Interactions of Arachidonic Acid with AAC1 and UCP1
Abstract
1. Introduction
2. Results
2.1. Arachidonic Acid Binds in a Variety of Interhelical Sites and in the AAC1 IMS-Facing Cavity
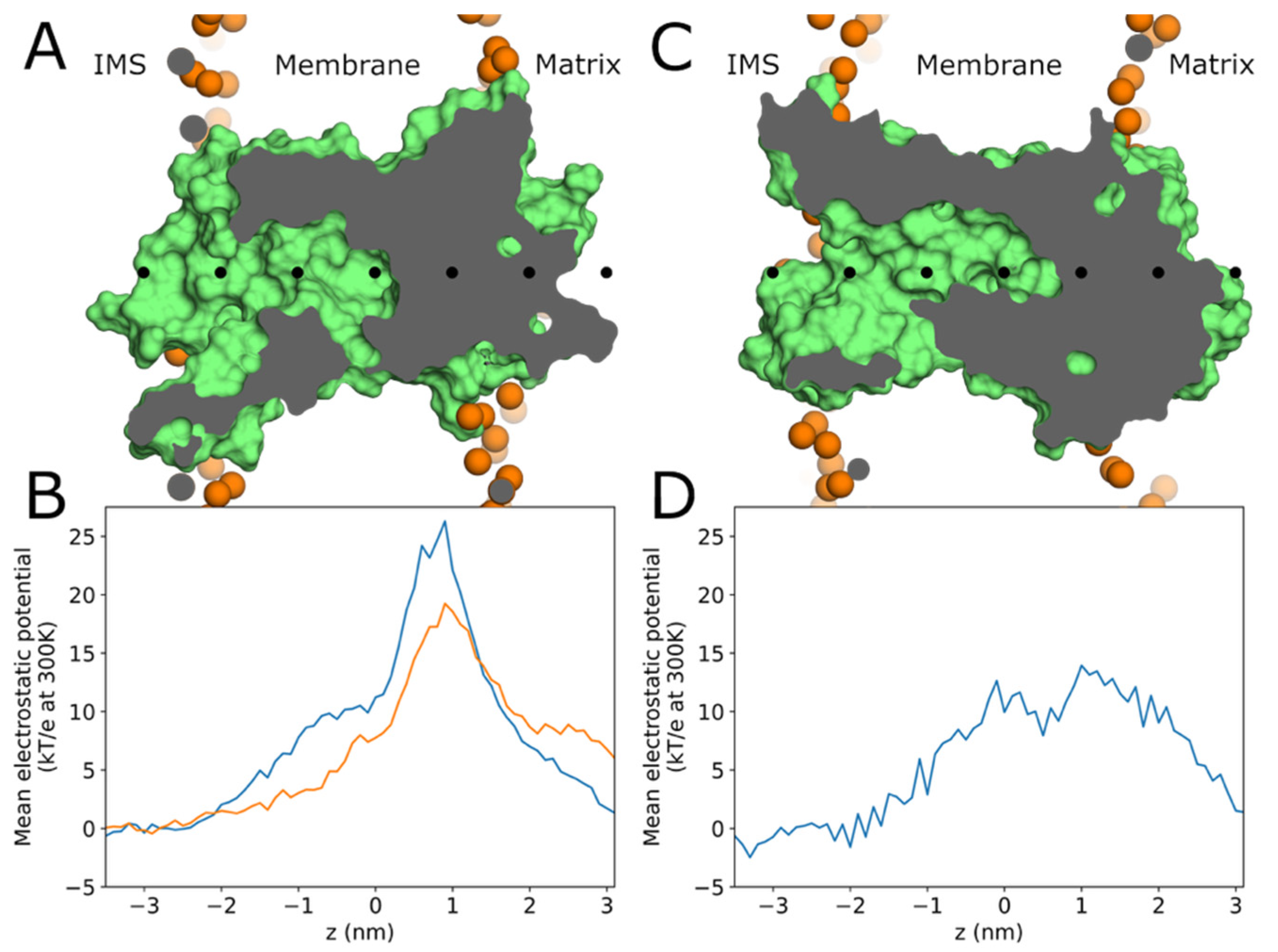
2.2. Arachidonic Acid Is Enriched in the Membrane Adjacent to AAC1 and UCP1
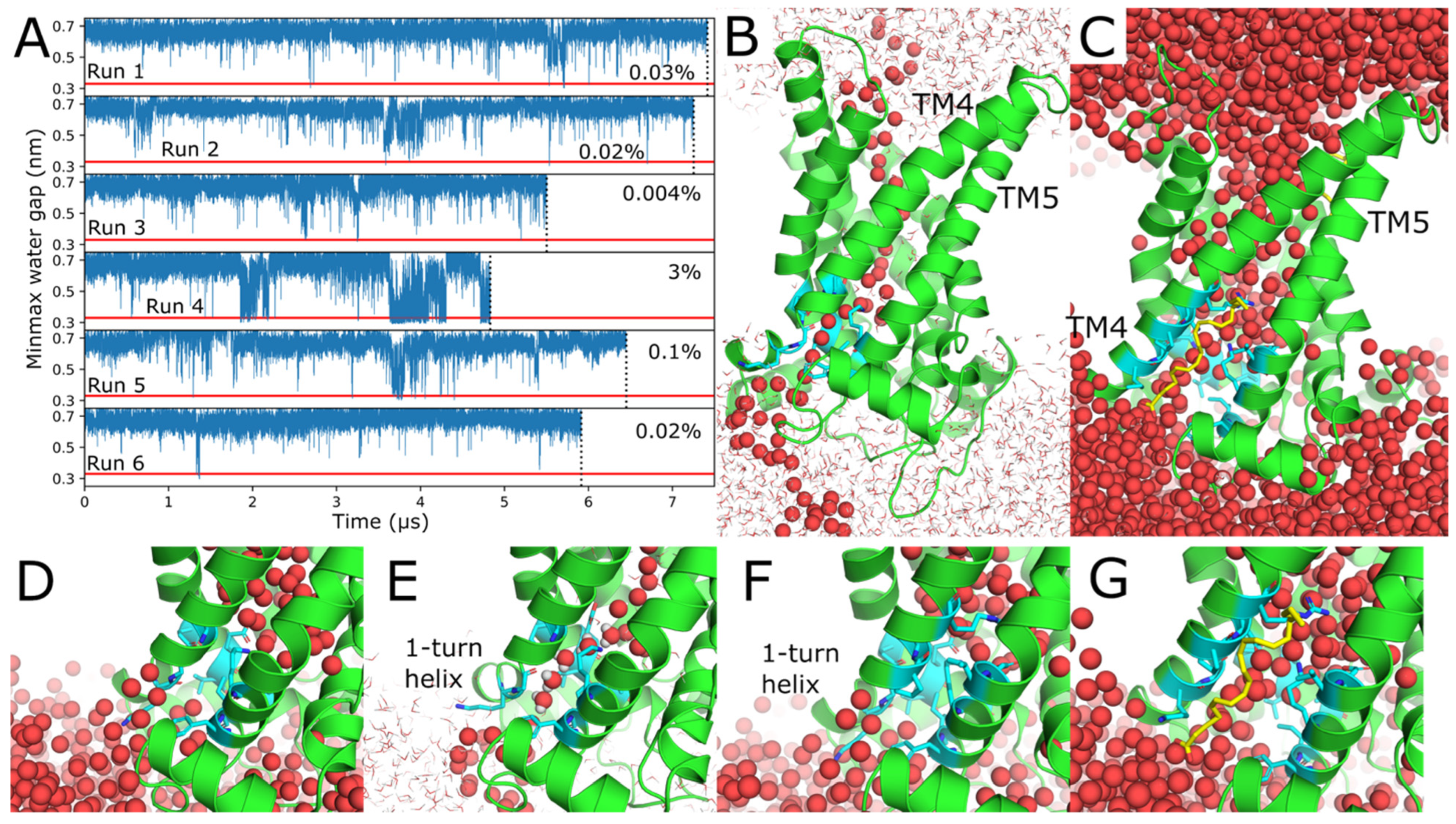
2.3. Water Wires Form Through AAC1 and UCP1
3. Discussion
4. Methods
4.1. Simulations
4.1.1. System Construction
4.1.2. MD Simulations
4.2. Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brustovetsky, N.; Klingenberg, M. The reconstituted ADP/ATP carrier can mediate H+ transport by free fatty acids, which is further stimulated by mersalyl. J. Biol. Chem. 1994, 269, 27329–27336. [Google Scholar] [CrossRef]
- Bertholet, A.M.; Chouchani, E.T.; Kazak, L.; Angelin, A.; Fedorenko, A.; Long, J.Z.; Vidoni, S.; Garrity, R.; Cho, J.; Terada, N.; et al. H+ transport is an integral function of the mitochondrial ADP/ATP carrier. Nature 2019, 571, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, M.; Winkler, E. The reconstituted isolated uncoupling protein is a membrane potential driven H+ translocator. EMBO J. 1985, 4, 3087–3092. [Google Scholar] [CrossRef]
