Beyond Antibiotics: Repurposing Non-Antibiotic Drugs as Novel Antibacterial Agents to Combat Resistance
Abstract
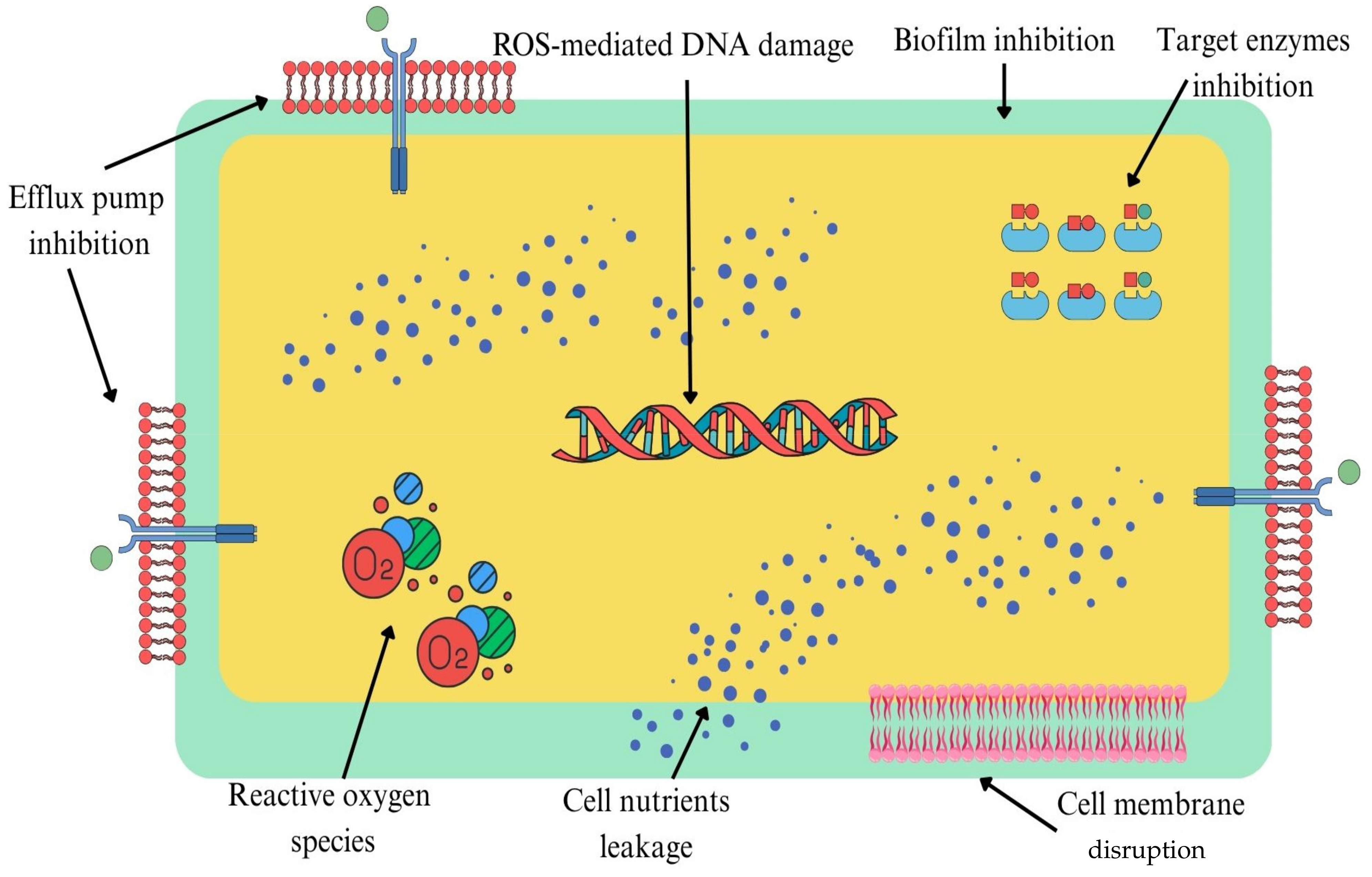
1. Introduction
2. Mechanisms of Non-Antibiotic Drug Antibacterial Activity
2.1. Efflux Pump Inhibition
2.1.1. Phenothiazine Antipsychotics
2.1.2. Selective Serotonin Reuptake Inhibitors
2.1.3. Calcium Channel Blockers
2.1.4. Statins
2.1.5. Non-Steroidal Anti-Inflammatory Drugs
2.2. Biofilm Inhibition and Disruption of Bacterial Membranes
2.2.1. Phenothiazine Antipsychotics
2.2.2. Selective Serotonin Reuptake Inhibitors
2.2.3. Calcium Channel Blockers
2.2.4. Statins
2.2.5. Non-Steroidal Anti-Inflammatory Drugs
2.3. DNA/Enzyme Inhibition, Disruption of Bacterial Nutrients and Immunomodulatory Effects
2.3.1. Phenothiazine Antipsychotics
2.3.2. Selective Serotonin Reuptake Inhibitors and Other Antidepressants
| Drug (Class) | Mechanism | Key Findings | Limitations | Reference |
|---|---|---|---|---|
| Thioridazine, Chlorpromazine (Phenothiazines) | ROS induction, DNA damage, iron homeostasis disruption, immunomodulation | In S. aureus ATCC 29213: ROS elevation, DNA damage, upregulation of katA/sodA; improved macrophage killing (THP-1); disturbed bacterial iron balance | In vitro only, one strain; no β-lactamase assays | [58] |
| Phenothiazines | ROS induction, metabolic stress | Increased susceptibility of E. coli, K. pneumoniae, P. aeruginosa to ROS; synergy with iron chelators | No enzyme assays; lab-based only | [59] |
| Phenothiazines | ROS accumulation, DNA strand breaks, interference with cell division | Confirmed DNA damage in MRSA and E. coli via comet assays | No β-lactamase assays; limited experimental validation | [9] |
| Phenothiazines | ROS generation, DNA damage, membrane potential disruption | Effects confirmed in A. baumannii, E. coli, K. pneumoniae | No β-lactamase or DNA gyrase assays | [60] |
| Chlorpromazine, Thioridazine (Phenothiazines) | ROS induction, impaired DNA repair | In E. coli: ROS increase, mutagenesis, reduced DNA repair, higher antibiotic susceptibility | In vitro only | [61] |
| SSRIs (Fluoxetine, Sertraline, Paroxetine, Citalopram, Escitalopram) | ROS induction, DNA damage, immunomodulation | In MSSA, MRSA, E. coli, K. pneumoniae, P. aeruginosa: ROS increase, DNA strand breaks; checkerboard assays showed ROS potentiates β-lactam/fluoroquinolone efficacy | No β-lactamase effects; in vitro testing only | [62] |
| SSRIs (Fluoxetine, Sertraline) | ROS induction, immunomodulation | In macrophages infected with S. aureus: increased oxidative burst, improved killing; also, activity vs. E. coli, K. pneumoniae | No β-lactamase assays; no in vivo testing | [15] |
| SSRIs | ROS induction, DNA damage | In S. aureus and E. coli: ROS accumulation, DNA strand breaks, replication impairment | No β-lactamase assays | [17] |
| TCAs | ROS induction, DNA damage | In S. aureus, E. coli, Klebsiella pneumoniae, Pseudomonas aeruginosa: increased ROS, DNA fragmentation, growth suppression | No β-lactamase assays | [63] |
| Amlodipine (CCB) | Membrane perturbation, PBP interference | In S. aureus: membrane leakage, PBP interference; enhanced β-lactam activity | No specific enzyme assays | [64] |
| CCBs | Alter cell wall metabolism | Potentiated β-lactams; confirmed bactericidal activity | Mechanism unclear; no enzyme or ROS assays | [65] |
| Amlodipine (CCB) | Antibacterial activity | Inhibition zones vs. S. aureus, E. coli, K. pneumoniae, P. aeruginosa | No MIC or synergy data | [66] |
| Amlodipine (CCB) | Membrane perturbation, energy disruption | Dose- and time-dependent killing; synergy with thioridazine/promethazine | No mechanistic assays | [67] |
| Amlodipine (CCB) | β-lactamase inhibition | In MRSA: reduced β-lactamase activity, restored cefuroxime activity; FIC < 0.1 confirmed strong synergy | Limited to MRSA; not tested in Gram-negative β-lactamase producers | [68] |
| Statins | FMM disruption | In MRSA: disrupted membrane microdomains, destabilized resistance proteins | No mechanistic details; no MICs | [69] |
| Simvastatin (Statin) | Membrane perturbation, virulence pathway inhibition | Direct bactericidal activity vs. Gram-positives and Gram-negatives | Mechanism not fully defined | [70] |
