The Photosynthetic Complexes of Thylakoid Membranes of Photoautotrophs and a Quartet of Their Polar Lipids
Abstract
1. Introduction
2. Thylakoid Membranes
3. Photosystems and Antenna Complexes for Light Harvesting
3.1. PSII and LHCII
3.2. Photosystem I and LHCI
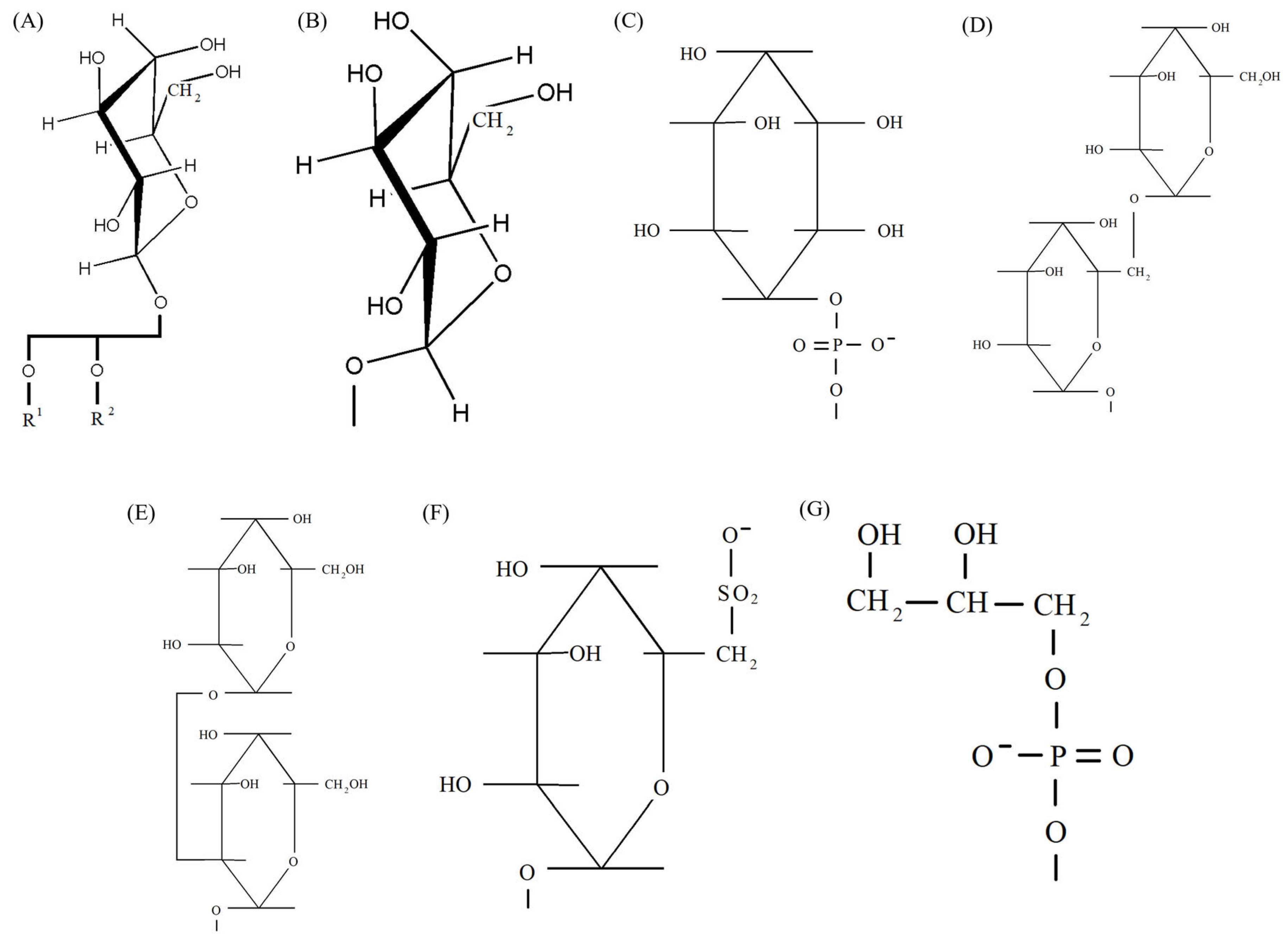
4. Participation of Polar Lipids in the Functioning of Photosynthetic Apparatus
4.1. The Need for Glycerolipids for Photosystems
4.2. Polar Lipids in PS II
| № | Object | Numbers of all Lipid Molecules in a Monomer and Number for Each of MGDG, DGDG, SQDG, and PG | References |
|---|---|---|---|
| 1. | PSII of Thermosynechococcus elongatus | A total of 25 molecules in a monomer; 11:7:5:2 | [61,90] |
| 2. | PSII of Synechocystis sp. PCC 6803 | Over 20 molecules | [60] |
| 3. | PSII of Pisum sativum. | A total of 25 molecules; 7:5:4:9 | based on [20] |
| 4. | C2S2 PSII of Chlamidomonas reinchardtii | A total of 21 molecules; 5:5:2:9 | based on [20] |
| 5. | LHCII of Spinacia oleracea | Four molecules in trimer; 0:3:0:1. DGDG at the border between monomers | [23,94] |
| 6. | PSII-LHCII of Spinacia oleracea | Calculated, 30 molecules per a monomer of PSII; 5:4:3:18 | [22] |
| 7. | PSI of Pisum sativum | All four lipids and phosphatidylcholine (PC) detected by high performance liquid chromatography/electrospray ionization mass spectrometry | [95] |
| 8. | Cyt b6f of Spinacia oleracea | A total of 12 molecules in dimer (2:0:3:4 and 3 PC, added exogenously) | [96] |
| 9. | NDH-1 Thermosynechococcus elongatus | A total of 13 molecules (2:2:3:6) or A total of 15 molecules (0:2:4:9) | [19,97] |
4.2.1. Neutral Glycolipids in PSII
4.2.2. Anionic Lipids in PSII
4.3. Polar Lipids in LHCII and PSII-LHCII
4.4. Involvement of Polar Lipids in the Assembly and Functioning of PSI
4.5. Involvement of Polar Lipids in the Assembly and Functioning of LHCI and PSI-LHCI
5. Cytochrome b6f Complex
6. NDH-1 Protein Complex
7. The Role of Bound Fatty Acids in Photosynthesis
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Chl | chlorophyll |
| Cyt b6f | cytochrome b6f |
| Cryo-EM | single-particle cryo-electron microscopy |
| DPG | diphosphatidylglycerols (cardiolipins) |
| FAs | fatty acids |
| ISP | iron-sulfur protein |
| LHCII and LHCI | major light-harvesting complexes II and I |
| LPO | lipids peroxide oxidation |
| MGDG and DGDG | mono- and digalactosyldiacylglycerols |
| NADP+ and NADPH | nicotinamide adenine dinucleotide phosphate in oxidized and reduced forms |
| PCs | phosphatidylcholines |
| PEs | phosphatidylethanolamines |
| PG | phosphatidylglycerols |
| PL | polar lipids |
| PSII and PSI | photosystems II and I |
| PQ (QA and QB) | plastoquinone electron acceptors (primary and secondary) |