- Winkler, E.; Klingenberg, M. Effect of fatty acids on H+ transport activity of the reconstituted uncoupling protein. J. Biol. Chem. 1994, 269, 2508–2515. [Google Scholar] [CrossRef]
- Strieleman, P.J.; Schalinske, K.L.; Shrago, E. Fatty acid activation of the reconstituted brown adipose tissue mitochondria uncoupling protein. J. Biol. Chem. 1985, 260, 13402–13405. [Google Scholar] [CrossRef] [PubMed]
- Žuna, K.; Jovanović, O.; Khailova, L.S.; Škulj, S.; Brkljača, Z.; Kreiter, J.; Kotova, E.A.; Vazdar, M.; Antonenko, Y.N.; Pohl, E.E. Mitochondrial Uncoupling Proteins (UCP1-UCP3) and Adenine Nucleotide Translocase (ANT1) Enhance the Protonophoric Action of 2,4-Dinitrophenol in Mitochondria and Planar Bilayer Membranes. Biomolecules 2021, 11, 1178. [Google Scholar] [CrossRef] [PubMed]
- Beck, V.; Jaburek, M.; Demina, T.; Rupprecht, A.; Porter, R.K.; Jezek, P.; Pohl, E.E. Polyunsaturated fatty acids activate human uncoupling proteins 1 and 2 in planar lipid bilayers. FASEB J. 2007, 21, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 25 September 2025).