| Statins | Antibacterial activity | Inhibited Staphylococcus and Streptococcus spp. from skin infections | In vitro testing only; no mechanism assays | [71] |
| Simvastatin (Statin) | Isoprenoid biosynthesis interference | Reduced S. aureus invasion of host cells via isoprenoid modulation | In vitro testing only | [72] |
| Simvastatin (Statin) | Host immunomodulation | In S. aureus skin wound infection: reduced burden, promoted healing | Animal model only; unclear direct vs. host effects | [73] |
| Simvastatin (Statin) | Mevalonate pathway disruption | In S. aureus: disrupted mevalonate metabolism, weakened cell wall integrity | In vitro testing only | [74] |
| Diclofenac (NSAID) | DNA replication interference | Suppressed DNA synthesis in Gram-positives/Gram-negatives; protected mice vs. S. typhimurium | Target undefined | [75] |
| NSAIDs (Carprofen, Bromfenac, Vedaprofen) | β-clamp inhibition (DNA Pol III) | In E. coli: inhibited β-clamp, disrupted replication | In vitro testing only | [76] |
| Diflunisal (NSAID) | β-clamp inhibition | In H. pylori: micromolar inhibitor, structurally confirmed binding at subsite I | In vitro only, limited testing scope | [77] |
| Naproxen (NSAID) | DNA intercalation + ROS (light-dependent) | On plasmid pBR322: ROS-mediated DNA nicking/cleavage under irradiation | Purified DNA only; physiological relevance unclears | [78] |
| Celecoxib (NSAID) | Host-directed immunomodulation (SIRT1 activation) | In S. aureus-infected macrophages: decrease in NF-κB and cytokines, increased oxidative enzymes; enhanced antibiotic killing | In vitro testing only | [79] |
| Celecoxib (NSAID) | Immunomodulation (microglia model) | In microglia infected with S. aureus: reduced bacteria, rebalanced cytokines, enhanced ciprofloxacin activity | In vitro testing only | [80] |
| Celecoxib (NSAID) | Direct synthesis inhibition (RNA, DNA, protein) | In S. aureus: inhibited RNA/DNA/protein synthesis; efficacy in C. elegans and murine MRSA skin model; synergistic with topical/systemic antibiotics | Gram-negative activity required colistin; topical testing only | [81] |
2.3.3. Calcium Channel Blockers
2.3.4. Statins
2.3.5. Non-Steroidal Anti-Inflammatory Drugs
3. Artificial Intelligence in Antimicrobial Discovery: Current Advances and Translational Opportunities
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAC | Aminoglycoside acetyltransferase |
| AMR | Antimicrobial resistance |
| ATCC | American Type Culture Collection |
| ATP | Adenosine triphosphate |
| BPEI | Branched polyethylenimine |
| CCB | Calcium channel blocker |
| CFU | Colony-forming unit |
| CNS | Central nervous system |
| EO | Essential oil |
| ESBL | Extended spectrum β-lactamase |
| FIC FMM | Fractional inhibitory concentration Functional membrane microdomains |
| HMG | Hydroxymethylglutaryl |
| IL | Interleukin |
| INF | Interferon |
| JNK | C-Jun N-terminal Kinase |
| MBEC | Minimum biofilm eradication concentration |
| MBIC | Minimum biofilm inhibitory concentration |
| MBC | Minimum bactericidal concentration |
| MIC | Minimum inhibitory concentration |
| MICC | Minimum inhibitory concentration of combination |
| MIP | Macrophage inflammatory protein |
| MOA | Mechanism of action |
| MDR | Multidrug resistance |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| MRSE | Methicillin-resistant Staphylococcus epidermidis |
| MSSA | Methicillin-susceptible Staphylococcus aureus |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NO | Nitric oxide |
| NSAID | Non-steroidal anti-inflammatory drug |
| PBP | Penicillin-binding protein |
| PD | Pharmacodynamics |
| PI | Propidium iodide |
| PIA | Polysaccharide intercellular adhesin |
| PK | Pharmacokinetics |
| RNA | Ribonucleic acid |
| ROS | Reactive oxygen species |
| SSRI | Selective serotonin reuptake inhibitor |
| TB | Tuberculosis |
| TEM | Transmission electron microscopy |
| USD | United States dollar |
| UTI | Urinary tract infection |
References
- World Health Organization. New Report Calls for Urgent Action to Avert Antimicrobial Resistance Crisis. 2019. Available online: https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis (accessed on 20 August 2025).
