Jumping Translocation Breakpoint Expression in Midgestation Mouse Embryos
Abstract
1. Introduction
2. Results and Discussion
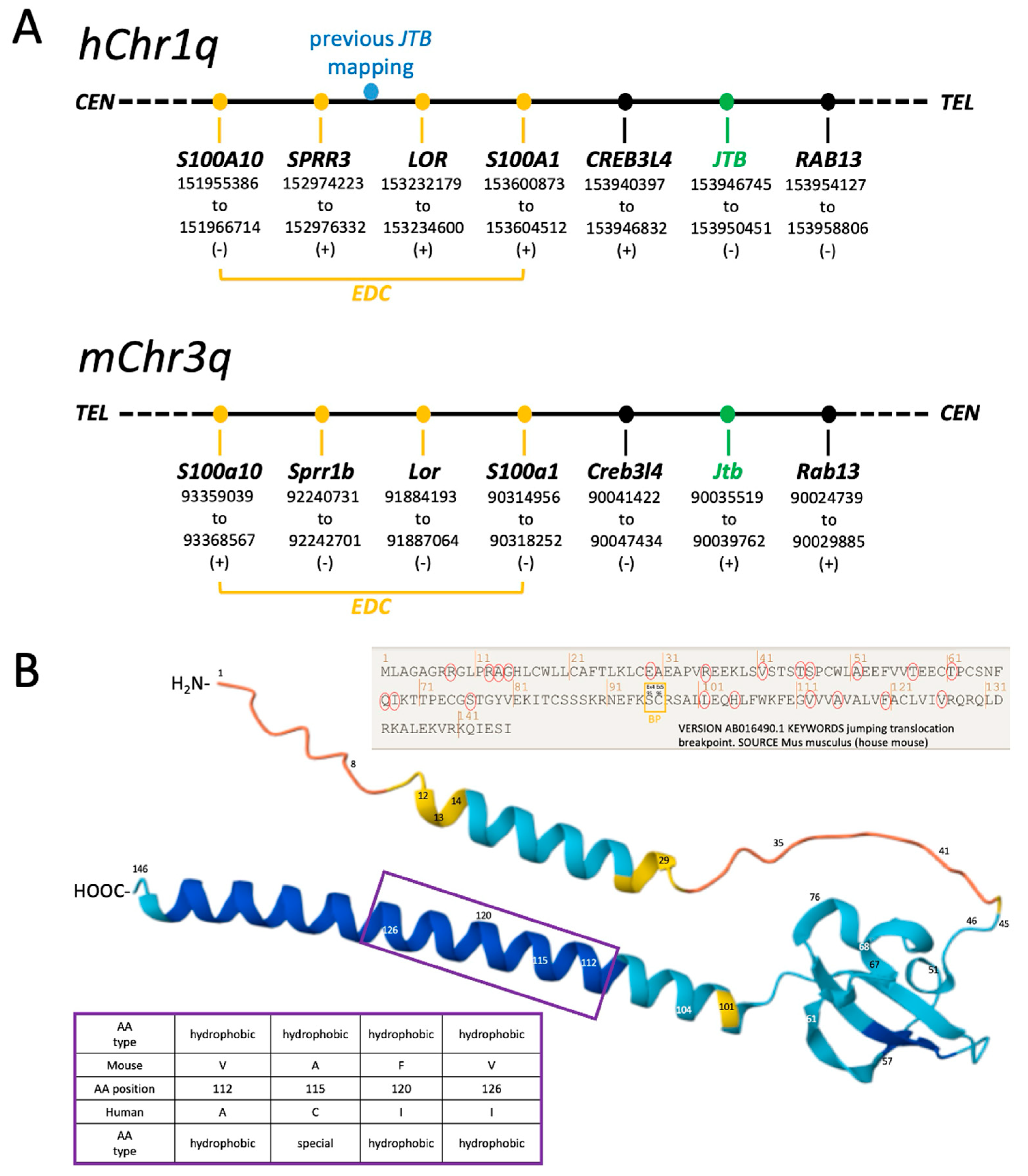
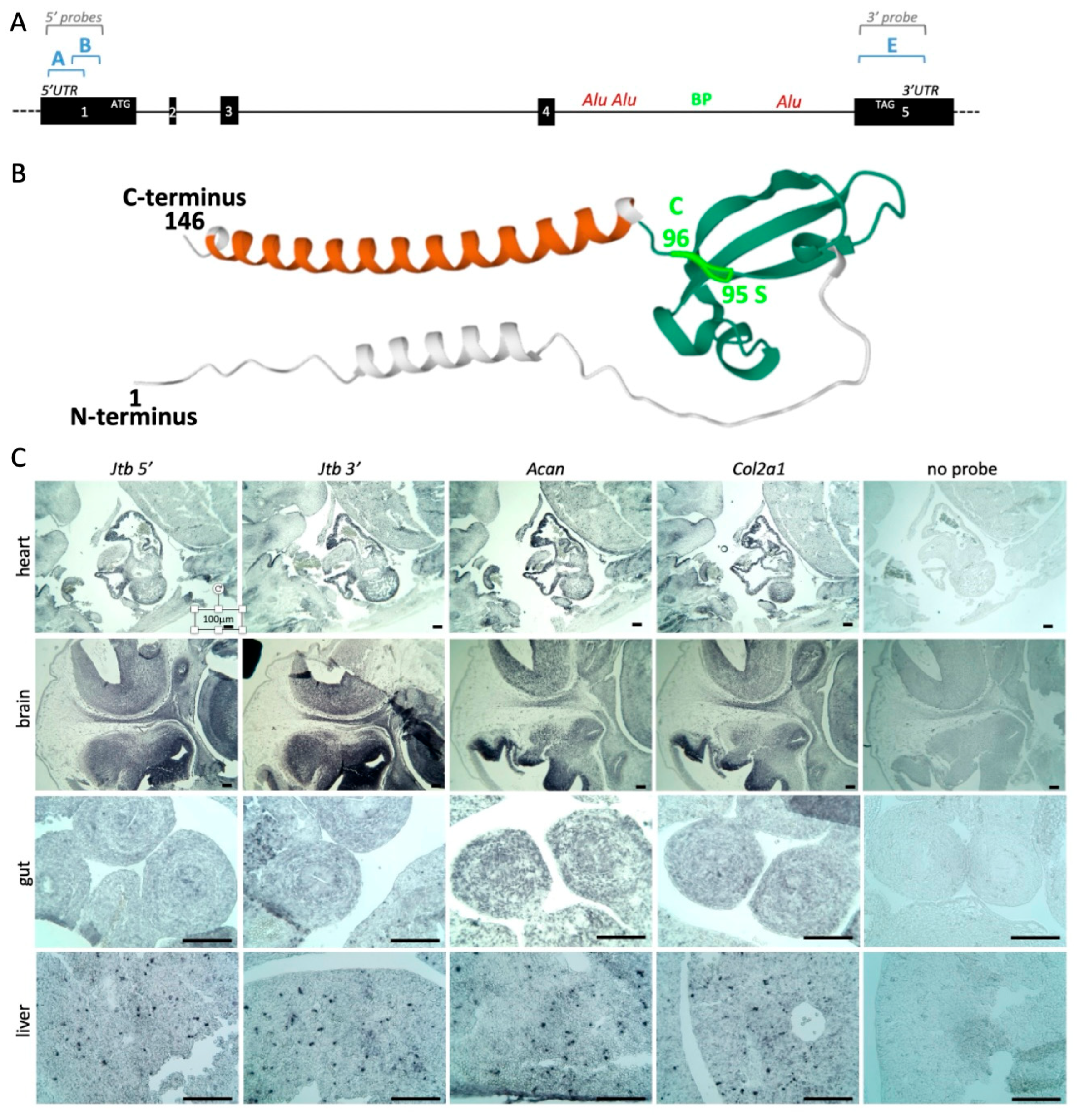
2.1. Revised Genomic Location of Human JTB and Murine Jtb in the Context of the EDC
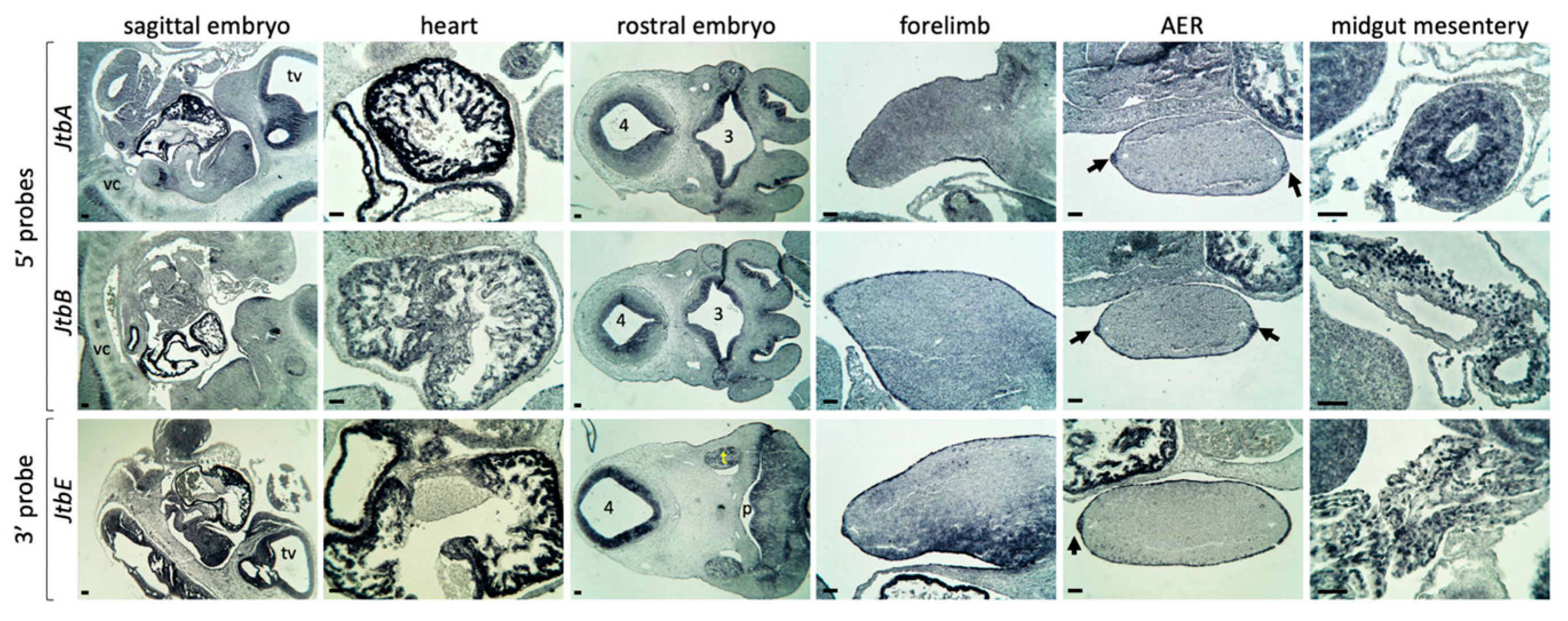
2.2. Jtb Is Present in Many Crucial Tissues During Mouse Embryonic Development
2.3. Jtb Neighbors
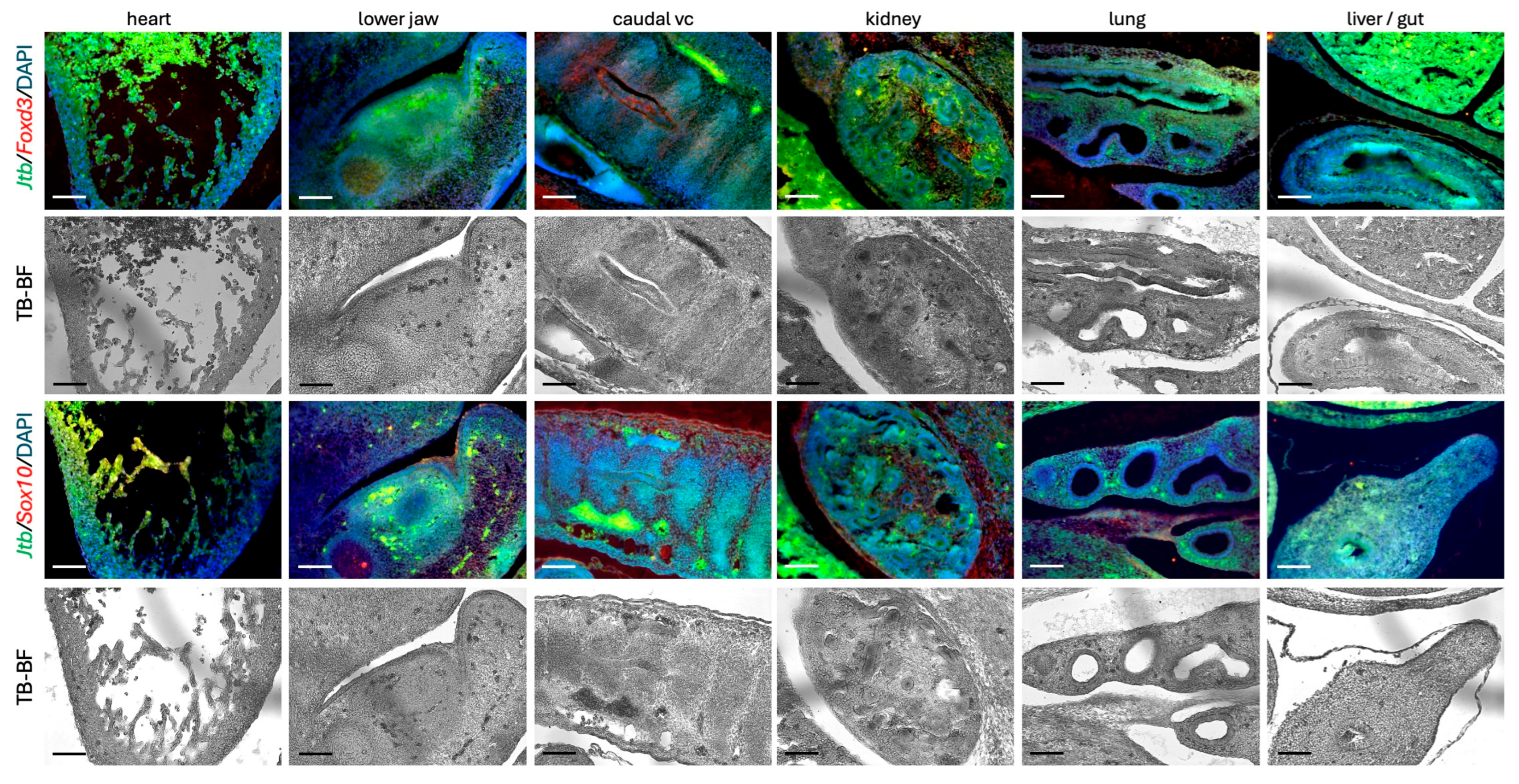
2.4. Jtb Expression in the Context of Migratory Cell Markers
2.5. Jtb Is Expressed in the Developing Vertebral Column
3. Materials and Methods
3.1. JTB/Jtb Mapping Refinement
3.2. Mouse Embryos
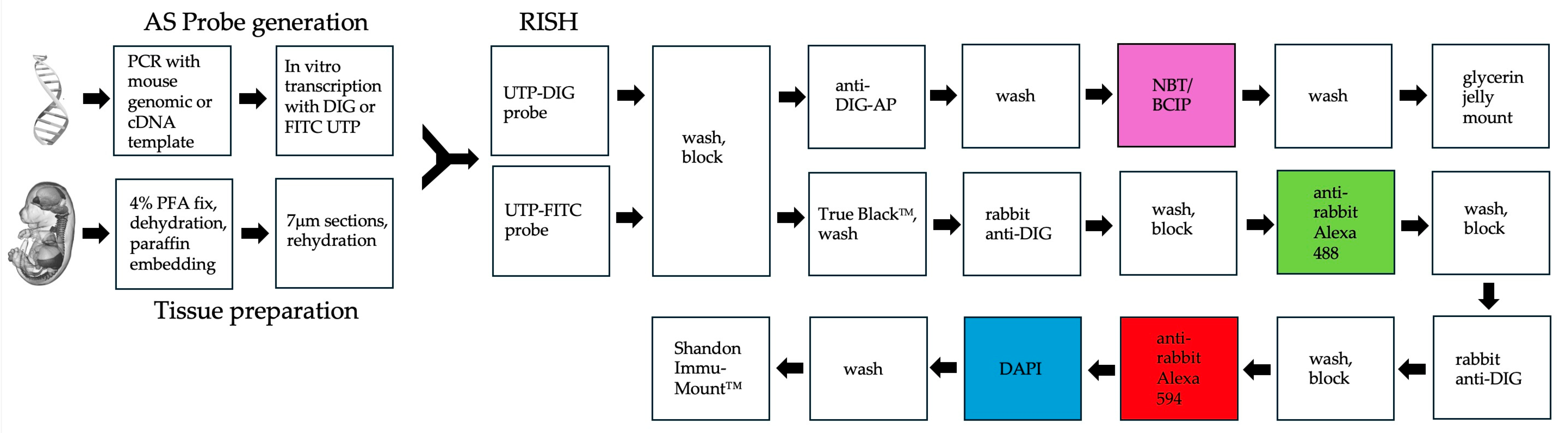
3.3. RNA In Situ Hybridization (RISH) for Gene Expression Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| JTB | Jumping Translocation Breakpoint |
| EDC | Epidermal Differentiation Complex |
| CPP/CPC | Chromosomal Passenger Proteins/Complex |
| PAR | Prostate Androgen-Regulated |
| AR | Androgen Receptor |
| AP | Alkaline Phosphatase |
| DAPI | 4′,6-diamidino-2-phenylindole |
| UTP | Uridine-Triphosphate Nucleotide |
| DIG | Digoxigenin |
| FITC | Fluorescein Isothiocyanate |
| LOR | Loricrin |
| SEDC | Single Coding Exon EDC |
| CREB3L4 | cAMP-responsive Element-Binding Protein 3-Like Protein 4 |
| LAR | Luminal Androgen Receptor subtype |
| TNBC | Triple-Negative Breast Cancer |
| OASIS | Old Astrocyte Specifically Induced Substance |
| RISH | RNA In Situ Hybridization |
| IACUC | Institutional Animal Care and Use Committee |
| NACLAR | National Advisory Committee for Laboratory Animal Research |
| PBS | Phosphate-Buffered Saline |
| PFA | Paraformaldehyde |
| NBT/BCIP | Nitro Blue Tetrazolium/5-bromo-4-chloro-3-indolyl phosphate substrate |
| TB | TrueBlackTM |
| TFS | Thermo Fisher Scientific |
References
- Jayaweera, T.M.; Jayathirtha, M.; Weraduwage, K.; Kraus, P.; Darie, C.C.; Neagu, A.N. From Jumping Gene to Cancer: Revisiting the Role of JTB Protein. Biomedicines 2025, 13, 1705. [Google Scholar] [CrossRef]
- Platica, O.; Chen, S.; Ivan, E.; Lopingco, M.C.; Holland, J.F.; Platica, M. PAR, a novel androgen regulated gene, ubiquitously expressed in normal and malignant cells. Int. J. Oncol. 2000, 16, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S.; Osawa, M.; Omine, M.; Ishikawa, F. JTB: A novel membrane protein gene at 1q21 rearranged in a jumping translocation. Oncogene 1999, 18, 2085–2090. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Ma, L.; Qin, L. Potential Role of the Epidermal Differentiation Complex in the Pathogenesis of Psoriasis. Front. Biosci. 2022, 27, 325. [Google Scholar] [CrossRef]