- Pfaff, E.; Klingenberg, M. Adenine Nucleotide Translocation of Mitochondria. Eur. J. Biochem. 1968, 6, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Krämer, R.; Klingenberg, M. Reconstitution of adenine nucleotide transport with purified ADP, ATP-carrier protein. FEBS Lett. 1977, 82, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Garlid, K.D.; Jabůrek, M.; Ježek, P.; Vařecha, M. How do uncoupling proteins uncouple? Biochim. Biophys. Acta BBA—Bioenerg. 2000, 1459, 383–389. [Google Scholar] [CrossRef]
- Jacobsen, L.; Menon, S.; Gaudry, M.J.; Hakami Zanjani, A.A.; Reinholdt, P.; Jastroch, M.; Khandelia, H. A Novel Model for Proton Transport Mediated by Uncoupling Protein 1. bioRxiv 2025. [Google Scholar] [CrossRef]
- Kreiter, J.; Rupprecht, A.; Škulj, S.; Brkljača, Z.; Žuna, K.; Knyazev, D.G.; Bardakji, S.; Vazdar, M.; Pohl, E.E. ANT1 Activation and Inhibition Patterns Support the Fatty Acid Cycling Mechanism for Proton Transport. Int. J. Mol. Sci. 2021, 22, 2490. [Google Scholar] [CrossRef]
- Kreiter, J.; Škulj, S.; Brkljača, Z.; Bardakji, S.; Vazdar, M.; Pohl, E.E. FA Sliding as the Mechanism for the ANT1-Mediated Fatty Acid Anion Transport in Lipid Bilayers. Int. J. Mol. Sci. 2023, 24, 13701. [Google Scholar] [CrossRef] [PubMed]
- Bertholet, A.M.; Natale, A.M.; Bisignano, P.; Suzuki, J.; Fedorenko, A.; Hamilton, J.; Brustovetsky, T.; Kazak, L.; Garrity, R.; Chouchani, E.T.; et al. Mitochondrial uncouplers induce proton leak by activating AAC and UCP1. Nature 2022, 606, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Ježek, P.; Žáčková, M.; Růžička, M.; Škobisová, E.; Jabůrek, M. Mitochondrial uncoupling proteins—Facts and fantasies. Physiol. Res. 2004, 53, S199–S211. [Google Scholar] [CrossRef] [PubMed]
- Vojvodić, S.; Roticiani, G.; Vazdar, M.; Pohl, E.E. Molecular Dynamics Simulations of a Putative Novel Mechanism for UCP1-Assisted FA Anion Transport. Acta Physiol. Oxf. Engl. 2025, 241, e70068. [Google Scholar] [CrossRef]
- Robinson, A.J.; Overy, C.; Kunji, E.R.S. The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc. Natl. Acad. Sci. USA 2008, 105, 17766–17771. [Google Scholar] [CrossRef]
- Crichton, P.G.; Lee, Y.; Kunji, E.R.S. The molecular features of uncoupling protein 1 support a conventional mitochondrial carrier-like mechanism. Biochimie 2017, 134, 35–50. [Google Scholar] [CrossRef]
- Bertholet, A.M.; Kirichok, Y. UCP1: A transporter for H+ and fatty acid anions. Biochimie 2017, 134, 28–34. [Google Scholar] [CrossRef]
- Kang, Y.; Chen, L. Structural basis for the binding of DNP and purine nucleotides onto UCP1. Nature 2023, 620, 226–231. [Google Scholar] [CrossRef]
- Nury, H.; Dahout-Gonzalez, C.; Trézéguet, V.; Lauquin, G.; Brandolin, G.; Pebay-Peyroula, E. Structural basis for lipid-mediated interactions between mitochondrial ADP/ATP carrier monomers. FEBS Lett. 2005, 579, 6031–6036. [Google Scholar] [CrossRef]
- Fedorenko, A.; Lishko, P.V.; Kirichok, Y. Mechanism of Fatty-Acid-Dependent UCP1 Uncoupling in Brown Fat Mitochondria. Cell 2012, 151, 400–413. [Google Scholar] [CrossRef] [PubMed]
- Gagelin, A.; Largeau, C.; Masscheleyn, S.; Piel, M.S.; Calderón-Mora, D.; Bouillaud, F.; Hénin, J.; Miroux, B. Molecular determinants of inhibition of UCP1-mediated respiratory uncoupling. Nat. Commun. 2023, 14, 2594. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, L.; Lydersen, L.; Khandelia, H. ATP-Bound State of the Uncoupling Protein 1 (UCP1) from Molecular Simulations. J. Phys. Chem. B 2023, 127, 9685–9696. [Google Scholar] [CrossRef] [PubMed]
- PubChem. Arachidonic Acid. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/444899 (accessed on 18 September 2025).