- Kariuki, S. Global burden of antimicrobial resistance and forecasts to 2050. Lancet 2024, 404, 1172–1173. [Google Scholar] [CrossRef]
- Barbarossa, A.; Rosato, A.; Corbo, F.; Clodoveo, M.L.; Fracchiolla, G.; Carrieri, A.; Carocci, A. Non-antibiotic drug repositioning as an alternative antimicrobial approach. Antibiotics 2022, 11, 816. [Google Scholar] [CrossRef]
- Bansal, K.K.; Goyal, R.; Sharma, A.; Sharma, P.C.; Goyal, R.K. Repurposing of drugs for the treatment of microbial diseases. In Drug Repurposing for Emerging Infectious Diseases and Cancer; Sobti, R.C., Lal, S.K., Goyal, R.K., Eds.; Springer: Singapore, 2023; pp. 265–279. [Google Scholar] [CrossRef]
- Camacho, D.M.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-generation machine learning for biological networks. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef] [PubMed]
- de la Lastra, J.M.P.; Wardell, S.J.T.; Pal, T.; de la Fuente-Nunez, C.; Pletzer, D. From data to decisions: Leveraging artificial intelligence and machine learning in combating antimicrobial resistance—A comprehensive review. J. Med. Syst. 2024, 48, 71. [Google Scholar] [CrossRef]
- Amaral, L.; Viveiros, M. Thioridazine: A non-antibiotic drug highly effective, in combination with first line anti-tuberculosis drugs, against any form of antibiotic resistance of Mycobacterium tuberculosis due to its multi-mechanisms of action. Antibiotics 2017, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Luna-Herrera, J.; Aleid Cornejo-Báez, A.; Delgadillo-Gutiérrez, K. Relevance of efflux pumps in the development of drug resistance in mycobacterial infections. In Mycobacteria–Comparative Genomics, Biomarker Identification, Laboratory Diagnosis and Clinical Treatment; IntechOpen: London, UK, 2025. [Google Scholar] [CrossRef]
- Zou, X.; Xie, B. Therapeutic mechanisms of phenothiazine drugs: A mini-review of advances in cancer treatment and antibiotic resistance. Iran. J. Pharm. Res. 2025, 24, e157923. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, L.; Machado, D.; Couto, I.; Amaral, L.; Viveiros, M. Thioridazine and chlorpromazine inhibition of ethidium bromide efflux in Mycobacterium avium and Mycobacterium smegmatis. J. Antimicrob. Chemother. 2008, 61, 1076–1082. [Google Scholar] [CrossRef]
- Kaatz, G.W.; Moudgal, V.V.; Seo, S.M.; Kristiansen, J.E. Phenothiazines and thioxanthenes inhibit multidrug efflux pump activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 2003, 47, 719–726. [Google Scholar] [CrossRef]
- Dastidar, S.G.; Kristiansen, J.E.; Molnar, J.; Amaral, L. Role of phenothiazines and structurally similar compounds of plant origin in the fight against infections by drug resistant bacteria. Antibiotics 2013, 2, 58–72. [Google Scholar] [CrossRef]
- Grimsey, E.M.; Piddock, L.J.V. Chlorpromazine reduces efflux and enhances biofilm formation in Escherichia coli. FEMS Microbiol. Rev. 2019, 43, 577–596. [Google Scholar] [CrossRef]
- Chan, Y.Y.; Ong, Y.M.; Chua, K.L. Synergistic interaction between phenothiazines and antimicrobial agents against Burkholderia pseudomallei. Antimicrob. Agents Chemother. 2007, 51, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Caldara, M.; Marmiroli, N. Antimicrobial properties of antidepressants and antipsychotics—Possibilities and implications. Pharmaceuticals 2021, 14, 915. [Google Scholar] [CrossRef]
- Endo, T.H.; Santos, M.H.d.M.; Scandorieiro, S.; Gonçalves, B.C.; Vespero, E.C.; Perugini, M.R.E.; Pavanelli, W.R.; Nakazato, G.; Kobayashi, R.K.T. Selective serotonin reuptake inhibitors: Antimicrobial activity against ESKAPEE bacteria and mechanisms of action. Antibiotics 2025, 14, 51. [Google Scholar] [CrossRef]
- Zolotareva, D.; Zazybin, A.; Belyankova, Y.; Bayazit, S.; Dauletbakov, A.; Seilkhanov, T.; Kemelbekov, U.; Aydemir, M. Heterocyclic antidepressants with antimicrobial and fungicide activity. Molecules 2025, 30, 1102. [Google Scholar] [CrossRef]
- Dong, Z.; Han, K.; Xie, Q.; Lin, C.; Shen, X.; Hao, Y.; Li, J.; Xu, H.; He, L.; Yu, T.; et al. Core antibiotic resistance genes mediate gut microbiota to intervene in the treatment of major depressive disorder. J. Affect. Disord. 2024, 363, 507–519. [Google Scholar] [CrossRef]
- da Rosa, T.F.; Machado, C.D.; Serafin, M.B.; Bottega, A.; Coelho, S.S.; Foletto, V.S.; Rampelotto, R.F.; Lorenzoni, V.V.; Mainardi, A.; Hörner, R. Repurposing of escitalopram oxalate and clonazepam in combination with ciprofloxacin and sulfamethoxazole–trimethoprim for treatment of multidrug-resistant microorganisms and evaluation of the cleavage capacity of plasmid DNA. Can. J. Microbiol. 2021, 67, 599–612. [Google Scholar] [CrossRef]
- Chandal, N.; Sharma, N.; Cernicchi, G.; Felicetti, T.; Rondini, T.; Acito, M.; Nandanwar, H.; Sabatini, S. In vitro and in vivo investigations into the potential of quinazoline and quinoline derivatives as NorA efflux pump inhibitors against resistant Staphylococcus aureus strains. Antibiotics 2025, 14, 339. [Google Scholar] [CrossRef]
- Costa, S.S.; Lopes, E.; Azzali, E.; Machado, D.; Coelho, T.; Da Silva, P.E.; Viveiros, M.; Pieroni, M.; Couto, I. An experimental model for the rapid screening of compounds with potential use against mycobacteria. Assay Drug Dev. Technol. 2016, 14, 524–534. [Google Scholar] [CrossRef]
- Amaral, R.C.; Caleffi-Ferracioli, K.R.; Demitto, F.D.; Almeida, A.L.; Siqueira, V.L.; Scodro, R.B.; Leite, C.Q.; Pavan, F.R.; Cardoso, R.F. Is the efflux pump inhibitor verapamil a potential booster for isoniazid against Mycobacterium tuberculosis? Braz. J. Pharm. Sci. 2020, 56, e18309. [Google Scholar] [CrossRef]