- Hatakeyama, S.; Fujita, K.; Mori, H.; Omine, M.; Ishikawa, F. Shortened telomeres involved in a case with a jumping translocation at 1q21. Blood 1998, 91, 1514–1519. [Google Scholar] [CrossRef]
- Solomon, E.; Borrow, J.; Goddard, A.D. Chromosome aberrations and cancer. Science 1991, 254, 1153–1160. [Google Scholar] [CrossRef]
- Platica, M.; Ivan, E.; Ionescu, A.; Holland, J.F.; Mora, G.; Tindall, D.J.; Mandeli, J.; Unger, P.D.; Platica, O. Transformation of NIH 3T3 cells by enhanced PAR expression. Biochem. Biophys. Res. Commun. 2004, 314, 891–896. [Google Scholar] [CrossRef]
- Rousseau, F.; Pan, B.; Fairbrother, W.J.; Bazan, J.F.; Lingel, A. The structure of the extracellular domain of the jumping translocation breakpoint protein reveals a variation of the midkine fold. J. Mol. Biol. 2012, 415, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Kanome, T.; Itoh, N.; Ishikawa, F.; Mori, K.; Kim-Kaneyama, J.R.; Nose, K.; Shibanuma, M. Characterization of Jumping translocation breakpoint (JTB) gene product isolated as a TGF-β1-inducible clone involved in regulation of mitochondrial function, cell growth and cell death. Oncogene 2007, 26, 5991–6001. [Google Scholar] [CrossRef]
- Liu, S.; Chen, S.; Zeng, J. TGF-β signaling: A complex role in tumorigenesis (Review). Mol. Med. Rep. 2018, 17, 699–704. [Google Scholar] [CrossRef]
- Jayathirtha, M.; Jayaweera, T.; Whitham, D.; Petre, B.A.; Neagu, A.-N.; Darie, C.C. Two-Dimensional Polyacrylamide Gel Electrophoresis Coupled with Nanoliquid Chromatography–Tandem Mass Spectrometry-Based Identification of Differentially Expressed Proteins and Tumorigenic Pathways in the MCF7 Breast Cancer Cell Line Transfected for Jumping Translocation Breakpoint Protein Overexpression. Int. J. Mol. Sci. 2023, 24, 14714. [Google Scholar]
- Platica, M.; Ionescu, A.; Ivan, E.; Holland, J.F.; Mandeli, J.; Platica, O. PAR, a protein involved in the cell cycle, is functionally related to chromosomal passenger proteins. Int. J. Oncol. 2011, 38, 777–785. [Google Scholar] [CrossRef]