- Echtay, K.S.; Bienengraeber, M.; Klingenberg, M. Role of Intrahelical Arginine Residues in Functional Properties of Uncoupling Protein (UCP1). Biochemistry 2001, 40, 5243–5248. [Google Scholar] [CrossRef]
- Horvath, L.I.; Drees, M.; Beyer, K.; Klingenberg, M.; Marsh, D. Lipid-protein interactions in ADP-ATP carrier/egg phosphatidylcholine recombinants studied by spin-label ESR spectroscopy. Biochemistry 1990, 29, 10664–10669. [Google Scholar] [CrossRef]
- Comte, J.; Maïsterrena, B.; Gautheron, D.C. Lipid composition and protein profiles of outer and inner membranes from pig heart mitochondria. Comparison with microsomes. Biochim. Biophys. Acta 1976, 419, 271–284. [Google Scholar] [CrossRef]
- Schiaffarino, O.; González, D.V.; García-Pérez, I.M.; Peñalva, D.A.; Almendro-Vedia, V.G.; Natale, P.; López-Montero, I. Mitochondrial membrane models built from native lipid extracts: Interfacial and transport properties. Front. Mol. Biosci. 2022, 9, 910936. [Google Scholar] [CrossRef]
- Pebay-Peyroula, E.; Dahout-Gonzalez, C.; Kahn, R.; Trézéguet, V.; Lauquin, G.J.-M.; Brandolin, G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 2003, 426, 39–44. [Google Scholar] [CrossRef]
- Jones, S.A.; Gogoi, P.; Ruprecht, J.J.; King, M.S.; Lee, Y.; Zögg, T.; Pardon, E.; Chand, D.; Steimle, S.; Copeman, D.M.; et al. Structural basis of purine nucleotide inhibition of human uncoupling protein 1. Sci. Adv. 2023, 9, eadh4251. [Google Scholar] [CrossRef]
- Agmon, N. The Grotthuss mechanism. Chem. Phys. Lett. 1995, 244, 456–462. [Google Scholar] [CrossRef]
- Adelman, J.L.; Sheng, Y.; Choe, S.; Abramson, J.; Wright, E.M.; Rosenberg, J.M.; Grabe, M. Structural Determinants of Water Permeation through the Sodium-Galactose Transporter vSGLT. Biophys. J. 2014, 106, 1280–1289. [Google Scholar] [CrossRef] [PubMed]
- Kratochvil, H.T.; Watkins, L.C.; Mravic, M.; Thomaston, J.L.; Nicoludis, J.M.; Somberg, N.H.; Liu, L.; Hong, M.; Voth, G.A.; DeGrado, W.F. Transient water wires mediate selective proton transport in designed channel proteins. Nat. Chem. 2023, 15, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Beyer, K.; Klingenberg, M. ADP/ATP carrier protein from beef heart mitochondria has high amounts of tightly bound cardiolipin, as revealed by phosphorus-31 nuclear magnetic resonance. Biochemistry 1985, 24, 3821–3826. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Willers, C.; Kunji, E.R.S.; Crichton, P.G. Uncoupling protein 1 binds one nucleotide per monomer and is stabilized by tightly bound cardiolipin. Proc. Natl. Acad. Sci. USA 2015, 112, 6973–6978. [Google Scholar] [CrossRef]
- VMD PMEPot Plugin, Version 1.0. Available online: https://www.ks.uiuc.edu/Research/vmd/plugins/pmepot/ (accessed on 28 September 2025).
- Aksimentiev, A.; Schulten, K. Imaging alpha-hemolysin with molecular dynamics: Ionic conductance, osmotic permeability, and the electrostatic potential map. Biophys. J. 2005, 88, 3745–3761. [Google Scholar] [CrossRef]
- Wang, Y.; Tajkhorshid, E. Electrostatic funneling of substrate in mitochondrial inner membrane carriers. Proc. Natl. Acad. Sci. USA 2008, 105, 9598–9603. [Google Scholar] [CrossRef]
- Moss, F.R.; Lincoff, J.; Tucker, M.; Mohammed, A.; Grabe, M.; Frost, A. Brominated lipid probes expose structural asymmetries in constricted membranes. Nat. Struct. Mol. Biol. 2023, 30, 167–175. [Google Scholar] [CrossRef]
- Jezek, P.; Modrianský, M.; Garlid, K.D. A structure-activity study of fatty acid interaction with mitochondrial uncoupling protein. FEBS Lett. 1997, 408, 166–170. [Google Scholar] [CrossRef]
- Klingenberg, M.; Echtay, K.S.; Bienengraeber, M.; Winkler, E.; Huang, S.G. Structure–Function Relationship in UCP1. Int. J. Obes. 1999, 23, S24–S29. [Google Scholar] [CrossRef]
- Li, C.; Voth, G.A. A quantitative paradigm for water-assisted proton transport through proteins and other confined spaces. Proc. Natl. Acad. Sci. USA 2021, 118, e2113141118. [Google Scholar] [CrossRef]
- Izadi, S.; Anandakrishnan, R.; Onufriev, A.V. Building Water Models: A Different Approach. J. Phys. Chem. Lett. 2014, 5, 3863–3871. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef]
- Schrödinger, LLC. The PyMOL Molecular Graphics System, Version 1.8; Schrödinger, LLC: New York, NY, USA, 2015.
