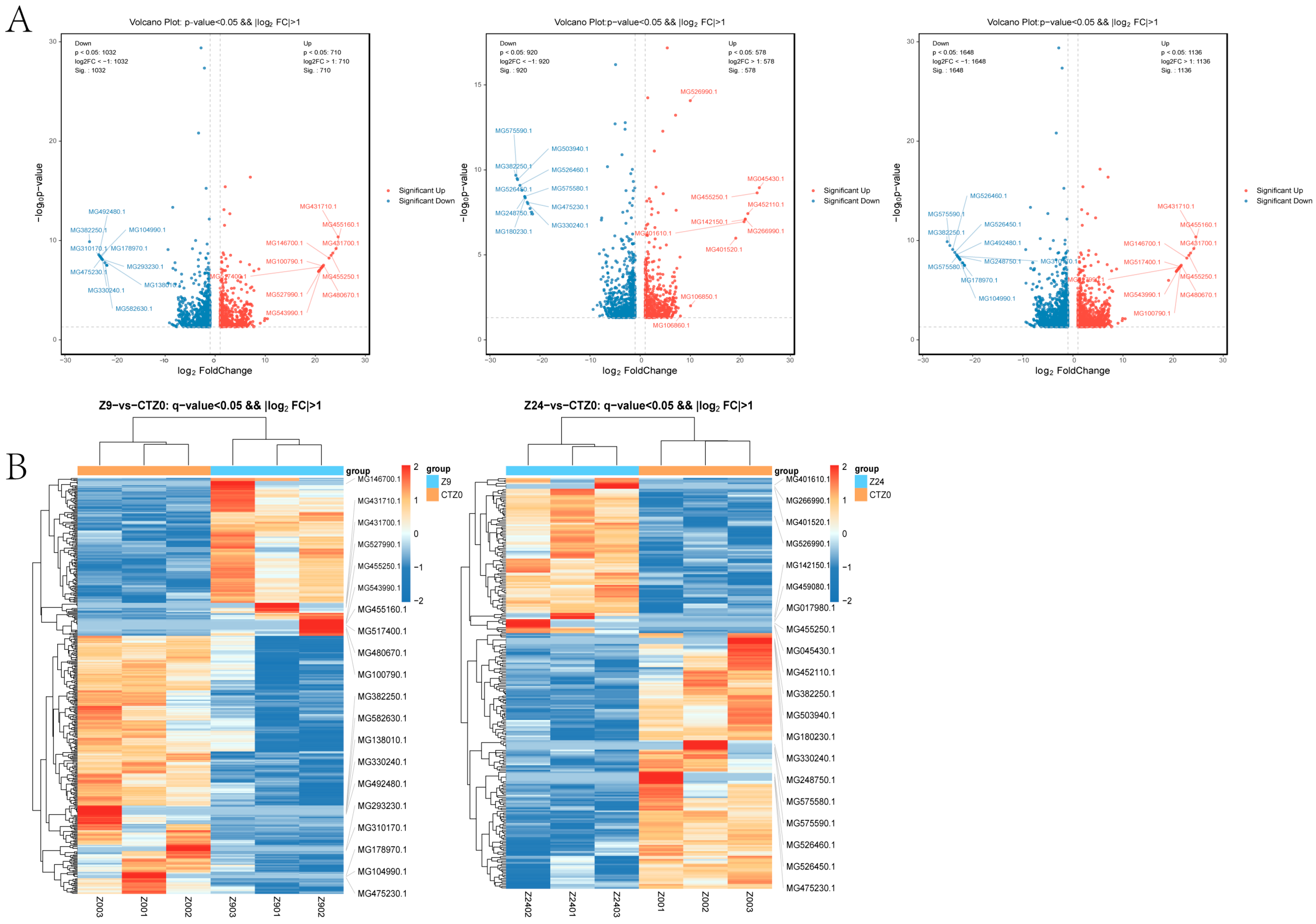
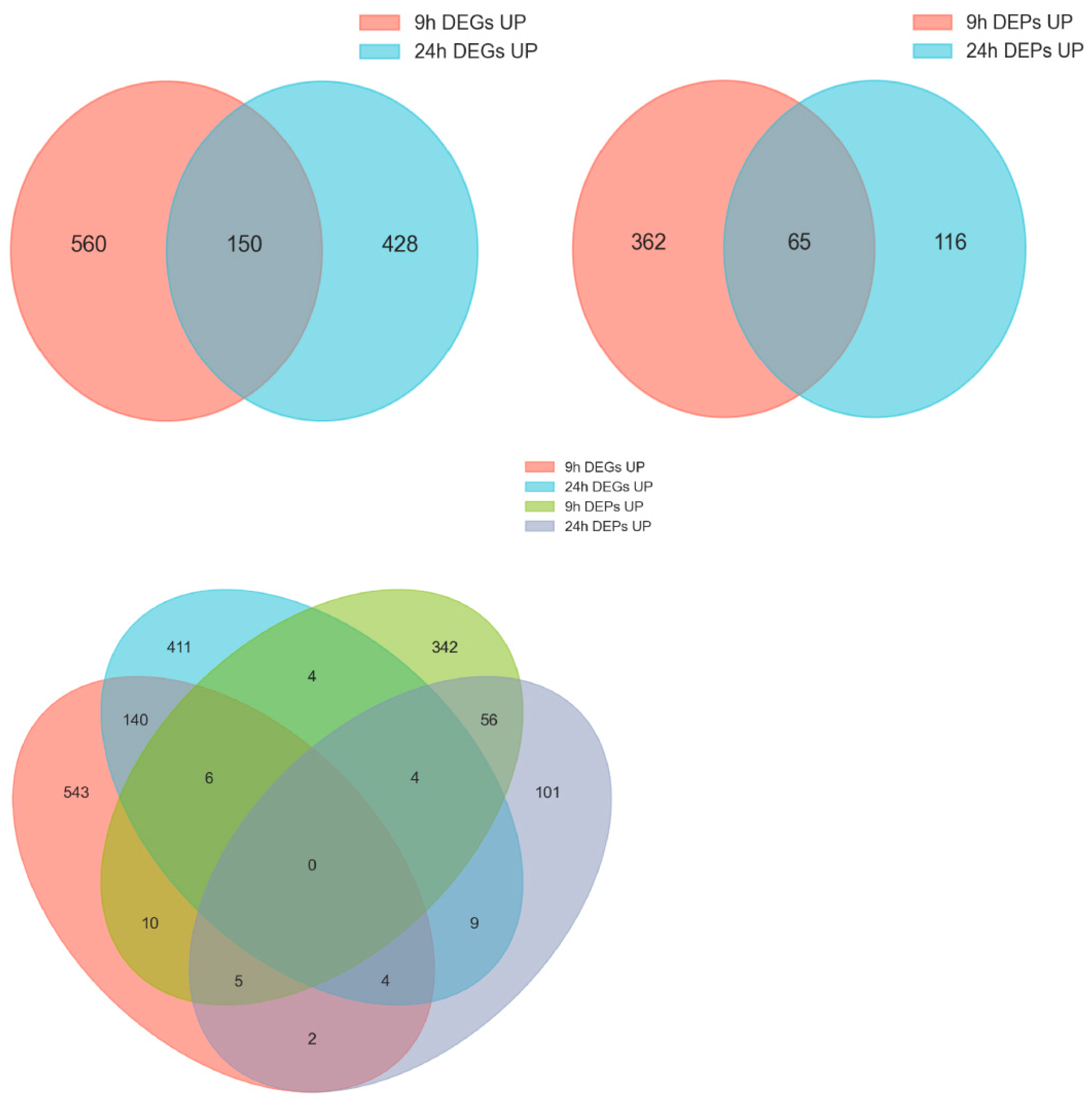
2.2. Analysis of Differentially Expressed Genes (DEGs)
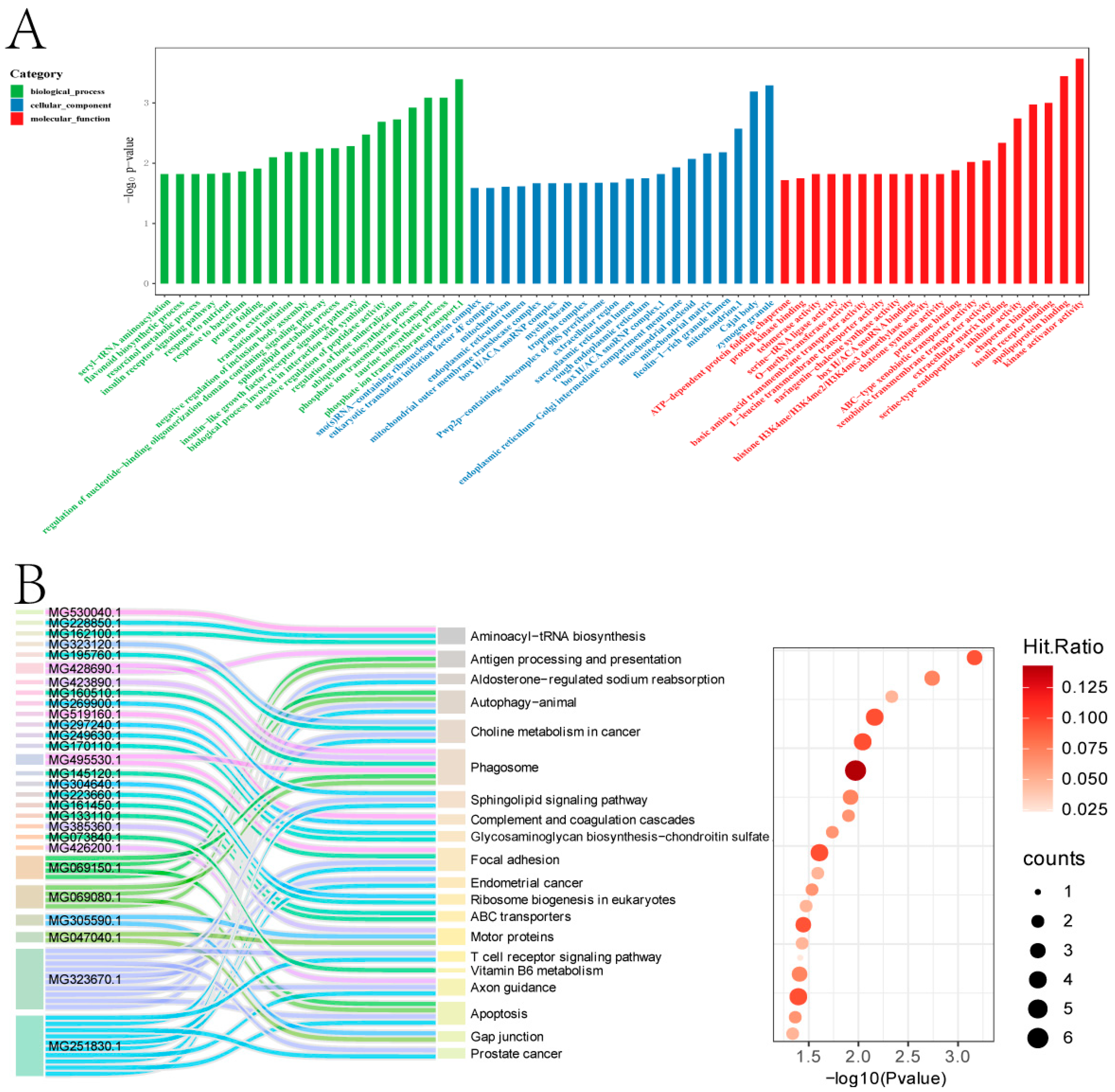
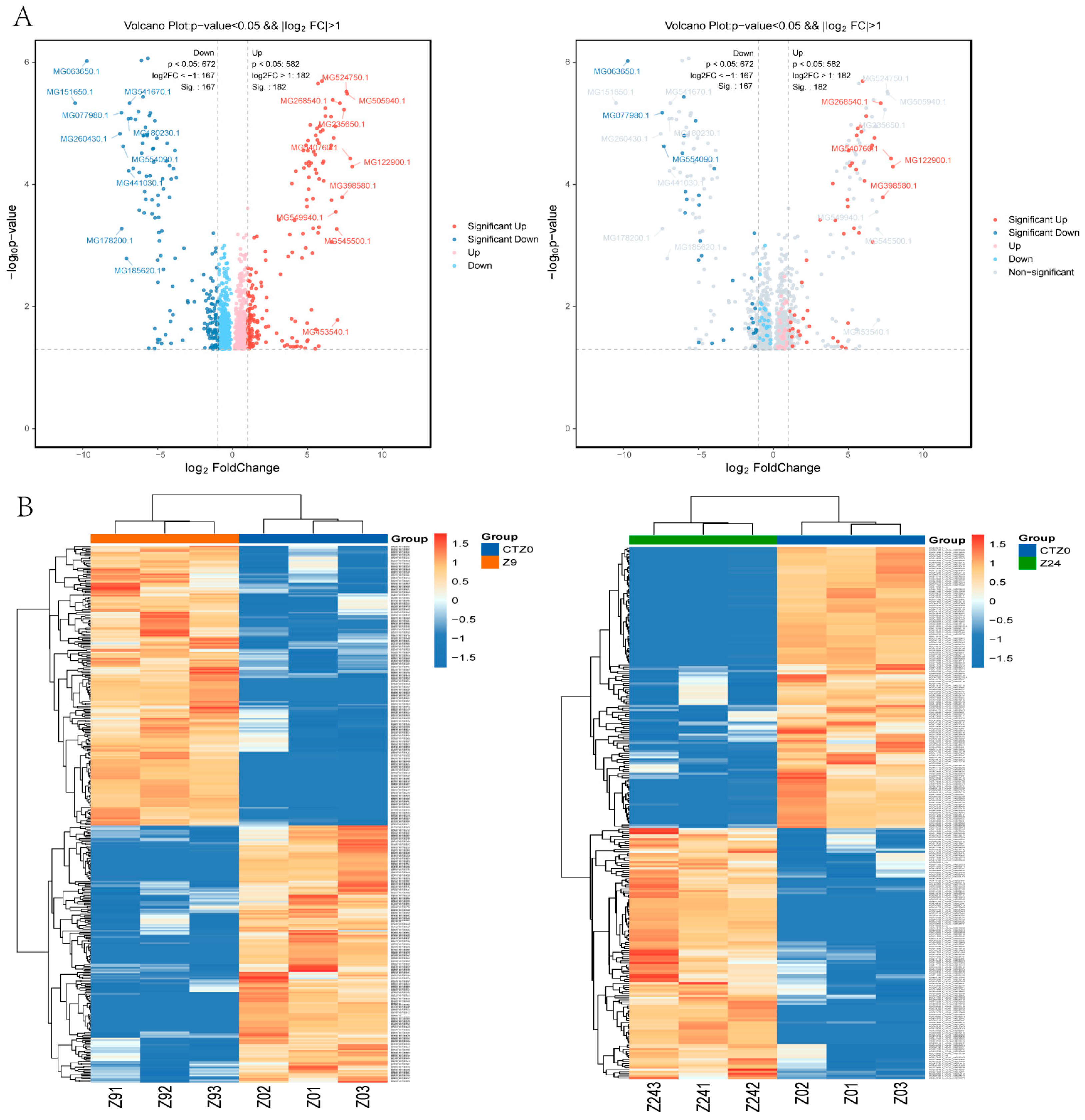
2.3. Analysis of Differentially Expressed Proteins (DEPs)
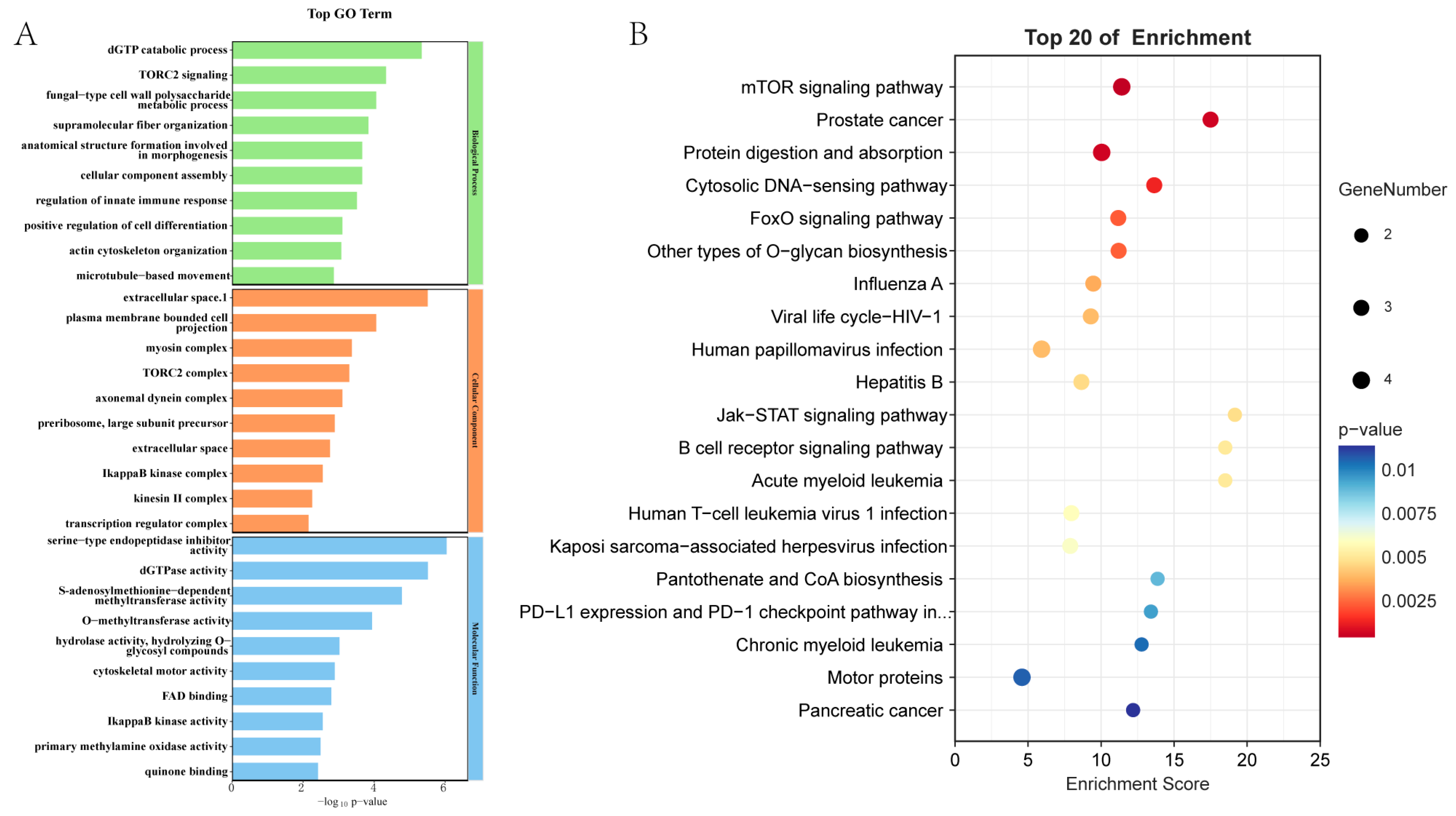
2.4. Integrated Transcriptome-Proteome Analysis
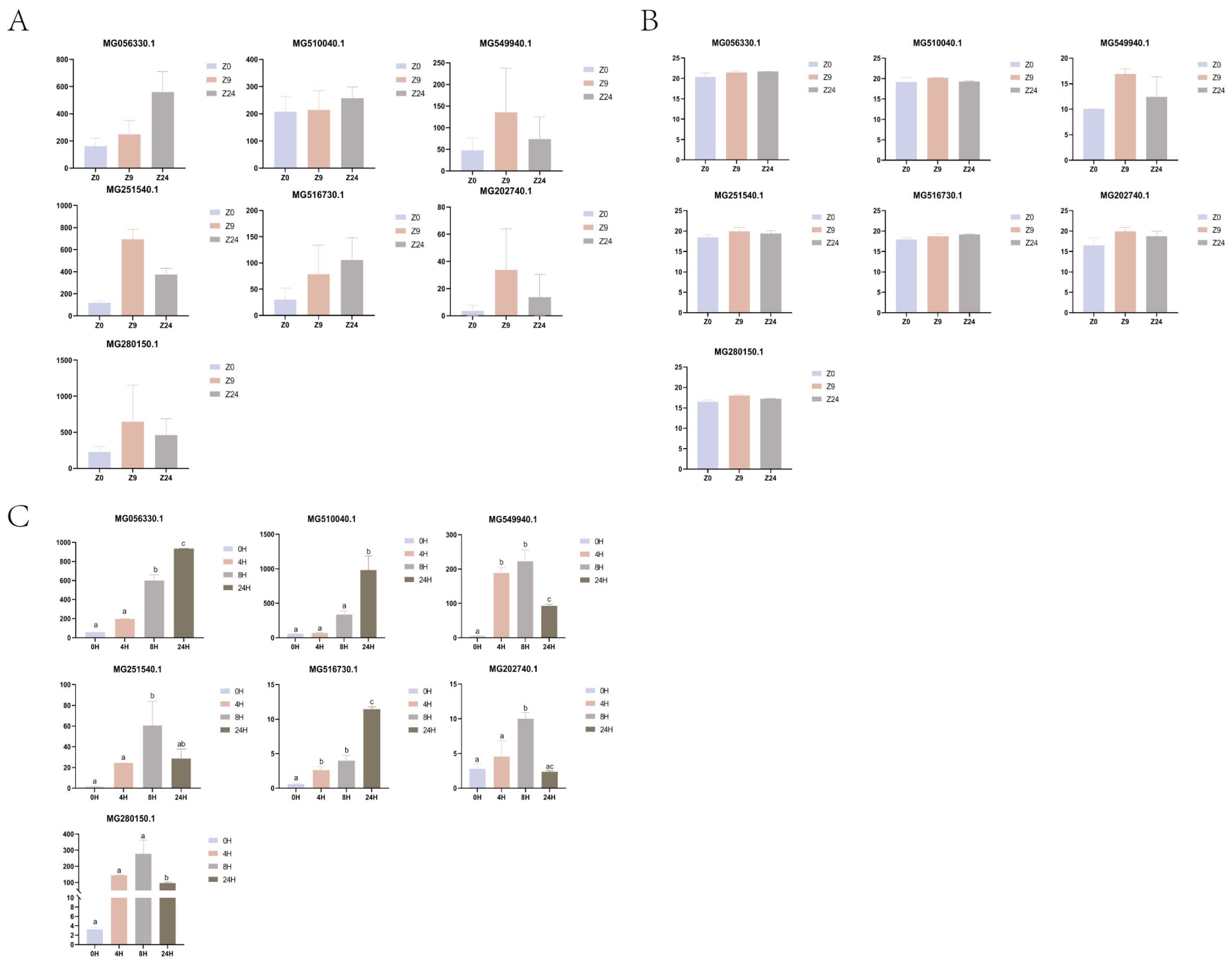
2.5. qPCR Validation of 7 Genes with DEGs
3. Discussion