- Xu, X.F.; Zhou, S.W.; Zhang, X.; Ye, Z.Q.; Zhang, J.H.; Ma, X.; Zheng, T.; Li, H.Z. Prostate androgen-regulated gene: A novel potential target for androgen-independent prostate cancer therapy. Asian J. Androl. 2006, 8, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Yuxiong, W.; Faping, L.; Bin, L.; Yanghe, Z.; Yao, L.; Yunkuo, L.; Yishu, W.; Honglan, Z. Regulatory mechanisms of the cAMP-responsive element binding protein 3 (CREB3) family in cancers. Biomed. Pharmacother. 2023, 166, 115335. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Saito, A.; Kamikawa, Y.; Nakazawa, N.; Imaizumi, K. AIbZIP/CREB3L4 Promotes Cell Proliferation via the SKP2-p27 Axis in Luminal Androgen Receptor Subtype Triple-Negative Breast Cancer. Mol. Cancer Res. 2024, 22, 373–385. [Google Scholar] [CrossRef]
- Kim, T.H.; Park, J.M.; Kim, M.Y.; Ahn, Y.H. The role of CREB3L4 in the proliferation of prostate cancer cells. Sci. Rep. 2017, 7, 45300. [Google Scholar] [CrossRef]
- Pawlowski, J.; Kraft, A.S. Bax-induced apoptotic cell death. Proc. Natl. Acad. Sci. USA 2000, 97, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Volz, A.; Korge, B.P.; Compton, J.G.; Ziegler, A.; Steinert, P.M.; Mischke, D. Physical mapping of a functional cluster of epidermal differentiation genes on chromosome 1q21. Genomics 1993, 18, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Holthaus, K.B.; Eckhart, L. Development-Associated Genes of the Epidermal Differentiation Complex (EDC). J. Dev. Biol. 2024, 12, 4. [Google Scholar] [CrossRef]
- Tyszkiewicz, T.; Jarzab, M.; Szymczyk, C.; Kowal, M.; Krajewska, J.; Jaworska, M.; Fraczek, M.; Krajewska, A.; Hadas, E.; Swierniak, M.; et al. Epidermal differentiation complex (locus 1q21) gene expression in head and neck cancer and normal mucosa. Folia Histochem. Cytobiol. 2014, 52, 79–89. [Google Scholar] [CrossRef]
- Abhishek, S.; Palamadai Krishnan, S. Epidermal Differentiation Complex: A Review on Its Epigenetic Regulation and Potential Drug Targets. Cell J. 2016, 18, 1–6. [Google Scholar] [PubMed]
- Gregory, S.; Sekhon, M.; Schein, J.; Zhao, S.; Osoegawa, K.; Scott, C.; Evans, R.; Burridge, P.; Cox, T.; Fox, C.; et al. A physical map of the mouse genome. Nature 2002, 418, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Jayathirtha, M.; Neagu, A.N.; Whitham, D.; Alwine, S.; Darie, C.C. Investigation of the effects of downregulation of jumping translocation breakpoint (JTB) protein expression in MCF7 cells for potential use as a biomarker in breast cancer. Am. J. Cancer Res. 2022, 12, 4373–4398. [Google Scholar]