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Park, S.; Choi, Y.K.; Im, W. CHARMM-GUI Membrane Builder: Past, Current, and Future Developments and Applications. J. Chem. Theory Comput. 2023, 19, 2161–2185. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D., Jr.; Pastor, R.W. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Beglov, D.; Roux, B. Finite representation of an infinite bulk system: Solvent boundary potential for computer simulations. J. Chem. Phys. 1994, 100, 9050–9063. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Bernetti, M.; Bussi, G. Pressure control using stochastic cell rescaling. J. Chem. Phys. 2020, 153, 114107. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- McGibbon, R.T.; Beauchamp, K.A.; Harrigan, M.P.; Klein, C.; Swails, J.M.; Hernández, C.X.; Schwantes, C.R.; Wang, L.-P.; Lane, T.J.; Pande, V.S. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015, 109, 1528–1532. [Google Scholar] [CrossRef] [PubMed]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]

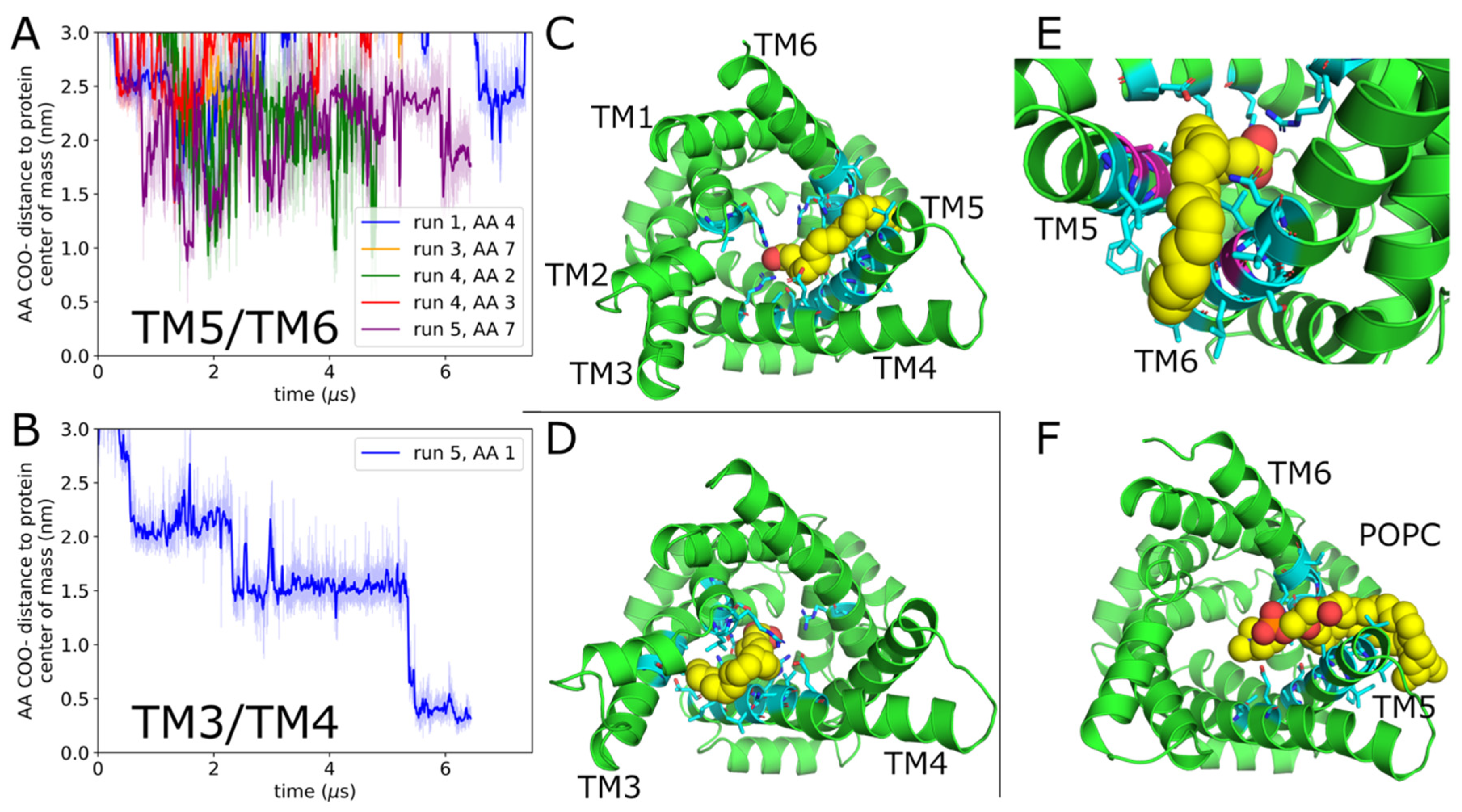