4. Materials and Methods
4.1. Animal Materials
4.2. Transcriptome Sequencing and Analysis
4.3. Proteome Sequencing and Analysis
4.4. Joint Analysis Transcriptome and Proteome Data
4.5. Quantitative Real-Time PCR (qPCR) Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, Z.; He, C.; Lin, Q.; Liang, X.; Yang, J. Effects of Mytilus galloprovincialis peptide on settlement of mussel juveniles induced by bacterial biofilms. J. Fish. China 2024, 48, 089614. [Google Scholar]
- Bjerknes, H.; Elvevoll, E.O.; Sundset, M.A.; Langdal, A.; Eilertsen, K.E. Farmed blue mussels (Mytilus edulis)-a nutrient-dense resource retaining nutritional value through processing. Front. Nutr. 2024, 11, 1443229. [Google Scholar] [CrossRef]
- Geller, J.B.; Carlton, J.T.; Powers, D.A. Interspecific genetic variation and the distribution of Mytilus galloprovincialis in the North Pacific. Biol. Invasions 1999, 1, 235–244. [Google Scholar]
- Branch, G.M.; Steffani, C.N. Can we predict the effects of alien species? A case-history of the invasion of South Africa by Mytilus galloprovincialis (Lamarck). J. Exp. Mar. Biol. Ecol. 2004, 300, 189–215. [Google Scholar] [CrossRef]
- Lupo, C.; Bougeard, S.; Le Bihan, V.; Blin, J.L.; Allain, G.; Azema, P.; Benoit, F.; Bechemin, C.; Bernard, I.; Blachier, P.; et al. Mortality of marine mussels Mytilus edulis and M. galloprovincialis: Systematic literature review of risk factors and recommendations for future research. Rev. Aquac. 2021, 13, 504–536. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, W.; Li, Y.; Zhao, W.; Peng, H.; Zou, J.; Song, A. Effects of body measurement and weight traits on byssus traits of Mytilus galloprovincialis. Mar. Sci. 2022, 46, 169–176. [Google Scholar]
- Ministry of Agriculture and Rural Affairs of the People’s Republic of China. 2024 China Fisheries Statistical Yearbook; China Agriculture Press: Beijing, China, 2024.