- Jayathirtha, M.; Whitham, D.; Alwine, S.; Donnelly, M.; Neagu, A.N.; Darie, C.C. Investigating the Function of Human Jumping Translocation Breakpoint Protein (hJTB) and Its Interacting Partners through In-Solution Proteomics of MCF7 Cells. Molecules 2022, 27, 8301. [Google Scholar] [CrossRef]
- Kraus, P.; Sivakamasundari, V.; Xing, X.; Lufkin, T. Generating Mouse Lines for Lineage Tracing and Knockout Studies; Methods in Molecular Biology; Coppola, V., Singh, S.R., Eds.; Humana Press: Clifton, NJ, USA; New York, NY, USA, 2014; Volume 1194, pp. 37–62. [Google Scholar]
- Ma, Y.; Zhang, P.; Wang, F.; Yang, J.; Yang, Z.; Qin, H. The relationship between early embryo development and tumourigenesis. J. Cell Mol. Med. 2010, 14, 2697–2701. [Google Scholar] [CrossRef]
- Harder, T.; Kube, E.; Gerke, V. Cloning and characterization of the human gene encoding p11: Structural similarity to other members of the S-100 gene family. Gene 1992, 113, 269–274. [Google Scholar] [CrossRef]
- Nishimura, N.; Sasaki, T. Rab family small G proteins in regulation of epithelial apical junctions. Front. Biosci. 2009, 14, 2115–2129. [Google Scholar] [CrossRef]
- Ioannou, M.S.; McPherson, P.S. Regulation of Cancer Cell Behavior by the Small GTPase Rab13. J. Biol. Chem. 2016, 291, 9929–9937. [Google Scholar] [CrossRef]
- Sleeboom, J.J.F.; van Tienderen, G.S.; Schenke-Layland, K.; van der Laan, L.J.W.; Khalil, A.A.; Verstegen, M.M.A. The extracellular matrix as hallmark of cancer and metastasis: From biomechanics to therapeutic targets. Sci. Transl. Med. 2024, 16, eadg3840. [Google Scholar] [CrossRef] [PubMed]
- Coffey, J.C.; O’Leary, D.P. The mesentery: Structure, function, and role in disease. Lancet Gastroenterol. Hepatol. 2016, 1, 238–247. [Google Scholar] [CrossRef]
- Byrnes, K.G.; McDermott, K.; Coffey, J.C. Development of mesenteric tissues. Semin. Cell Dev. Biol. 2019, 92, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Du, M.; Zhang, W.; Liu, L.; Gao, Z.; Chen, W.; Gu, Y.; Zhu, K.; Niu, X.; Sun, Q.; et al. Mesenteric Neural Crest Cells Are the Embryological Basis of Skip Segment Hirschsprung’s Disease. Cell Mol. Gastroenterol. Hepatol. 2021, 12, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Celia-Terrassa, T.; Kang, Y. How important is EMT for cancer metastasis? PLoS Biol. 2024, 22, e3002487. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhang, Z.; Zhou, C.; Liu, L.; Huang, C. Epithelial-mesenchymal transition: The history, regulatory mechanism, and cancer therapeutic opportunities. MedComm 2022, 3, e144. [Google Scholar] [CrossRef]