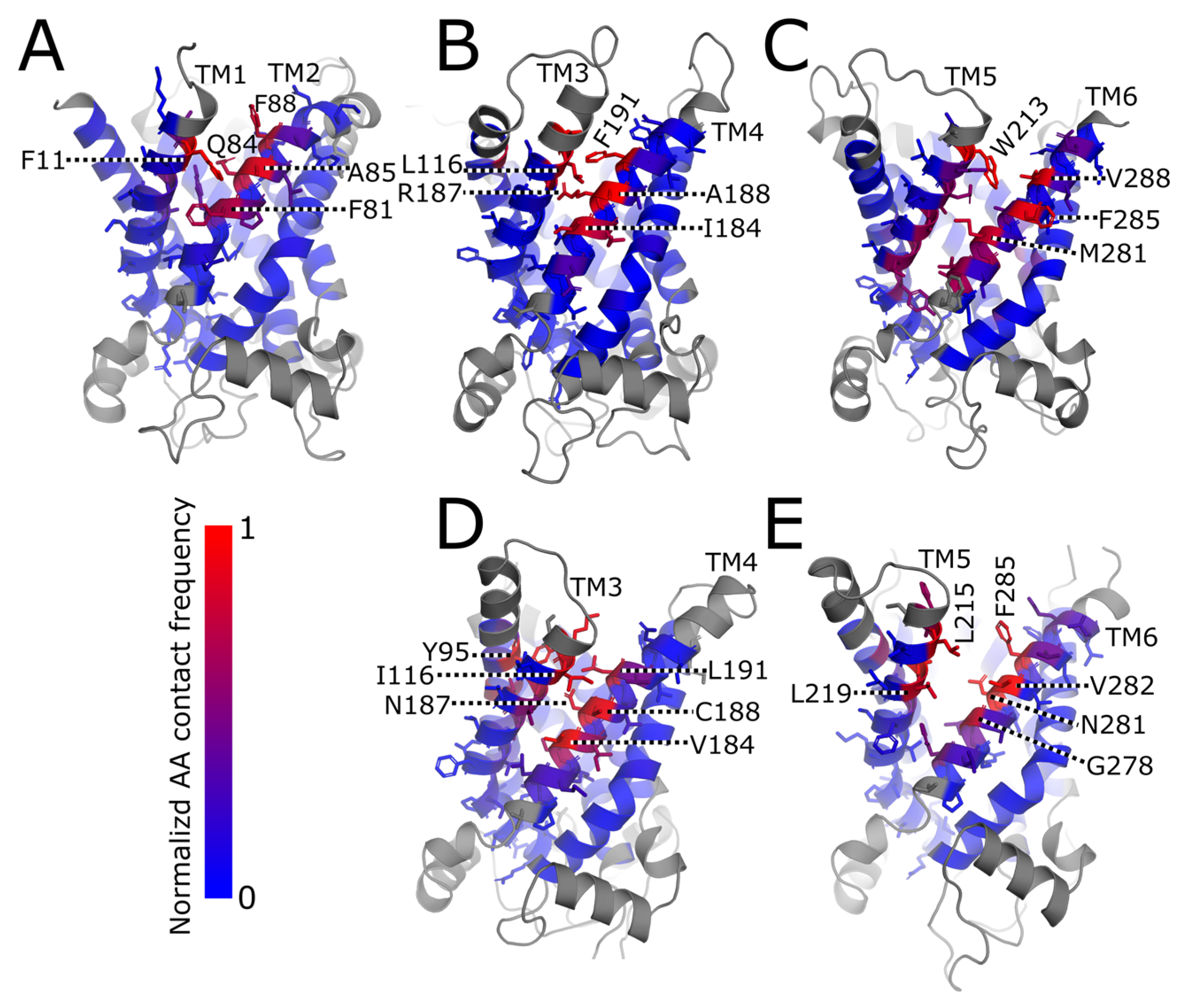
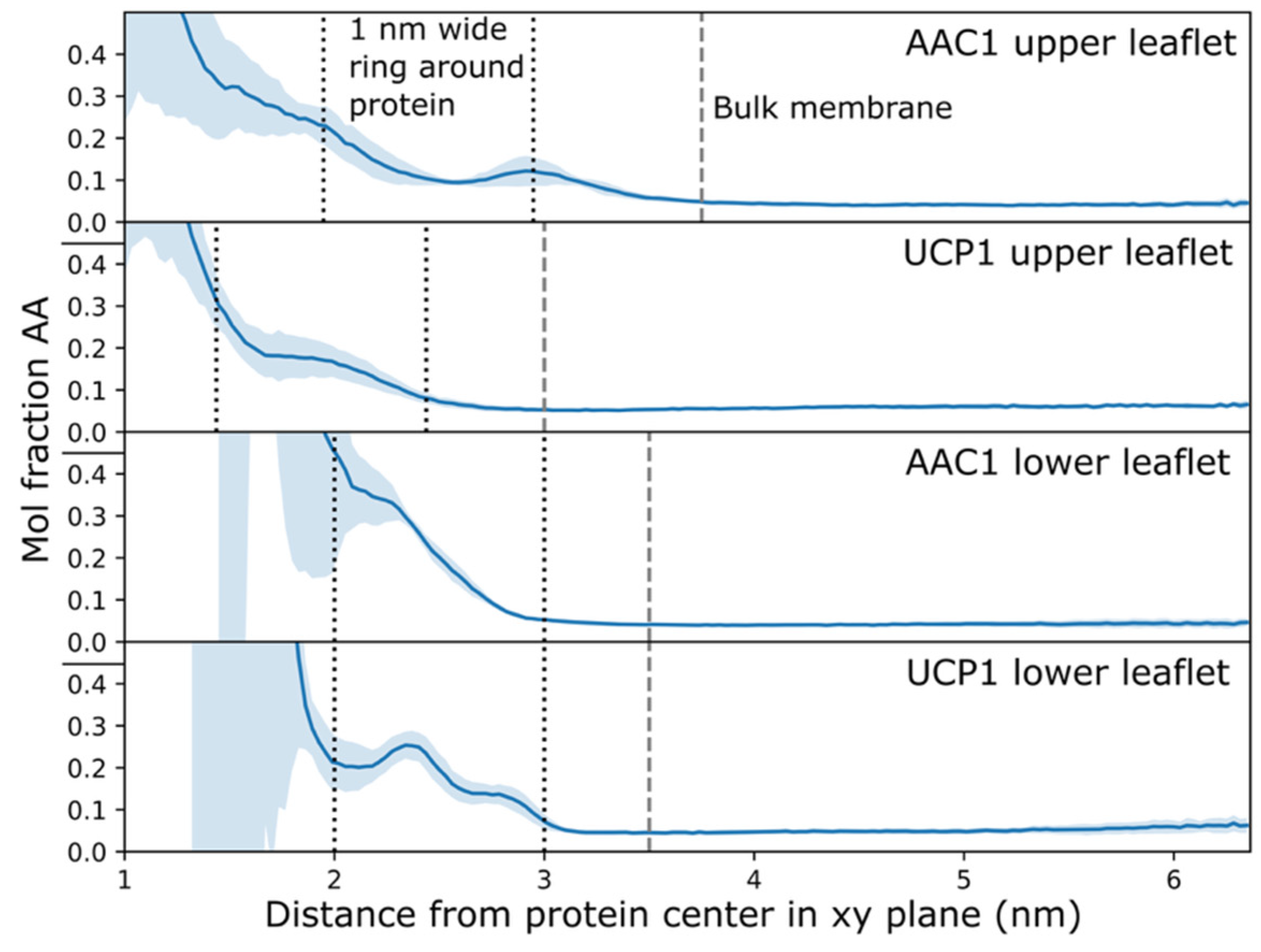
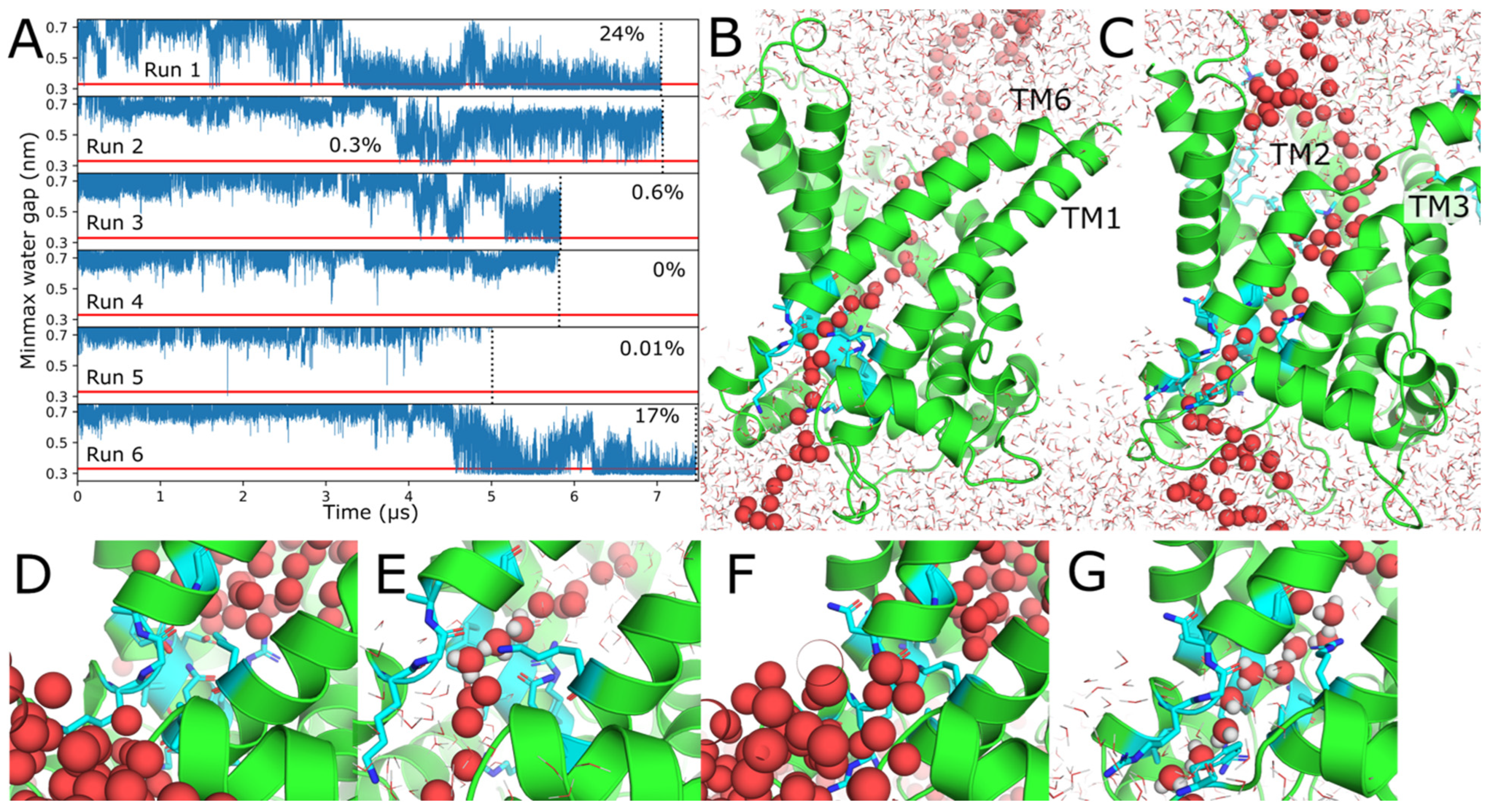
| Protein | Run | Timestep (fs) | Save Frequency (ps) | Steps | Time (μs) |
|---|---|---|---|---|---|
| AAC1 | 1 | 4 | 200 | 35,242 | 7.0484 |
| AAC1 | 2 | 4 | 200 | 35,336 | 7.0672 |
| AAC1 | 3 | 2 | 100 | 58,325 | 5.8325 |
| AAC1 | 4 | 2 | 100 | 58,169 | 5.8169 |
| AAC1 | 5 | 4 | 200 | 25,043 | 5.0086 |
| AAC1 | 6 | 4 | 200 | 37,351 | 7.4702 |
| UCP1 | 1 | 4 | 200 | 37,084 | 7.4168 |
| UCP1 | 2 | 4 | 200 | 36,265 | 7.253 |
| UCP1 | 3 | 2 | 100 | 55,026 | 5.5026 |
| UCP1 | 4 | 2 | 100 | 48,309 | 4.8309 |
| UCP1 | 5 | 4 | 200 | 32,256 | 6.4512 |
| UCP1 | 6 | 4 | 200 | 29,577 | 5.9154 |
| AA | POPC | |||
|---|---|---|---|---|
| Site | AAC1 | UCP1 | AAC1 | UCP1 |
| TM1/TM2 | 5 | 0 | 8 * | 0 |
| TM3/TM4 | 2 * | 1 | 0 | 0 |
| TM5/TM6 | 4 * | 5 * | 6 | 7 ** |
| TM6/TM1 | 0 | 0 | 4 | 0 |
| IMS cavity | 3 | 0 | 0 | 0 |
| Molecule | Number in AAC1 System | Number in UCP1 System |
|---|---|---|
| Protein (AAC1 or UCP1, respectively) | 1 | 1 |