| RC | reaction centers of photosystems |
| XRC | Xray crystallography |
| SQDG | sulfoquinovosyl diacylglycerols |
| TM | transmembrane (proteins or helices) |
References
- Yu, H.; Hamaguchi, T.; Nakajima, Y.; Kato, K.; Kawakami, K.; Akita, F.; Yonekura, K.; Shen, J.-R. Cryo-EM structure of monomeric photosystem II at 2.78 Å resolution reveals factors important for the formation of dimer. Biochim. Biophys. Acta BBA Bioenerg. 2021, 1862, 148471. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Zhao, L.; Wang, W.; Pi, X.; Han, G.; Wang, J.; Cheng, L.; He, Y.-K.; Kuang, T.; Qin, X.; et al. Antenna arrangement and energy-transfer pathways of PSI–LHCI from the moss Physcomitrella patens. Cell Discov. 2021, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.-R. The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu. Rev. Plant Biol. 2015, 66, 23–48. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Cao, P.; Su, X.; Liu, Z.; Li, M. Structural analysis and comparison of light-harvesting complexes I and II. Biochim. Biophys. Acta BBA Bioenerg. 2020, 1861, 148038. [Google Scholar] [CrossRef] [PubMed]
- Sato, N. Are Cyanobacteria an Ancestor of Chloroplasts or Just One of the Gene Donors for Plants and Algae? Genes 2021, 12, 823. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K. Role of membrane glycerolipids in photosynthesis, thylakoid biogenesis and chloroplast development. J. Plant Res. 2016, 129, 565–580, Correction in J. Plant Res. 2018, 131, 563. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Wada, H. Lipid Biosynthesis and its Regulation in Cyanobacteria. In Lipids in Photosynthesis: Essential and Regulatory Functions; Advances in Photosynthesis and Respiration; Wada, H., Murata, N., Eds.; Springer: Dordrecht, The Netherlands, 2009; Volume 30. [Google Scholar] [CrossRef]
- Johnson, M.P.; Vasilev, C.; Olsen, J.D.; Hunter, C.N. Nanodomains of cytochrome b6f and photosystem II complexes in spinach grana thylakoid membranes. Plant Cell 2014, 26, 3051–3061. [Google Scholar] [CrossRef]
- Zhukov, A.; Vereshchagin, M. Polar Glycerolipids and Membrane Lipid Rafts. Int. J. Mol. Sci. 2024, 25, 8325. [Google Scholar] [CrossRef]
- Kobayashi, K.; Yoshihara, A.; Kubota-Kawai, H. Evolutionary implications from lipids in membrane bilayers and photosynthetic complexes in cyanobacteria and chloroplasts. J. Biochem. 2023, 174, 399–408. [Google Scholar] [CrossRef]
- Kobayashi, K.; Osawa, Y.; Yoshihara, A.; Shimojima, M.; Awai, K. Relationship between glycerolipids and photosynthetic components during recovery of thylakoid membranes from nitrogen starvation-induced attenuation in Synechocystis sp. PCC 6803. Front. Plant Sci. 2020, 11, 432. [Google Scholar] [CrossRef]