- FAO. The State of World Fisheries and Aquaculture (SOFIA) 2022; FAO: Rome, Italy, 2022. [Google Scholar]
- Gilg, M.R.; Hilbish, T.J. The geography of marine larval dispersal: Coupling genetics with fine-scale physical oceanography. Ecology 2003, 84, 2989–2998. [Google Scholar] [CrossRef]
- Yu, M.E.; Hwang, J.Y.; Deming, T.J. Role of L-3,4-dihydroxyphenylalanine in mussel adhesive proteins. J. Am. Chem. Soc. 1999, 121, 5825–5826. [Google Scholar] [CrossRef]
- Wang, J.; Scheibel, T. Recombinant Production of Mussel Byssus Inspired Proteins. Biotechnol. J. 2018, 13, 1800146. [Google Scholar] [CrossRef]
- Anderson, K.E.; Waite, J.H. Immunolocalization of Dpfp1, a byssal protein of the zebra mussel. J. Exp. Biol. 2000, 203, 3065–3076. [Google Scholar] [CrossRef]
- Tamarin, A.; Lewis, P.; Askey, J. Structure and formation of byssus attachment plaque in Mytilus. J. Morphol. 1976, 149, 199–221. [Google Scholar] [CrossRef]
- Suhre, M.H.; Gertz, M.; Steegborn, C.; Scheibel, T. Structural and functional features of a collagen-binding matrix protein from the mussel byssus. Nat. Commun. 2014, 5, 3392. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Fang, Y.; Zhou, W.; Yan, L.; Xu, Y.; Zhu, H.; Liu, H. Mussel foot protein inspired tough tissue-selective underwater adhesive hydrogel. Mater. Horiz. 2021, 8, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Gurry, T.; Cheng, A.A.; Downey, J.; Deng, Z.T.; Stultz, C.M.; Lu, T.K. Strong underwater adhesives made by self-assembling multi-protein nanofibres. Nat. Nanotechnol. 2014, 9, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Achazi, K.; Liebe, H.; Schulz, A.; Noeske, P.L.M.; Grunwald, I.; Haag, R. Mussel-Inspired Dendritic Polymers as Universal Multifunctional Coatings. Angew. Chem.-Int. Ed. 2014, 53, 11650–11655. [Google Scholar] [CrossRef]
- Wang, X.-e.; Liao, Z.; Yang, Q.-m.; Ye, Y.-y.; Shen, W.; Liu, H.-h.; Yan, X.-j.; Li, Y.-f.; Zhang, X.-l. Characterization of a novel antioxidant byssal protein from Mytilus coruscus foot. Int. J. Biol. Macromol. 2024, 273, 133095. [Google Scholar] [CrossRef]
- Akdogan, Y.; Wei, W.; Huang, K.Y.; Kageyama, Y.; Danner, E.W.; Miller, D.R.; Rodriguez, N.R.M.; Waite, J.H.; Han, S. Intrinsic Surface-Drying Properties of Bioadhesive Proteins. Angew. Chem.-Int. Ed. 2014, 53, 11253–11256. [Google Scholar] [CrossRef]
- Jiang, Z.; Liu, J.; Jin, L.; Zhang, Q. Extraction and application of Perna viridis foot protein as bioadhesive. J. Biomed. Eng. 2010, 27, 1266–1273. [Google Scholar]
- Clancy, S.K.; Sodano, A.; Cunningham, D.J.; Huang, S.S.; Zalicki, P.J.; Shin, S.; Ahn, B.K. Marine Bioinspired Underwater Contact Adhesion. Biomacromolecules 2016, 17, 1869–1874. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, H.; Liu, F.H.; Chen, T.; Hu, G.X.; Guo, D.H.; Hou, Q.F.; Wu, X.; Su, Y.; Wang, J.B. Molecular interactions between DOPA and surfaces with different functional groups: A chemical force microscopy study. RSC Adv. 2017, 7, 32518–32527. [Google Scholar] [CrossRef]
- Aich, P.; An, J.; Yang, B.; Ko, Y.H.; Kim, J.; Murray, J.; Cha, H.J.; Roh, J.H.; Park, K.M.; Kim, K. Self-assembled adhesive biomaterials formed by a genetically designed fusion protein. Chem. Commun. 2018, 54, 12642–12645. [Google Scholar] [CrossRef]
- Hao, D.; Wei, W.; Zhou, H.; Li, M.; Qiao, S.; Zhao, S.; Hou, Z. Preparation of recombinant mussel mucin Mfp-3P and its promotion of wound healing. Chin. J. Biotechnol. 2024, 40, 1498–1508. [Google Scholar]
- Zhong, Z.X.; Chen, G.Z.; Tu, H.H.; Yao, X.Y.; Peng, X.; Lan, X.; Tang, Q.Y.; Yi, S.K.; Xia, Z.L.; Cai, M.Y.; et al. Transcriptomic Analysis and Functional Gene Expression in Different Stages of Gonadal Development of Macrobrachium rosenbergii. Fishes 2023, 8, 94. [Google Scholar] [CrossRef]
- Sun, X.J.; Liu, Z.H.; Wu, B.; Zhou, L.Q.; Wang, Q.; Wu, W.; Yang, A.G. Differences between fast and slow muscles in scallops revealed through proteomics and transcriptomics. BMC Genom. 2018, 19, 377. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Huang, J.; Shi, Y.; Zhang, H.; He, M.X. Comparative transcriptomic and proteomic analysis of yellow shell and black shell pearl oysters, Pinctada fucata martensii. BMC Genom. 2019, 20, 469. [Google Scholar] [CrossRef]
- Cheong, S.; Peng, Y.; Lu, F.; He, Y. Structural extracellular matrix-mediated molecular signaling in wound repair and tissue regeneration. J. Funct. Biomater. 2023, 14, 123. [Google Scholar] [CrossRef]