- Chen, C.; Gardete, S.; Jansen, R.S.; Shetty, A.; Dick, T.; Rhee, K.Y.; Dartois, V. Verapamil targets membrane energetics in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2018, 62, e02107-17. [Google Scholar] [CrossRef] [PubMed]
- Viljoen, A.; Raynaud, C.; Johansen, M.D.; Roquet-Banères, F.; Herrmann, J.L.; Daher, W.; Kremer, L. Verapamil improves the activity of bedaquiline against Mycobacterium abscessus in vitro and in macrophages. Antimicrob. Agents Chemother. 2019, 63, e00705-19. [Google Scholar] [CrossRef]
- Ko, H.H.T.; Lareu, R.R.; Dix, B.R.; Hughes, J.D. Statins: Antimicrobial resistance breakers or makers? PeerJ 2017, 5, e3952. [Google Scholar] [CrossRef] [PubMed]
- Schelz, Z.; Muddather, H.F.; Zupkó, I. Repositioning of HMG-CoA reductase inhibitors as adjuvants in the modulation of efflux pump-mediated bacterial and tumor resistance. Antibiotics 2023, 12, 1468. [Google Scholar] [CrossRef]
- Rampelotto, R.F.; Lorenzoni, V.V.; Silva, D.D.; Moraes, G.A.; Serafin, M.B.; Tizotti, M.K.; Coelho, S.; Zambiazi, P.; Hörner, M.; Hörner, R. Synergistic antibacterial effect of statins with the complex {[1-(4-bromophenyl)-3-phenyltriazene N3-oxide-κ2 N1, O4](dimethylbenzylamine-κ1 C1, N4) palladium (II)}. Braz. J. Pharm. Sci. 2018, 54, e17369. [Google Scholar] [CrossRef]
- Abdelaziz, A.A.; El-Barrawy, M.A.; El-Nagar, R.A. Potent synergistic combination of rosuvastatin and levofloxacin against Staphylococcus aureus: In vitro and in vivo study. J. Appl. Microbiol. 2021, 131, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Al-Kuraishy, H.M.; Al-Gareeb, A.I.; Al-Buhadily, A.K. Rosuvastatin as forthcoming antibiotic or as adjuvant additive agent: In vitro novel antibacterial study. J. Lab. Physicians 2018, 10, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Halliday, C.; Kim, H.Y.; Tay, E.; Chen, S.C.; Alffenaar, J.W. Exploring synergy between azole antifungal drugs and statins for Candida auris. J. Antimicrob. Chemother. 2023, 78, 2824–2829. [Google Scholar] [CrossRef]
- Nyilasi, I.; Kocsubé, S.; Krizsán, K.; Galgóczy, L.; Papp, T.; Pesti, M.; Nagy, K.; Vágvölgyi, C. Susceptibility of clinically important dermatophytes against statins and different statin-antifungal combinations. Sabouraudia 2013, 52, 140–148. [Google Scholar] [CrossRef]
- Ahmed, E.F.; El-Baky, R.M.; Ahmed, A.B.; Waly, N.G.; Gad, G.F. Antibacterial activity of some non-steroidal anti-inflammatory drugs against bacteria causing urinary tract infection. Am. J. Infect. Dis. Microbiol. 2017, 5, 66–73. [Google Scholar]
- Chan, E.W.L.; Yee, Z.Y.; Raja, I.; Yap, J.K. Synergistic effect of non-steroidal anti-inflammatory drugs (NSAIDs) on antibacterial activity of cefuroxime and chloramphenicol against methicillin-resistant Staphylococcus aureus. J. Glob. Antimicrob. Resist. 2017, 10, 70–74. [Google Scholar] [CrossRef]
- Okpala, O.E.; Rondevaldova, J.; Osei-Owusu, H.; Kudera, T.; Kokoskova, T.; Kokoska, L. Susceptibility of Staphylococcus aureus to anti-inflammatory drugs with a focus on the combinatory effect of celecoxib with oxacillin in vitro. Molecules 2024, 29, 3665. [Google Scholar] [CrossRef]
- Altaf, M.; Ijaz, M.; Ghaffar, A.; Rehman, A.; Avais, M. Antibiotic susceptibility profile and synergistic effect of non-steroidal anti-inflammatory drugs on antibacterial activity of resistant antibiotics (Oxytetracycline and Gentamicin) against methicillin resistant Staphylococcus aureus (MRSA). Microb. Pathog. 2019, 136, 103755. [Google Scholar] [CrossRef]
- Mazumdar, K.; Dastidar, S.G.; Park, J.H.; Dutta, N.K. The anti-inflammatory non-antibiotic helper compound diclofenac: An antibacterial drug target. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 881–891. [Google Scholar] [CrossRef]
- Rosato, A.; Altini, E.; Sblano, S.; Salvagno, L.; Maggi, F.; de Michele, G.; Carocci, A.; Clodoveo, M.L.; Corbo, F.; Fracchiolla, G. Synergistic activity of new diclofenac and essential oils combinations against different Candida spp. Antibiotics 2021, 10, 688. [Google Scholar] [CrossRef]
- Khodaparast, S.; Ghanbari, F.; Zamani, H. Evaluation of the effect of ibuprofen in combination with ciprofloxacin on the virulence-associated traits, and efflux pump genes of Pseudomonas aeruginosa. World J. Microbiol. Biotechnol. 2022, 38, 125. [Google Scholar] [CrossRef] [PubMed]
- Sekar, A.; Gil, D.; Tierney, P.; McCanne, M.; Daesety, V.; Trendafilova, D.; Muratoglu, O.K.; Oral, E. Synergistic use of anti-inflammatory ketorolac and gentamicin to target staphylococcal biofilms. J. Transl. Med. 2024, 22, 102. [Google Scholar] [CrossRef] [PubMed]
- Carilo, I. Investigations into the Mechanisms of Anti-Mycobacterial Drug Resistance Using Antipsychotic Compounds. Ph.D. Thesis, University of Ghana, Accra, Ghana, 2018. Available online: https://ugspace.ug.edu.gh/bitstreams/9115607a-ea58-418b-be50-665481d313be/download (accessed on 21 August 2025).
- Bozóki-Nové, M. Efflux Pump Inhibitors and Potential Adjuvants to Reverse Multidrug Resistance in Bacteria and Tumor Cells. Ph.D. Thesis, University of Szeged, Szeged, Hungary, 2021. Available online: https://doktori.bibl.u-szeged.hu/11230/ (accessed on 21 August 2025).
- Grimsey, E.M. Understanding the Mode of Action of Compounds That Are Suggested to Possess Efflux Inhibitory Properties. Ph.D. Thesis, University of Birmingham, Birmingham, UK, 2021. Available online: https://etheses.bham.ac.uk/id/eprint/11554/ (accessed on 21 August 2025).
- Lam, A. Antibiotic Combination Therapy Against Multidrug-Resistant Staphylococcus epidermidis Biofilms and Broadening Antibiotic Spectrum Using Polyethylenimine. Ph.D. Thesis, University of Oklahoma, Norman, OK, USA, 2020. Available online: https://shareok.org/handle/11244/325287 (accessed on 21 August 2025).