- Kato, K.; Nagao, R.; Jiang, T.-Y.; Ueno, Y.; Yokono, M.; Chan, S.K.; Watanabe, M.; Ikeuchi, M.; Shen, J.-R.; Akimoto, S.; et al. Structure of a cyanobacterial photosystem I tetramer revealed by cryo-electron microscopy. Nat. Commun. 2019, 10, 4929. [Google Scholar] [CrossRef] [PubMed]
- Mizusawa, N.; Wada, H. The role of lipids in photosystem II. Biochim. Biophys. Acta BBA Bioenerg. 2012, 1817, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Jordan, P.; Fromme, P.; Witt, H.T.; Klukas, O.; Saenger, W.; Krauß, N. Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 2001, 411, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Suga, M.; Akita, F.; Hirata, K.; Ueno, G.; Murakami, H.; Nakajima, Y.; Shimizu, T.; Yamashita, K.; Yamamoto, M.; Ago, H.; et al. Native structure of photosystem II at 1.95 Å resolution viewed by femtosecond X-ray pulses. Nature 2015, 517, 99–103. [Google Scholar] [CrossRef]
- Burrows, P.A.; Sazanov, L.A.; Svab, Z.; Maliga, P.; Nixon, P.J. Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 1998, 17, 868–876. [Google Scholar] [CrossRef]
- Nixon, P.J. Chlororespiration. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2000, 355, 1541–1547. [Google Scholar] [CrossRef]
- Peltier, G.; Cournac, L. Chlororespiration. Annu. Rev. Plant Biol. 2002, 53, 523–550. [Google Scholar] [CrossRef]
- Zhang, C.; Shuai, J.; Ran, Z.; Zhao, J.; Wu, Z.; Liao, R.; Wu, J.; Ma, W.; Lei, M. Structural insights into NDH-1 mediated cyclic electron transfer. Nat. Commun. 2020, 11, 888. [Google Scholar] [CrossRef]
- Yoshihara, A.; Kobayashi, K. Lipids in photosynthetic protein complexes in the thylakoid membrane of plants, algae, and cyanobacteria. J. Exp. Bot. 2022, 73, 2735–2750. [Google Scholar] [CrossRef] [PubMed]
- Loll, B.; Kern, J.; Saenger, W.; Zouni, A.; Biesiadka, J. Lipids in photosystem II: Interactions with protein and cofactors. Biochim. Biophys. Acta BBA Bioenerg. 2007, 1767, 509–519. [Google Scholar] [CrossRef]
- Kobayashi, K.; Endo, K.; Wada, H. Specific Distribution of Phosphatidylglycerol to Photosystem Complexes in the Thylakoid Membrane. Front. Plant Sci. 2017, 8, 1991. [Google Scholar] [CrossRef]
- Sheng, X.; Liu, X.; Cao, P.; Li, M.; Liu, Z. Structural roles of lipid molecules in the assembly of plant PSII-LHCII supercomplex. Biophys. Rep. 2018, 4, 189–203. [Google Scholar] [CrossRef]
- Wei, X.; Su, X.; Cao, P.; Liu, X.; Chang, W.; Li, M.; Zhang, X.; Liu, Z. Structure of spinach photosystem II–LHCII supercomplex at 3.2 Å resolution. Nature 2016, 534, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.R. Lipids in photosynthetic reaction centres: Structural roles and functional holes. Prog. Lipid Res. 2007, 46, 56–87. [Google Scholar] [CrossRef] [PubMed]