- Liu, F.; Wang, X.; Zhang, X.; Ren, C.; Xin, J. Role of Serum Cartilage Oligomeric Matrix Protein (COMP) in the Diagnosis of Rheumatoid Arthritis (RA): A Case-Control Study. J. Int. Med. Res. 2016, 44, 940–949. [Google Scholar] [CrossRef]
- Tan, K.; Lawler, J. The Interaction of Thrombospondins with Extracellular Matrix Proteins. J. Cell Commun. Signal. 2009, 3, 177–187. [Google Scholar] [CrossRef]
- Cui, J.; Zhang, J. Cartilage Oligomeric Matrix Protein, Diseases, and Therapeutic Opportunities. Int. J. Mol. Sci. 2022, 23, 9253. [Google Scholar] [CrossRef]
- Li, S.; Liu, D.; Cui, Y.; Tong, H.; Li, S.; Yan, Y. Podocan Promotes Differentiation of Bovine Skeletal Muscle Satellite Cells by Regulating the Wnt4-β-Catenin Signaling Pathway. Front. Physiol. 2019, 10, 1010. [Google Scholar] [CrossRef]
- Zhang, F. Podocan Unraveled: Understanding Its Role in Tumor Proliferation and Smooth Muscle Regulation. Biomed. Pharmacother. 2024, 179, 117416. [Google Scholar] [CrossRef]
- Krapež, G.; Šamec, N.; Zottel, A.; Katrašnik, M.; Kump, A.; Šribar, J.; Križaj, I.; Stojan, J.; Romih, R.; Bajc, G.; et al. In Vitro Functional Validation of an Anti-FREM2 Nanobody for Glioblastoma Cell Targeting. Antibodies 2025, 14, 8. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, Y.; Wang, F.; Qi, J. Correlation analysis of COL6A3 gene and immune infiltration level and clinical prognosis of gastric cancer. Dig. Oncol. Electron. Version 2020, 12, 4. [Google Scholar]
- Wen, J.; Cheng, G.; Zhang, Y.; Liu, S. Hippo signaling pathway in polycystic ovary syndrome. Front. Endocrinol. 2025, 16, 1623143. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Yin, S.; Shu, Y. NF2: An underestimated player in cancer metabolic reprogramming and tumor immunity. npj Precis. Oncol. 2024, 8, 133. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M. Function and cancer genomics of FAT family genes (Review). Int. J. Oncol. 2012, 41, 1913–1924. [Google Scholar] [CrossRef] [PubMed]
- Kanamori, A.; Nakayama, J.; Fukuda, M.N.; Stallcup, W.B.; Sasaki, K.; Fukuda, M.; Hirabayashi, Y. Expression cloning and characterization of a cDNA encoding a novel membrane protein required for the formation of O-acetylated ganglioside: A putative acetyl -CoA transporter. Proc. Natl. Acad. Sci. USA 1997, 94, 2897–2902. [Google Scholar] [CrossRef]
- Jonas, M.C.; Pehar, M.; Puglielli, L. AT-1 is the ER membrane acetyl-CoA transporter and is essential for cell viability. J. Cell Sci. 2010, 123, 3378–3388. [Google Scholar] [CrossRef]
- Zhou, D.; Chen, N.H.; Huang, S.T.; Song, C.; Zhang, Z. Mechanistic insights into the acetyl-CoA recognition by SLC33A1. Cell Discov. 2025, 11, 36. [Google Scholar] [CrossRef]
- Xie, J.M.; Chen, X.Y.; Zheng, M.; Zhu, J.Z.; Mao, H. The Metabolism of Coenzyme A and Its Derivatives Plays a Crucial Role in Diseases. Front. Biosci.-Landmark 2024, 29, 143. [Google Scholar] [CrossRef]
- Fournier, A.E.; GrandPre, T.; Strittmatter, S.M. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature 2001, 409, 341–346. [Google Scholar] [CrossRef]
- Wang, K.C.; Koprivica, V.; Kim, J.A.; Sivasankaran, R.; Guo, Y.; Neve, R.L.; He, Z.G. Oligodendrocyte-myelin glycoprotein is a Nogo receptor ligand that inhibits neurite outgrowth. Nature 2002, 417, 941–944. [Google Scholar] [CrossRef] [PubMed]
- Dickendesher, T.L.; Baldwin, K.T.; Mironova, Y.A.; Koriyama, Y.; Raiker, S.J.; Askew, K.L.; Wood, A.; Geoffroy, C.G.; Zheng, B.H.; Liepmann, C.D.; et al. NgR1 and NgR3 are receptors for chondroitin sulfate proteoglycans. Nat. Neurosci. 2012, 15, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Ersdal-Badju, E.; Lu, A.Q.; Zuo, Y.C.; Picard, V.; Bock, S.C. Identification of the antithrombin III heparin binding site. J. Biol. Chem. 1997, 272, 19393–19400. [Google Scholar] [CrossRef] [PubMed]
- Perry, D.J. Antithrombin and Its Inherited Deficiencies. Blood Rev. 1994, 8, 37–55. [Google Scholar] [CrossRef]
- Kelly-Robinson, G.A.; Reihill, J.A.; Lundy, F.T.; McGarvey, L.P.; Lockhart, J.C.; Litherland, G.J.; Thornbury, K.D.; Martin, S.L. The Serpin Superfamily and Their Role in the Regulation and Dysfunction of Serine Protease Activity in COPD and Other Chronic Lung Diseases. Int. J. Mol. Sci. 2021, 22, 6351. [Google Scholar] [CrossRef]
- Conrad, D.H.; Ford, J.W.; Sturgill, J.L.; Gibb, D.R. CD23: An overlooked regulator of allergic disease. Curr. Allergy Asthma Rep. 2007, 7, 331–337. [Google Scholar] [CrossRef]