- Ahmed, S.A.; Jordan, R.L.; Isseroff, R.R.; Lenhard, J.R. Potential synergy of fluoxetine and antibacterial agents against skin and soft tissue pathogens and drug-resistant organisms. Antibiotics 2024, 13, 1165. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, L.L.; de Farias Cabral, V.P.; Ferreira, T.L.; Coutinho, T.N.P.; Barbosa, L.B.; Gomes, A.O.C.V.; Barroso, F.D.D.; Valente Sá, L.G.A.; da Silva, C.R.; Nobre Júnior, H.V.; et al. Antimicrobial activity of selective serotonin reuptake inhibitors in bacteria and fungi: A systematic review. J. Health Biol. Sci. 2022, 10, 1–12. [Google Scholar]
- Pinto, I.C.C.M. The Effects of Emerging Contaminants on the Behaviour of Drinking Water-Isolated Bacteria. Ph.D. Thesis, University of Porto, Porto, Portugal, 2022. Available online: https://sigarra.up.pt/feup/en/pub_geral.pub_view?pi_pub_base_id=565442 (accessed on 21 August 2025).
- Graziano, T.S.; Cuzzullin, M.C.; Franco, G.C.; Schwartz-Filho, H.O.; de Andrade, E.D.; Groppo, F.C.; Cogo-Müller, K. Statins and antimicrobial effects: Simvastatin as a potential drug against Staphylococcus aureus biofilm. PLoS ONE 2015, 10, e0128098. [Google Scholar] [CrossRef]
- Trombetta, R.P.; Lowak, K.; Albach, A.M.; Lopez, R.A.; Romano, D.; Sanchez, C.J.; Wenke, J.C. The in vitro evaluation of statins as antimicrobials for trauma-related chronic infection. Res. Sq. 2020; preprint. [Google Scholar] [CrossRef]
- Baldiris, R.; Teherán, V.; Vivas-Reyes, R.; Montes, A.; Arzuza, O. Anti-biofilm activity of ibuprofen and diclofenac against some biofilm-producing Escherichia coli and Klebsiella pneumoniae uropathogens. Afr. J. Microbiol. Res. 2016, 10, 1675–1684. [Google Scholar] [CrossRef]
- Reśliński, A.; Dąbrowiecki, S.; Głowacka, K. The impact of diclofenac and ibuprofen on biofilm formation on the surface of polypropylene mesh. Hernia 2015, 19, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Leão, C.; Borges, A.; Simões, M. NSAIDs as a drug repurposing strategy for biofilm control. Antibiotics 2020, 9, 591. [Google Scholar] [CrossRef] [PubMed]
- Paes Leme, R.C.; da Silva, R.B. Antimicrobial activity of non-steroidal anti-inflammatory drugs on biofilm: Current evidence and potential for drug repurposing. Front. Microbiol. 2021, 12, 707629. [Google Scholar] [CrossRef] [PubMed]
- Dotto, C.; Lombarte Serrat, A.; Cattelan, N.; Barbagelata, M.S.; Yantorno, O.M.; Sordelli, D.O.; Ehling-Schulz, M.; Grunert, T.; Buzzola, F.R. The active component of aspirin, salicylic acid, promotes Staphylococcus aureus biofilm formation in a PIA-dependent manner. Front. Microbiol. 2017, 8, 4. [Google Scholar] [CrossRef]
- Ding, P.; Lu, J.; Wang, Y.; Schembri, M.A.; Guo, J. Antidepressants promote the spread of antibiotic resistance via horizontally conjugative gene transfer. Environ. Microbiol. 2022, 24, 5261–5276. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Z.; Ding, P.; Lu, J.; Mao, L.; Ngiam, L.; Yuan, Z.; Engelstädter, J.; Schembri, M.A.; Guo, J. Antidepressants can induce mutation and enhance persistence toward multiple antibiotics. Proc. Natl. Acad. Sci. USA 2023, 120, e2208344120. [Google Scholar] [CrossRef]
- Lu, J.; Ding, P.; Wang, Y.; Guo, J. Antidepressants promote the spread of extracellular antibiotic resistance genes via transformation. ISME Commun. 2022, 2, 47. [Google Scholar] [CrossRef]
- Händel, N.; Hoeksema, M.; Freijo Mata, M.; Brul, S.; ter Kuile, B.H. Effects of stress, reactive oxygen species, and the SOS response on de novo acquisition of antibiotic resistance in Escherichia coli. Antimicrob. Agents Chemother. 2016, 60, 3113–3119. [Google Scholar] [CrossRef]
- Lorente-Torres, B.; Llano-Verdeja, J.; Castañera, P.; Ferrero, H.Á.; Fernández-Martínez, S.; Javadimarand, F.; Mateos, L.M.; Letek, M.; Mourenza, Á. Innovative strategies in drug repurposing to tackle intracellular bacterial pathogens. Antibiotics 2024, 13, 834. [Google Scholar] [CrossRef]
- Sousa, S.A.; Feliciano, J.R.; Pita, T.; Soeiro, C.F.; Mendes, B.L.; Alves, L.G.; Leitão, J.H. Bacterial nosocomial infections: Multidrug resistance as a trigger for the development of novel antimicrobials. Antibiotics 2021, 10, 942. [Google Scholar] [CrossRef]
- Aguilar-Vega, L.; López-Jácome, L.E.; Franco, B.; Muñoz-Carranza, S.; Vargas-Maya, N.; Franco-Cendejas, R.; Hernández-Durán, M.; Otero-Zúñiga, M.; Campo-Beleño, C.; Jiménez-Cortés, J.G.; et al. Antibacterial properties of phenothiazine derivatives against multidrug-resistant Acinetobacter baumannii strains. J. Appl. Microbiol. 2021, 131, 2235–2243. [Google Scholar] [CrossRef]
- Ghosh, S.; Orman, M.A. Exploring the links between SOS response, mutagenesis, and resistance during the recovery period. Antimicrob. Agents Chemother. 2024, 68, e0146223. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Chen, X.; Lv, Y.; Peng, Q.; Wang, R.; Yang, B.; Wei, L. Research progress on the synergistic effect and its mechanisms of antidepressants and antibiotics against resistant pathogens. Arch. Microbiol. 2025, 207, 157. [Google Scholar] [CrossRef]
- de Farias Cabral, V.P.; Rodrigues, D.S.; do Amaral Valente Sá, L.G.; Moreira, L.E.A.; da Silva, C.R.; de Andrade Neto, J.B.; da Costa, É.R.M.; Ferreira, T.L.; de Oliveira, L.C.; de Souza, B.O.; et al. Antibacterial evaluation of tricyclic antidepressants against S. aureus and the possible pathways of the mechanism of action. Pathogens 2025, 14, 613. [Google Scholar] [CrossRef]