- van Eerden, F.J.; de Jong, D.H.; de Vries, A.H.; Wassenaar, T.A.; Marrink, S.J. Characterization of thylakoid lipid membranes from cyanobacteria and higher plants by molecular dynamics simulations. Biochim. Biophys. Acta BBA Biomembr. 2015, 1848, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Li, Y.; Li, X.; Zhong, Q.; Li, N.; Zhang, K.; Zhang, Y.; Chu, H.; Ma, C.; Li, G.; et al. Structural and functional insights into the tetrameric photosystem I from heterocyst-forming cyanobacteria. Nat. Plants 2019, 5, 1087–1097. [Google Scholar] [CrossRef] [PubMed]
- Zhukov, A.; Popov, V. Eukaryotic Cell Membranes: Structure, Composition, Research Methods and Computational Modelling. Int. J. Mol. Sci. 2023, 24, 11226. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, Y.; Umena, Y.; Nagao, R.; Endo, K.; Kobayashi, K.; Akita, F.; Suga, M.; Wada, H.; Noguchi, T.; Shen, J.-R. Thylakoid membrane lipid sulfoquinovosyl-diacylglycerol (SQDG) is required for full functioning of photosystem II in Thermosynechococcus elongatus. J. Biol. Chem. 2018, 293, 14786–14797. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.F. Curvature forces in membrane lipid-protein interactions. Biochemistry 2012, 51, 9782–9795. [Google Scholar] [CrossRef]
- Quinn, P.J. Lipid phase behaviour and lipid-protein interactions in the chloroplast photosynthetic membrane. Biochem. Soc. Trans. 1987, 15, 86–91. [Google Scholar] [CrossRef]
- Schaller-Laudel, S.; Latowski, D.; Jemioła-Rzemińska, M.; Strzałka, K.; Daum, S.; Bacia, K.; Wilhelm, C.; Goss, R. Influence of thylakoid membrane lipids on the structure of aggregated light-harvesting complexes of the diatom Thalassiosira pseudonana and the green alga Mantoniella squamata. Physiol. Plant. 2017, 160, 339–358. [Google Scholar] [CrossRef]
- Wang, J.; Yu, L.; Wang, W.; Yan, Q.; Kuang, T.; Qin, X.; Shen, J. Structure of plant photosystem I−light harvesting complex I supercomplex at 2.4 Å resolution. J. Integr. Plant Biol. 2021, 63, 1367–1381. [Google Scholar] [CrossRef]
- Wietrzynski, W.; Lamm, L.; Wood, W.H.; Loukeri, M.J.; Malone, L.; Peng, T.; Johnson, M.P.; Engel, B.D. Molecular architecture of thylakoid membranes within intact spinach chloroplasts. eLife 2024, 14, RP105496. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, H.; Mukherjee, U.; Galla, H.-J. Molecular architecture of the thylakoid membrane: Lipid diffusion space for plastoquinone. Biochemistry 2002, 41, 4872–4882. [Google Scholar] [CrossRef] [PubMed]
- Quinn, P.J. Lipid unsaturation and the organization of photosynthetic complexes in higher-plant chloroplasts. Biochem Soc. Trans. 1997, 25, 1080–1088. [Google Scholar] [CrossRef]