| Tetralinoleoylcardiolipin (TLCL2; charge−2) | 3 | 3 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) | 211 (107 in M leaflet, 104 in C leaflet) | 217 (107 in M leaflet, 110 in C leaflet) |
| Arachidonic acid (ARAN; charge−1) | 8 per leaflet (16 total) | 8 per leaflet (16 total) |
| Water | 14,375 | 14,595 |
| Potassium (K+) | 41 | 49 |
| Chloride (Cl−) | 38 | 38 |
| Step | Length (ns) | Protein Backbone Restraints | Protein Sidechain Restraints | Restraints on All Other Molecules | Thermostat | Barostat |
|---|---|---|---|---|---|---|
| 1 (minimization; no dynamics) | n/a; up to 500,000 steps; emtol = 1000 kJ·mol−1·nm−1 | 4000 | 4000 | 400 | n/a | n/a |
| 2 (NVT) | 0.1 | 4000 | 4000 | 400 | Berendsen [58] | none |
| 3 | 0.1 | 4000 | 4000 | 400 | Berendsen | Berendsen |
| 4 | 0.1 | 2000 | 2000 | 80 | Berendsen | Berendsen |
| 5 | 0.1 | 2000 | 2000 | 16 | Berendsen | Berendsen |
| 6 | 20 | 2000 | 2000 | 0 | Berendsen | Berendsen |
| 7 | 2 | 2000 | 1000 | 0 | Berendsen | Berendsen |
| 8 | 2 | 2000 | 400 | 0 | Berendsen | Berendsen |