- Bronner-Fraser, M. Neural crest cell migration in the developing embryo. Trends Cell Biol. 1993, 3, 392–397. [Google Scholar] [CrossRef]
- Simoes-Costa, M.; Bronner, M.E. Establishing neural crest identity: A gene regulatory recipe. Development 2015, 142, 242–257. [Google Scholar] [CrossRef]
- Wahlbuhl, M.; Reiprich, S.; Vogl, M.R.; Bosl, M.R.; Wegner, M. Transcription factor Sox10 orchestrates activity of a neural crest-specific enhancer in the vicinity of its gene. Nucleic Acids Res. 2012, 40, 88–101. [Google Scholar] [CrossRef]
- Wu, Z.; Li, Y.; Niu, Y.; Lu, J.; Yan, Z.; Xu, T.; Guo, Y.; Dong, Z.; Guo, W. FOXD3 suppresses epithelial-mesenchymal transition through direct transcriptional promotion of SMAD7 in esophageal squamous cell carcinoma. Mol. Carcinog. 2021, 60, 859–873. [Google Scholar] [CrossRef]
- Neeb, Z.; Lajiness, J.D.; Bolanis, E.; Conway, S.J. Cardiac outflow tract anomalies. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 499–530. [Google Scholar] [CrossRef]
- Grow, M.W.; Krieg, P.A. Tinman function is essential for vertebrate heart development: Elimination of cardiac differentiation by dominant inhibitory mutants of the tinman-related genes, XNkx2-3 and XNkx2-5. Dev. Biol. 1998, 204, 187–196. [Google Scholar] [CrossRef]
- Murphy, K.; Lufkin, T.; Kraus, P. Development and Degeneration of the Intervertebral Disc-Insights from Across Species. Vet. Sci. 2023, 10, 540. [Google Scholar] [CrossRef]
- Kraus, P.; Sivakamasundari, V.; Olsen, V.; Villeneuve, V.; Hinds, A.; Lufkin, T. Klhl14 Antisense RNA is a Target of Key Skeletogenic Transcription Factors in the Developing Intervertebral Disc. Spine 2019, 44, E260–E268. [Google Scholar] [CrossRef]
- Kraus, P.; Sivakamasundari, V.; Yu, H.B.; Xing, X.; Lim, S.L.; Adler, T.; Pimentel, J.A.; Becker, L.; Bohla, A.; Garrett, L.; et al. Pleiotropic functions for transcription factor zscan10. PLoS ONE 2014, 9, e104568, Erratum in PLoS ONE 2015, 10, e0118766. [Google Scholar] [CrossRef] [PubMed]