- Wilson, S.; Clarke, C.D.; Carbajal, M.A.; Buccafusca, R.; Fleck, R.A.; Daskalakis, V.; Ruban, A.V. Hydrophobic Mismatch in the Thylakoid Membrane Regulates Photosynthetic Light Harvesting. J. Am. Chem. Soc. 2024, 146, 14905–14914. [Google Scholar] [CrossRef] [PubMed]
- Ago, H.; Adachi, H.; Umena, Y.; Tashiro, T.; Kawakami, K.; Kamiya, N.; Tian, L.; Han, G.; Kuang, T.; Liu, Z.; et al. Novel features of eukaryotic photosystem II revealed by its crystal structure analysis from a red alga. J. Biol. Chem. 2016, 291, 5676–5687. [Google Scholar] [CrossRef]
- Kopečná, J.; Pilný, J.; Krynická, V.; Tomčala, A.; Kis, M.; Gombos, Z.; Komenda, J.; Sobotka, R. Lack of phosphatidylglycerol inhibits chlorophyll biosynthesis at multiple sites and limits chlorophyllide reutilization in Synechocystis sp. strain PCC 6803. Plant Physiol. 2015, 169, 1307–1317. [Google Scholar] [CrossRef]
- Sheng, X.; Watanabe, A.; Li, A.; Kim, E.; Song, C.; Murata, K.; Song, D.; Minagawa, J.; Liu, Z. Structural insight into light harvesting for photosystem II in green algae. Nat. Plants 2019, 5, 1320–1330. [Google Scholar] [CrossRef]
- Perez-Boerema, A.; Klaiman, D.; Caspy, I.; Netzer-El, S.Y.; Amunts, A.; Nelson, N. Structure of a minimal photosystem I from the green alga Dunaliella salina. Nat. Plants 2020, 6, 321–327. [Google Scholar] [CrossRef]
- Järvi, S.; Suorsa, M.; Aro, E.-M. Photosystem II repair in plant chloroplasts—Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta BBA Bioenerg. 2015, 1847, 900–909. [Google Scholar] [CrossRef]
- Qin, X.; Pi, X.; Wang, W.; Han, G.; Zhu, L.; Liu, M.; Cheng, L.; Shen, J.-R.; Kuang, T.; Sui, S.-F. Structure of a green algal photosystem I in complex with a large number of light-harvesting complex I subunits. Nat. Plants 2019, 5, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Fromme, P.; Yu, H.; DeRuyter, Y.S.; Jolley, C.; Chauhan, D.K.; Melkozernov, A.; Grotjohann, I. Structure of photosystems I and II. Comptes Rendus Chim. 2006, 9, 188–200. [Google Scholar] [CrossRef]
- Sakurai, I.; Mizusawa, N.; Wada, H.; Sato, N. Digalactosyldiacylglycerol is required for stabilization of the oxygen-evolving complex in photosystem II. Plant Physiol. 2007, 145, 1361–1370. [Google Scholar] [CrossRef]
- Seiwert, D.; Witt, H.; Janshoff, A.; Paulsen, H. The non-bilayer lipid MGDG stabilizes the major light-harvesting complex (LHCII) against unfolding. Sci. Rep. 2017, 7, 5158. [Google Scholar] [CrossRef] [PubMed]
- Garab, G.; Ughy, B.; Goss, R. Role of MGDG and Non-bilayer Lipid Phases in the Structure and Dynamics of Chloroplast Thylakoid Membranes. Subcell. Biochem. 2016, 86, 127–157. [Google Scholar] [CrossRef] [PubMed]