| 9 | 2 | 1000 | 400 | 0 | Berendsen | Berendsen |
| 10 | 2 | 400 | 200 | 0 | Berendsen | Berendsen |
| 11 | 2 | 200 | 20 | 0 | Berendsen | Berendsen |
| 12 | 10 | 20 | 0 | 0 | Berendsen | Berendsen |
| 13 | 20 | 0 | 0 | 0 | v-rescale [59] | c-rescale [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borowsky, J.H.; Grabe, M. Interactions of Arachidonic Acid with AAC1 and UCP1. Int. J. Mol. Sci. 2025, 26, 10504. https://doi.org/10.3390/ijms262110504
Borowsky JH, Grabe M. Interactions of Arachidonic Acid with AAC1 and UCP1. International Journal of Molecular Sciences. 2025; 26(21):10504. https://doi.org/10.3390/ijms262110504
Chicago/Turabian StyleBorowsky, Jonathan H., and Michael Grabe. 2025. "Interactions of Arachidonic Acid with AAC1 and UCP1" International Journal of Molecular Sciences 26, no. 21: 10504. https://doi.org/10.3390/ijms262110504
APA StyleBorowsky, J. H., & Grabe, M. (2025). Interactions of Arachidonic Acid with AAC1 and UCP1. International Journal of Molecular Sciences, 26(21), 10504. https://doi.org/10.3390/ijms262110504