- Kraus, P.; Sivakamasundari, V.; Lim, S.L.; Xing, X.; Lipovich, L.; Lufkin, T. Making sense of Dlx1 antisense RNA. Dev. Biol. 2013, 376, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Kraus, P.; Yerden, R.; Sipes, D.; Sur, S.; Lufkin, T. A quantitative and qualitative RNA expression profiling assay for cell culture with single cell resolution. Cytotechnology 2018, 70, 185–192. [Google Scholar] [CrossRef]
- Li, K.; Varden, L.; Henderson, A.; Lufkin, T.; Kraus, P. Simultaneous detection of multiple mRNAs and proteins in bovine IVD cells and tissue with single cell resolution. Biotechnol. Lett. 2021, 43, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Lema Fernandez, A.G.; Nardelli, C.; Pierini, V.; Crescenzi, B.; Pellanera, F.; Matteucci, C.; Crocioni, M.; Arniani, S.; Di Battista, V.; Quintini, M.; et al. Epigenetic Modeling of Jumping Translocations of 1q Heterochromatin in Acute Myeloid Leukemia After 5′-Azacytidine Treatment. Genes Chromosomes Cancer 2024, 63, e70013. [Google Scholar] [CrossRef]



| Probe | Forward Primer | Reverse Primer | Reference Sequence | Promoter | Product Length [bp] | Label/Fluorochrome for Double RISH |
|---|---|---|---|---|---|---|
| Jtb A | AGCCTCCGCGAGCAAGATG | GCGCTATAGGAACGGCCTCG | NM_206924.2 | T7 | 174 | |
| Jtb B | GACGCCTGCGTCCCACAA | AGCAGCCAGATGCCAGAGTTC | NM_206924.2 | T7 | 130 | |
| Jtb E | TGCCGTTCGGCTCTACTGGA | GCCGCTGAACTTTCTCCATCTGA | NM_206924.2 | T7 | 222 | DIG/Alexa Fluor 488 |
| Col2a1 | CCATTGCGAACCCAAAGGAC | CACCATTGTGTAGGACACGC | NM_031163.4 | T3 | 326 | |
| Acan | TGAGAGAGGCGAATGGAACG | CCAGTCCAGCCGAGAAATGA | NM_001361500.1 | T3 | 998 | |
| S100a1 | TATGTTGTGCTGGTGGCTGCT | CCTTGGTGCACGTCGAGACT | AF368423.1 | T7 | 288 | |
| S100a10 | CACCCACGGGGTCACTTGAG | TGAGGGCAATGGGATGCAAACA | NM_009112.2 | T7 | 171 | |
| Lor | TGGCAAGGGTGTGCCAGTCT | GGGGGAAGGGGCGCTTAAAAT | NM_008508.3 | T7 | 291 | |
| Rab13 | TGGCACCTCAAGGGGAGATGA | GGCAAGGTTCCGTCCACTCT | NM_026677.4 | T7 | 519 | |
| Sox10 | TCCAGCCAGGGTGTTTGGTG | CTCGTGAAGAGCCCAACGCC | NM_011437.1 | T7 | 217 | FITC/Alexa Fluor 594 |
| Nkx2.5 | GTGACGCAGAACTGCCCGT | TGGCGACGCAGGTTTCACAG | AF083133.1 | T7 | 251 | |
| FoxD3 | AACTCAACCCGTCCGCTGG | ATAAAACTGCGCAGAGTGAACCTT | AF067421.2 | T7 | 255 | FITC/Alexa Fluor 594 |
| RISH-Type | AP-RISH | FL-Double-RISH | Comments | |
|---|---|---|---|---|
| Steps | Repeats/Timing/Temperature | |||
| Deparaffinization | 3 × 20 min at RT | Histochoice | ||
| Rehydration | each step 10 min at RT | ethanol gradient: 100%/100%/90%/70%/50%/30%/PBS | ||