- Böde, K.; Javornik, U.; Dlouhý, O.; Zsíros, O.; Biswas, A.; Domonkos, I.; Šket, P.; Karlický, V.; Ughy, B.; Lambrev, P.H.; et al. Role of isotropic lipid phase in the fusion of photosystem II membranes. Photosynth. Res. 2024, 161, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Mazor, Y.; Borovikova, A.; Caspy, I.; Nelson, N. Structure of the plant photosystem I supercomplex at 2.6 Å resolution. Nat. Plants 2017, 3, 17014. [Google Scholar] [CrossRef]
- Kansy, M.; Wilhelm, C.; Goss, R. Influence of thylakoid membrane lipids on the structure and function if the plant photosystem II core complex. Planta 2014, 240, 81–796. [Google Scholar] [CrossRef]
- Kates, M. Techniques of Lipidology. Isolation, Analysis and Identification of Lipids; Elsevier: Amsterdam, The Netherlands, 1972. [Google Scholar]
- Zhou, F.; Liu, S.; Hu, Z.; Kuang, T.; Paulsen, H.; Yang, C. Effect of monogalactosyldiacylglycerol on the interaction between photosystem II core complex and its antenna complexes in liposomes of thylakoid lipids. Photosynth. Res. 2009, 99, 185–193. [Google Scholar] [CrossRef]
- Pottosin, I.I.; Schönknecht, G. Patch clamp study of the voltage-dependent anion channel in the thylakoid membrane. J. Membr. Biol. 1996, 148, 143–156. [Google Scholar] [CrossRef]
- Pottosin, I.; Schönknecht, G. Ion channel permeable for divalent and monovalent cations in native spinach thylakoid membranes. J. Membr. Biol. 1996, 152, 223–233. [Google Scholar] [CrossRef]
- Pottosin, I.; Dobrovinskaya, O. Ion Channels in Native Chloroplast Membranes: Challenges and Potential for Direct Patch-Clamp Studies. Front. Physiol. 2015, 6, 396. [Google Scholar] [CrossRef] [PubMed]
- Komenda, J.; Sobotka, R.; Nixon, P.J. The biogenesis and maintenance of photosystem II: Recent advances and current challenges. Plant Cell 2024, 36, 3997–4013. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Ma, J.; Su, X.; Cao, P.; Chang, W.; Liu, Z.; Zhang, X.; Li, M. Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II. Science 2018, 360, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Komenda, J.; Knoppová, J.; Kopečná, J.; Sobotka, R.; Halada, P.; Yu, J.; Nickelsen, J.; Boehm, M.; Nixon, P.J. The Psb27 assembly factor binds to the CP43 complex of photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2012, 158, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Pi, X.; Zhao, S.; Wang, W.; Liu, D.; Xu, C.; Han, G.; Kuang, T.; Sui, S.-F.; Shen, J.-R. The pigment–protein network of a diatom photosystem II–light-harvesting antenna supercomplex. Science 2019, 365, eaax4406. [Google Scholar] [CrossRef] [PubMed]
- Umena, Y.; Kawakami, K.; Shen, J.-R.; Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 2011, 473, 55–60. [Google Scholar] [CrossRef]