- Acharya, M.; Borland, G.; Edkins, A.L.; MacLellan, L.M.; Matheson, J.; Ozanne, B.W.; Cushley, W. CD23/FcεRII: Molecular multi-tasking. Clin. Exp. Immunol. 2010, 162, 12–23. [Google Scholar] [CrossRef]
- Gould, H.J.; Sutton, B.J. IgE in allergy and asthma today. Nat. Rev. Immunol. 2008, 8, 205–217. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, H.; Han, C.; Ou, H.; Zhan, X. The structure and proteomic analysis of byssus in Pteria penguin: Insights into byssus evolution and formation. J. Proteom. 2024, 307, 105267. [Google Scholar] [CrossRef]
- Pillaiyar, T.; Namasivayam, V.; Manickam, M.; Jung, S.-H. Inhibitors of Melanogenesis: An Updated Review. J. Med. Chem. 2018, 61, 7395–7418. [Google Scholar] [CrossRef]
- Ito, S.; Wakamatsu, K.; Ozeki, H. Chemical analysis of melanins and its application to the study of the regulation of melanogenesis. Pigment Cell Res. 2000, 13, 103–109. [Google Scholar] [CrossRef]
- Shafiq, Z.; Cui, J.X.; Pastor-Pérez, L.; San Miguel, V.; Gropeanu, R.A.; Serrano, C.; del Campo, A. Bioinspired Underwater Bonding and Debonding on Demand. Angew. Chem.-Int. Ed. 2012, 51, 4332–4335. [Google Scholar] [CrossRef]
- Li, N.N.; Shi, G.; Yang, X.X.; Wang, Z.P.; Liao, Z. Detection of Dihydroxyphenylalanine in Native Foot Proteins from Mytilus coruscus Byssus. Chin. J. Biochem. Mol. Biol. 2011, 27, 148–153. [Google Scholar]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–U121. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Carbon, S.; Douglass, E.; Dunn, N.; Good, B.; Harris, N.L.; Lewis, S.E.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; et al. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Song, S.; Fu, Z.; Guan, R. Intracellular hydroxyproline imprinting following resolution of bleomycin-induced pulmonary fibrosis. Eur. Respir. J. 2022, 59, 2100864. [Google Scholar] [CrossRef]
- Demichev, V.; Messner, C.B.; Vernardis, S.I.; Lilley, K.S.; Ralser, M. DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 2020, 17, 41–44. [Google Scholar] [CrossRef]
- Brandão, F.; Pinto, I.; Guilhermino, M. Impact of bisphenol A (BPA) on early embryo development in the marine mussel Mytilus galloprovincialis: Effects on gene transcription. Mar. Drugs 2017, 15, 210. [Google Scholar]
| Gene | Primer | Sequences (5′–3′) | Amplicon Length |
|---|---|---|---|
| EF1α | F | CGTTTTGCTGTCCGAGACATG | 92 bp |
| R | CCACGCCTCACATCATTTCTTG | ||
| MG056330.1 | F | TGAGACAGATAGCAATGGTT | 172 bp |
| R | ACAGTAGATAGGACAAGATGG | ||
| MG251540.1 | F | TTCTCTGCGTCTCATTATCA | 191 bp |
| R | GTTATTGCCTCTGTCATAGC | ||
| MG510040 | F | GATTGTGTTGGACCTAGTATG | 140 bp |
| R | TGCCATTGTCTTCTGCTAT | ||
| MG549940.1 | F | TCTTCGTTGATTCACATCCA | 167 bp |
| R | TTCCACGCAACTTCCATAT | ||
| MG516730.1 | F | GATAGACGATGCCACTGT | 145 bp |
| R | TATGCTCATCATGTCAACTG | ||
| MG202740.1 | F | TTGACAATGAAGAACGGAAC | 109 bp |
| R | GCACACCAGCATCTATACA | ||
| MG280150.1 | F | TGGATATGGTGGTTACAGAA | 186 bp |
| R | GTTAAGGTACTACTTCGTATGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhen, X.; Chen, Y.; Zhang, W.; Li, Y.; Li, L.; Qi, H.; Zhang, S. Comparative Transcriptomic and Proteomic Analyses Identify Byssogenesis-Associated Genes in the Mediterranean Mussel Mytilus galloprovincialis Lamarck, 1819. Int. J. Mol. Sci. 2025, 26, 10511. https://doi.org/10.3390/ijms262110511
Zhen X, Chen Y, Zhang W, Li Y, Li L, Qi H, Zhang S. Comparative Transcriptomic and Proteomic Analyses Identify Byssogenesis-Associated Genes in the Mediterranean Mussel Mytilus galloprovincialis Lamarck, 1819. International Journal of Molecular Sciences. 2025; 26(21):10511. https://doi.org/10.3390/ijms262110511
Chicago/Turabian StyleZhen, Xiuwei, Yiwen Chen, Wei Zhang, Yongren Li, Li Li, Haigang Qi, and Shoudu Zhang. 2025. "Comparative Transcriptomic and Proteomic Analyses Identify Byssogenesis-Associated Genes in the Mediterranean Mussel Mytilus galloprovincialis Lamarck, 1819" International Journal of Molecular Sciences 26, no. 21: 10511. https://doi.org/10.3390/ijms262110511
APA StyleZhen, X., Chen, Y., Zhang, W., Li, Y., Li, L., Qi, H., & Zhang, S. (2025). Comparative Transcriptomic and Proteomic Analyses Identify Byssogenesis-Associated Genes in the Mediterranean Mussel Mytilus galloprovincialis Lamarck, 1819. International Journal of Molecular Sciences, 26(21), 10511. https://doi.org/10.3390/ijms262110511