| Fixation | 4% (w/v) PFA in PBS | |||
| Wash | 3 × 5 min at RT | PBS | ||
| Prehybridization | 2–3 h at 62 °C | prehybridization solution | prevent dehydration | |
| Hybridization | o/n at 62 °C | prehybridization solution with DIG-labeled probe | prehybridization solution with DIG- and/or FITC labeled probe(s) | |
| Post-hybridizationwashes | 3 × 20 min at 62 °C | solution I | ||
| 3 × 5 min at RT | TNT | |||
| 10 min at RT | TNT/solution II (1:1) | |||
| 3 × 20 min at 58 °C | solution II | prevent dehydration | ||
| 3 × 5 min at RT | PBS | |||
| Blocking | 2 h at RT | SuperBlockTM (PBS, TFS, Waltham, MA, USA) | ||
| Primary antibody | o/n at 4 °C | anti-DIG-AP (1:2000) | rabbit anti-DIG (1:100) | |
| Wash | 3 × 10 min at RT | PBS | ||
| 6× hourly at RT | PBS | NA | ||
| 3 × 10 min at RT | NTMT | |||
| Color development | 24–48 h at 4 °C in the dark | NBT/BCIP | ||
| Blocking | 2 h at RT | NA | SuperBlockTM (PBS) | prevent dehydration |
| Secondary antibody | 3 h at RT in the dark | goat anti-rabbit Alexa Fluor 488 (1:1000) | ||
| Wash | 3 × 10 min at RT away from light | PBS | ||
| Blocking | 2 h at RT away from light | SuperBlockTM (PBS) | prevent dehydration | |
| Primary antibody | 3 h at 4 °C in the dark | rabbit anti-FITC (1:100) | ||
| Wash | 3 × 10 min at RT away from light | PBS | ||
| Blocking | 2 h at RT away from light | SuperBlockTM (PBS) | prevent dehydration | |
| Secondary antibody | o/n at 4 °C in the dark | goat anti-rabbit Alexa Fluor 594 (1:1000) | ||
| Counterstaining | 1 × 10 min at RT away from light | DAPI in PBS (1:1000) | ||
| Wash | 3 × 10 min at RT away from light | PBS | ||
| Mounting | At RT away from light | glycerin jelly | Shandon Immuno MountTM(TFS, Waltham, MA, USA) | |
| Imaging | Motic BA310 | Zeiss Axiovert | ||
| Type | Chemical/Solution | Composition/Concentration | Supplier/ Order Number |
|---|---|---|---|
| Antibodies | anti-DIG-AP Fab fragments | 1:2000 in SuperBlockTM (PBS) | Roche #11093274910 |
| rabbit anti-DIG | 1:100 in SuperBlockTM (PBS) | TFS #9H27L19 | |
| rabbit anti-FITC | 1:100 in SuperBlockTM (PBS) | TFS #71-1900 | |
| goat anti-rabbit-Alexa Fluor 488 | 1:1000 in SuperBlockTM (PBS) | TFS #A11008 | |
| goat anti-rabbit-Alexa Fluor 594 | 1:1000 in SuperBlockTM (PBS) | TFS #A11012 | |
| Solutions and stains | DAPI | 1:1000 in water | TFS #62248 |
| DEPC-water | 1:1000 | Agilent | |
| glycerin jelly | undiluted | Ted Paella | |