- Barbosa, A.D.; Sá, L.G.; Neto, J.B.A.; Rodrigues, D.S.; Cabral, V.P.; Moreira, L.E.A.; Aguiar, I.G.; Santos, H.S.; Cavalcanti, B.C.; Marinho, E.S.; et al. Activity of amlodipine against Staphylococcus aureus: Association with oxacillin and mechanism of action. Future Microbiol. 2023, 18, 505–519. [Google Scholar] [CrossRef] [PubMed]
- Alduaij, O.K.; Hussein, R.K.; Abu Alrub, S.; Zidan, S.A.H. Antimicrobial activities of diltiazem hydrochloride: Drug repurposing approach. PeerJ 2024, 12, e17809. [Google Scholar] [CrossRef] [PubMed]
- Boyd, N.K.; Lee, G.C.; Teng, C.; Frei, C.R. In vitro activity of non-antibiotic drugs against Staphylococcus aureus clinical strains. J. Glob. Antimicrob. Resist. 2021, 27, 167–171. [Google Scholar] [CrossRef]
- Akinjogunla, O.J.; Umo, A.N.; Alozie, M.F.; Oshosanya, G.O.; Saturday, G.I. Antibacterial potentiality and time-kill kinetics of amlodipine, thioridazine and promethazine against pathogenic clinical bacterial isolates. Afr. J. Clin. Exp. Microbiol. 2021, 22, 397–406. [Google Scholar] [CrossRef]
- Yi, Z.; Pei, Z.; Xiaoyan, M. Evaluation of amlodipine inhibition and antimicrobial effects. Int. J. Pharm. Chem. 2019, 5, 12–14. [Google Scholar] [CrossRef]
- García-Fernández, E.; Koch, G.; Wagner, R.M.; Fekete, A.; Stengel, S.T.; Schneider, J.; Mielich-Süss, B.; Geibel, S.; Markert, S.M.; Stigloher, C.; et al. Membrane microdomain disassembly inhibits MRSA antibiotic resistance. Cell 2017, 171, 1354–1367. [Google Scholar] [CrossRef]
- Thangamani, S.; Mohammad, H.; Abushahba, M.F.N.; Hamed, M.I.A.; Sobreira, T.J.P.; Hedrick, V.E.; Paul, L.N.; Seleem, M.N. Exploring simvastatin, an antihyperlipidemic drug, as a novel antibacterial agent. Sci. Rep. 2015, 5, 16407. [Google Scholar] [CrossRef]
- Ko, H.H.T.; Lareu, R.R.; Dix, B.R.; Hughes, J.D. In vitro antibacterial effects of statins against bacterial pathogens causing skin infections. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1125–1135. [Google Scholar] [CrossRef]
- Horn, M.P.; Knecht, S.M.; Rushing, F.L.; Birdsong, J.; Siddall, C.P.; Johnson, C.M.; Abraham, T.N.; Brown, A.; Volk, C.B.; Gammon, K.; et al. Simvastatin inhibits Staphylococcus aureus host cell invasion through modulation of isoprenoid intermediates. J. Pharmacol. Exp. Ther. 2008, 326, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, X.; Li, J.; Zhang, L.; Cao, X.; Wang, H.; Yang, B.; Tang, H.; Xie, J.; Liang, H. Topical simvastatin accelerates wound healing in Staphylococcus aureus-contaminated cutaneous wounds. Int. Wound J. 2016, 13, 1150–1157. [Google Scholar] [CrossRef]
- Cortês, I.T.; de Pádua Silva, K.; Cogo-Müller, K. Effects of simvastatin on the mevalonate pathway and cell wall integrity of Staphylococcus aureus. J. Appl. Microbiol. 2025, 136, lxafo12. [Google Scholar] [CrossRef]
- Dastidar, S.G.; Ganguly, K.; Chaudhuri, K.; Chakrabarty, A.N. The anti-bacterial action of diclofenac shown by inhibition of DNA synthesis. Int. J. Antimicrob. Agents. 2000, 14, 249–251. [Google Scholar] [CrossRef]
- Yin, Z.; Wang, Y.; Whittell, L.R.; Jergic, S.; Liu, M.; Harry, E.; Dixon, N.E.; Kelso, M.J.; Beck, J.L.; Oakley, A.J. DNA replication is the target for the antibacterial effects of nonsteroidal anti-inflammatory drugs. Chem. Biol. 2014, 21, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Verma, V.; Gautam, G.; Kumari, N.; Dhar, S.K.; Gourinath, S. Targeting the β-clamp in Helicobacter pylori with FDA-approved drugs reveals micromolar inhibition by diflunisal. FEBS Lett. 2017, 591, 2311–2322. [Google Scholar] [CrossRef]
- Husain, M.A.; Yaseen, Z.; Rehman, S.U.; Sarwar, T.; Tabish, M. Naproxen intercalates with DNA and causes photocleavage through ROS generation. FEBS J. 2013, 280, 6569–6580. [Google Scholar] [CrossRef]
- Annamanedi, M.; Kalle, A.M. Celecoxib sensitizes Staphylococcus aureus to antibiotics in macrophages by modulating SIRT1. PLoS ONE 2014, 9, e99285. [Google Scholar] [CrossRef] [PubMed]
- Dey, R.; Sultana, S.; Bishayi, B. Combination treatment of celecoxib and ciprofloxacin attenuates live S. aureus–induced oxidative damage and inflammation in murine microglia via regulation of cytokine balance. J. Neuroimmunol. 2018, 316, 23–39. [Google Scholar] [CrossRef]
- Thangamani, S.; Younis, W.; Seleem, M.N. Repurposing celecoxib as a topical antimicrobial agent. Front. Microbiol. 2015, 6, 750. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xue, X.; Jia, J.; Zou, X.; Guan, Y.; Zhu, L.; Wang, Z. Nonsteroidal anti-inflammatory drug diclofenac accelerates the emergence of antibiotic resistance via mutagenesis. Environ. Pollut. 2023, 326, 121457. [Google Scholar] [CrossRef]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep learning approach to antibiotic discovery. Cell 2020, 181, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Catacutan, D.B.; Rathod, K.; Swanson, K.; Jin, W.; Mohammed, J.C.; Chiappino-Pepe, A.; Syed, S.A.; Fragis, M.; Rachwalski, K.; et al. Deep Learning-Guided Discovery of an Antibiotic Targeting Acinetobacter baumannii. Nat. Chem. Biol. 2023, 19, 1342–1350. [Google Scholar] [CrossRef]
- Santos Júnior, C.D.; Torres, M.D.T.; Duan, Y.; Rodríguez del Río, Á.; Schmidt, T.S.B.; Chong, H.; Fullam, A.; Kuhn, M.; Zhu, C.; Houseman, A.; et al. Discovery of antimicrobial peptides in the global microbiome with machine learning. Cell 2024, 187, 3184–3201. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, J.L.; Dupont, C.L.; O’Rourke, A.; Beyhan, S.; Morales, P.; Spoering, A.; Meyer, K.J.; Chan, A.P.; Choi, Y.; Nierman, W.C.; et al. Predicting antimicrobial mechanism-of-action from transcriptomes: A generalizable explainable artificial intelligence approach. PLoS Comput. Biol. 2021, 17, e1008857. [Google Scholar] [CrossRef]