| Histochoice | undiluted | VWR | |
| NBT/BCIP | 1:50 in 100 mM Tris/HCl pH9.5, 100 mMNaCl | Roche/VWR | |
| NTMT | 100 mM Tris/HCl pH9.5, 50 mM MgCl2, 100 mM NaCl, 0.1% Tween20 | VWR | |
| phosphate-buffered saline (PBS) | 1× | Gibco | |
| prehybidization solution | 50% formamide 5× SSC (0.75 M sodium chloride/ 0.075 M sodium citrate dehydrate), 1× Denhardt’s (0.02% (w/v) each ficoll, polyvinylpyrrolidone, BSA), 0.1% (v/v) Tween 20 (Sigma), 0.1 mg/mL tRNA (Roche), 0.05 mg/mL Heparin | Amresco VWR VWR VWR Sigma Roche Alfa Aesar | |
| Shandon Immuno MountTM | undiluted | TFS | |
| solution I | 50% (v/v) formamide, 5× SSC (see above) 1% (v/v) sodium dodecylsulphate (SDS) | Amresco VWR VWR | |
| solution II | 50% (v/v) formamide 2× SSC 0.2% (v/v) SDS | Amresco | |
| SuperBlockTM (PBS) | undiluted | PBS | |
| TNT | 0.5 M NaCl 0.01 M Tris/HCl pH 7.5 0.1% (v/v) Tween 20 | VWR VWR Sigma | |
| True BlackTM (DMF) | 1:20 in 70% Ethanol | Biotium | |
| Probe generation | in vitro transcription | UTP-DIG labeling Mix | Roche #11277073910 |
| UTP-FITC labeling mix | Roche #11685619910 | ||
| T7-RNA polymerase 40 U/reaction | Promega | ||
| RNA Protector 40 U/reaction | Roche | ||
| template PCR | GoTaq Flexi 0.5 U/reaction | Promega | |
| dNTP mix | Promega |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McGrath, C.; Jayaweera, T.M.; Lufkin, T.; Darie, C.C.; Neagu, A.-N.; Kraus, P. Jumping Translocation Breakpoint Expression in Midgestation Mouse Embryos. Int. J. Mol. Sci. 2025, 26, 9952. https://doi.org/10.3390/ijms26209952
McGrath C, Jayaweera TM, Lufkin T, Darie CC, Neagu A-N, Kraus P. Jumping Translocation Breakpoint Expression in Midgestation Mouse Embryos. International Journal of Molecular Sciences. 2025; 26(20):9952. https://doi.org/10.3390/ijms26209952
Chicago/Turabian StyleMcGrath, Carley, Taniya M Jayaweera, Thomas Lufkin, Costel C. Darie, Anca-Narcisa Neagu, and Petra Kraus. 2025. "Jumping Translocation Breakpoint Expression in Midgestation Mouse Embryos" International Journal of Molecular Sciences 26, no. 20: 9952. https://doi.org/10.3390/ijms26209952
APA StyleMcGrath, C., Jayaweera, T. M., Lufkin, T., Darie, C. C., Neagu, A.-N., & Kraus, P. (2025). Jumping Translocation Breakpoint Expression in Midgestation Mouse Embryos. International Journal of Molecular Sciences, 26(20), 9952. https://doi.org/10.3390/ijms26209952