- Quach, D.; Sharp, M.; Ahmed, S.; Ames, L.; Bhagwat, A.; Deshpande, A.; Parish, T.; Pogliano, J.; Sugie, J. Deep learning-driven bacterial cytological profiling to determine antimicrobial mechanisms in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2025, 122, e2419813122. [Google Scholar] [CrossRef]
| Drug (Class) | Possible Mechanism | Key Findings | Limitations | Ref |
|---|---|---|---|---|
| Thioridazine (AP, phenothiazine) | Efflux pump inhibition in M. tuberculosis | Reversed resistance to isoniazid and rifampicin; synergistic with first-line TB drugs | No MIC or FIC values provided | [7] |
| Thioridazine & Chlorpromazine (APs, phenothiazines) | Efflux pump inhibition in M. tuberculosis | Reduced clarithromycin and isoniazid MICs | No MIC/FIC data reported | [8] |
| Phenothiazines (APs) | Efflux pump inhibition in S. aureus | Enhanced oxacillin efficacy | No MIC data reported | [9] |
| Thioridazine (AP) | Efflux pump inhibition in M. avium | Reduced ethambutol MIC in (8 → 2 µg/mL); described as synergistic | No FIC index calculated | [10] |
| Chlorpromazine (AP) | Efflux inhibition in S. aureus | Reduced norfloxacin MIC (4 → 1 µg/mL); synergy inferred | No checkerboard synergy assays performed | [11] |
| Phenothiazines (APs) | Efflux pump inhibition in Gram-negatives and -positives | Synergy with penicillin against resistant bacteria; MIC as low as 2 µg/ml | No FIC indices calculated | [12] |
| Chlorpromazine (AP) | AcrAB-TolC efflux inhibition in E. coli | Reduced tetracycline MIC | No MIC/FIC values reported | [13] |
| Chlorpromazine (AP) | Efflux inhibition in B. pseudomallei | Synergy with erythromycin confirmed (FIC ≤ 0.5) | - | [14] |
| Sertraline (SSRI) | Efflux pump inhibition in C. albicans | Synergy with fluconazole; MIC90 = 3 µM; FIC < 0.5 | Methods for FIC derivation not detailed | [15] |
| Fluoxetine (SSRI) | Possible efflux pump inhibition in Gram-negatives | MICs 15–126 µg/mL | No FIC values; synergy not assessed | [16] |
| Paroxetine (SSRI) | Efflux pump inhibition in S. aureus | MIC 64 µg/mL; enhanced aminoglycoside efficacy | No FIC values; synergy inferred only from MIC shift | [17] |
| Citalopram (SSRI) | Efflux pump inhibition (predicted) | Increased antibiotic susceptibility | No MIC/FIC data; based on pathway predictions, not direct assays | [18] |
| Escitalopram (SSRI) | Efflux pump inhibition in multidrug-resistant strains | Synergy with sulfamethoxazole–trimethoprim; MIC reductions observed | No FIC indices provided; synergy inferred from MIC shift | [19] |
| Verapamil (CCB) | Efflux pump inhibition (NorA) in S. aureus | Enhanced ciprofloxacin activity | No precise MIC or FIC values detailed | [20] |
| Verapamil (CCB) | Efflux pump inhibition in M. tuberculosis | Synergy with rifampicin | Detailed MIC reductions not provided | [21] |
| Verapamil (CCB) | Efflux pump inhibition in M. tuberculosis H37Rv | No significant synergy with isoniazid (FIC ≥ 0.5; additive/indifferent) | - | [22] |
| Verapamil (CCB) | Efflux inhibition ± membrane energetics disruption M. tuberculosis | Bedaquiline MIC 0.5 → 0.025 µM (20×); FIC 0.06; synergy with clofazimine (MIC 1.0 → 0.25 µM; FIC 0.19) | Mechanistic ambiguity; cardiotoxicity risk | [23] |
| Verapamil (CCB) | Likely membrane destabilisation/energy depletion rather than specific pump targeting in M. abscessus | Potentiated bedaquiline activity | No MIC/FIC numbers reported; mechanism not confirmed as direct efflux inhibition | [24] |
| Simvastatin (Statin) | Efflux pump inhibition + membrane disruption in S. aureus | MIC 15.6–31.25 µg/mL; synergistic with tetracycline (FIC < 0.5) | No in vivo validation | [25] |
| Simvastatin & Atorvastatin (Statins) | Efflux pump inhibition in Gram-positives | MIC 64–128 µg/mL; synergy with gentamicin/ciprofloxacin mostly additive (FIC > 0.5 ≤ 1) | Limited potentiation | [3] |
| Simvastatin (Statin) | Efflux inhibition in P. aeruginosa | MIC 32 µg/mL; synergy with levofloxacin (FIC 0.31) | Mechanistic details are limited | [26] |
| Atorvastatin (Statin) | Efflux pump inhibition (with Ru-complex) in S. aureus | MIC >128 → 4 µg/mL in combination; FIC 0.17–0.5 | Mechanism not studied; ruthenium complex role unclear | [27] |
| Rosuvastatin (Statin) | Efflux/membrane modulation in S. aureus | With levofloxacin: MIC 4 → 0.5 µg/mL; FIC 0.3 | No in vivo PK/Tox data | [28] |
| Rosuvastatin (Statin) | Efflux/membrane affects Gram-positives and Gram-negatives | With cefixime: 2–4-fold MIC reduction; FIC 0.37–0.49 | Lack of in vivo validation | [29] |
| Statins + Azoles | Fungal efflux inhibition in C. auris | MIC reductions; FIC 0.5–1.0 (additive) | Mechanism hypothesised | [30] |
| Atorvastatin (Statin) | Efflux inhibition in T. rubrum | MIC 64 µg/mL; synergy with terbinafine (FIC 0.45) | Limited mechanistic validation | [31] |
| Ibuprofen & Diclofenac (NSAIDs) | Proposed efflux pump inhibition | MICs 64–512 µg/mL; synergy with gentamicin/ciprofloxacin (FIC < 0.5) | No efflux assays performed | [32] |
| Ibuprofen, Diclofenac, Aspirin (NSAIDs) | Proposed efflux pump inhibition in S. aureus | MICs 125–500 µg/mL; synergy with cefuroxime/chloramphenicol (FIC 0.313–0.625) | Mechanism inferred; no direct efflux assays | [33] |
| Celecoxib (NSAID) | Membrane disruption/efflux pump in S. aureus | MIC 32 µg/mL; synergy with oxacillin (FIC 0.25) | No efflux assays performed | [34] |
| Ibuprofen, Aspirin (NSAIDs) | Proposed efflux inhibition in S. aureus | Decreased oxytetracycline MIC 8 → 1 µg/mL; FIC 0.25 | No validation of efflux inhibition | [35] |
| Diclofenac (NSAID) | Proposed efflux inhibition in Gram-positives and Gram-negatives | MIC 64 µg/mL; synergy with gentamicin (FIC 0.38) | No direct efflux assays | [36] |
| Diclofenac + Essential oils (NSAID + EO) | Membrane disruption, ROS in Candida spp. | Synergy (FIC < 0.5) | No efflux assays | [37] |
| Ibuprofen (NSAID) | Biofilm formation and efflux pump gene expression in P. aeruginosa | MIC 2048 µg/mL; synergy with ciprofloxacin (FIC 0.4) | Suppressed efflux pump gene expression and virulence factors | [38] |
| Ketorolac (NSAID) | Biofilm matrix modulation S. aureus biofilms | Synergy with gentamicin (FIC 0.31) | No efflux assays performed | [39] |
| Drug (Class) | Possible Mechanism | Key Findings | Limitations | Reference |
|---|---|---|---|---|
| Thioridazine (Antipsychotic, phenothiazine) | Biofilm disruption via efflux inhibition | Disrupted biofilms of M. tuberculosis and M. ulcerans; potentiated antibiotics | No MBIC/MBEC, MIC/FIC data; no membrane assays (PI, SEM, TEM) | [40] |
| Chlorpromazine (Antipsychotic, phenothiazine) | Biofilm reduction via efflux inhibition | Reported activity in S. aureus biofilms (NorA-related) | No MBIC/MBEC; no membrane assays | [41] |
| Chlorpromazine (Antipsychotic, phenothiazine) | Biofilm reduction via efflux inhibition | Reduced biofilm resilience in S. aureus | No MBIC/MBEC; no membrane assays | [42] |
| Branched polyethylenimine (BPEI, polymer) | PBP2a inhibition, biofilm disruption | Restored S. epidermidis (MRSE) susceptibility to amoxicillin; also affected MRSA, P. aeruginosa, E. coli | No MIC/MBIC reductions reported; limited synergy quantification | [43] |
| SSRIs (Fluoxetine, Sertraline, Paroxetine, Fluvoxamine) | Biofilm/membrane disruption | Increased membrane permeability vs. ESKAPEE pathogens | No MBIC/MBEC or % biofilm data; no membrane assays beyond dye uptake | [16] |
| Fluoxetine (SSRI) | Proposed biofilm inhibition via membrane disruption | With ampicillin, vancomycin, linezolid: synergy vs. S. aureus (incl. MRSA) and E. faecalis | No MIC/MBIC, FIC data | [44] |
| SSRIs (Fluoxetine, Sertraline) | Antimicrobial activity | Reported effects in E. coli and fungi (Candida albicans, C. glabrata, C. parapsilosis, C. tropicalis) | No MBIC/MBEC, no quantitative biofilm data, no membrane assays | [45] |
| SSRIs | Biofilm disruption | Reported vs. Pseudomonas, Aeromonas, Enterobacter spp. | No quantitative biofilm or mechanistic data | [46] |
| Simvastatin (Statin) | Antibiofilm effect, membrane interference | Inhibited biofilm formation and reduced viability of S. aureus (crystal violet, CFU) | No MBIC/MBEC; no membrane assays; no biofilm synergy data | [47] |
| Statins | Antibiofilm effect | Tested vs. S. aureus UAMS-1 biofilms (MBEC assay) | No MBEC numbers; no synergy testing; no membrane assays | [48] |
| Ibuprofen & Diclofenac (NSAIDs) | Biofilm inhibition | Reduced biofilm in E. coli and K. pneumoniae UTI isolates; diclofenac ~50% reduction at 50 mg/L; ibuprofen suppressed morphotypes | No MBIC/MBEC defined; mechanistic evidence indirect | [49] |
| Ibuprofen & Diclofenac (NSAIDs) | Biofilm inhibition | Reduced S. aureus and E. coli biofilms on mesh; SEM confirmed fewer adherent cells | No MBIC/MBEC defined | [50] |
| Piroxicam, Diclofenac, Acetylsalicylic acid (NSAIDs) | Biofilm inhibition | Piroxicam reduced metabolic activity and removed biofilm mass in E. coli and S. aureus preformed biofilms | No MBIC/MBEC; MBC >2000 µg/mL; limited mechanistic data | [51] |
| NSAIDs (general) | Biofilm disruption via QS and virulence interference | Downregulated AgrA in S. aureus; interfered with quorum sensing, altered surface properties | No MBIC/MBEC; no membrane assays | [52] |
| Salicylic acid (NSAID metabolite) | Iron chelation, biofilm modulation | Enhanced PIA-mediated biofilm synthesis in S. aureus via Fe2+ limitation | Adverse effect; no MBIC/MBEC values | [53] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiwana, G.; Cock, I.E.; Taylor, S.M.; Cheesman, M.J. Beyond Antibiotics: Repurposing Non-Antibiotic Drugs as Novel Antibacterial Agents to Combat Resistance. Int. J. Mol. Sci. 2025, 26, 9880. https://doi.org/10.3390/ijms26209880
Tiwana G, Cock IE, Taylor SM, Cheesman MJ. Beyond Antibiotics: Repurposing Non-Antibiotic Drugs as Novel Antibacterial Agents to Combat Resistance. International Journal of Molecular Sciences. 2025; 26(20):9880. https://doi.org/10.3390/ijms26209880
Chicago/Turabian StyleTiwana, Gagan, Ian Edwin Cock, Stephen Maxwell Taylor, and Matthew James Cheesman. 2025. "Beyond Antibiotics: Repurposing Non-Antibiotic Drugs as Novel Antibacterial Agents to Combat Resistance" International Journal of Molecular Sciences 26, no. 20: 9880. https://doi.org/10.3390/ijms26209880
APA StyleTiwana, G., Cock, I. E., Taylor, S. M., & Cheesman, M. J. (2025). Beyond Antibiotics: Repurposing Non-Antibiotic Drugs as Novel Antibacterial Agents to Combat Resistance. International Journal of Molecular Sciences, 26(20), 9880. https://doi.org/10.3390/ijms26209880