- Kern, J.; Guskov, A. Lipids in photosystem II: Multifunctional cofactors. J. Photochem. Photobiol. B Biol. 2011, 104, 19–34. [Google Scholar] [CrossRef]
- Endo, K.; Mizusawa, N.; Shen, J.-R.; Yamada, M.; Tomo, T.; Komatsu, H.; Kobayashi, M.; Kobayashi, K.; Wada, H. Site-directed mutagenesis of amino acid residues of D1 protein interacting with phosphatidylglycerol affects the function of plastoquinone QB in photosystem II. Photosynth. Res. 2015, 126, 385–397. [Google Scholar] [CrossRef]
- Huang, Z.; Shen, L.; Wang, W.; Mao, Z.; Yi, X.; Kuang, T.; Shen, J.-R.; Zhang, X.; Han, G. Structure of photosystem I–LHCI–LHCII from the green alga Chlamydomonas reinhardtii in State 2. Nat. Commun. 2021, 12, 1100. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Ma, J.; Wei, X.; Cao, P.; Zhu, D.; Chang, W.; Liu, Z.; Zhang, X.; Li, M. Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. Science 2017, 357, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Nagao, R.; Kato, K.; Suzuki, T.; Ifuku, K.; Uchiyama, I.; Kashino, Y.; Dohmae, N.; Akimoto, S.; Shen, J.-R.; Miyazaki, N.; et al. Structural basis for energy harvesting and dissipation in a diatom PSII–FCPII supercomplex. Nat. Plants 2019, 5, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Huang, Z.; Chang, S.; Wang, W.; Wang, J.; Kuang, T.; Han, G.; Shen, J.R.; Zhang, X. Structure of a C2S2M2N2-type PSII–LHCII supercomplex from the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2019, 116, 21246–21255. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhu, Q.; Chen, J.; Shen, L.; Yi, X.; Huang, Z.; Wang, W.; Chen, M.; Kuang, T.; Shen, J.; et al. A unique photosystem I reaction center from a chlorophyll d-containing cyanobacterium Acaryochloris marina. J. Integr. Plant Biol. 2021, 63, 1740–1752. [Google Scholar] [CrossRef]
- Chen, M.; Perez-Boerema, A.; Zhang, L.; Li, Y.; Yang, M.; Li, S.; Amunts, A. Distinct structural modulation of photosystem I and lipid environment stabilizes its tetrameric assembly. Nat. Plants 2020, 6, 314–320. [Google Scholar] [CrossRef]
- Murata, N.; Nishiyama, Y. ATP is a driving force in the repair of photosystem II during photoinhibition. Plant Cell Environ. 2017, 41, 285–299. [Google Scholar] [CrossRef]
- Suga, M.; Ozawa, S.-I.; Yoshida-Motomura, K.; Akita, F.; Miyazaki, N.; Takahashi, Y. Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I. Nat. Plants 2019, 5, 626–636. [Google Scholar] [CrossRef]
- Su, X.; Ma, J.; Pan, X.; Zhao, X.; Chang, W.; Liu, Z.; Zhang, X.; Li, M. Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex. Nat. Plants 2019, 5, 273–281. [Google Scholar] [CrossRef]
- Awai, K.; Ohta, H.; Sato, N. Oxygenic photosynthesis without galactolipids. Proc. Natl. Acad. Sci. USA 2014, 111, 13571–13575. [Google Scholar] [CrossRef]
- Sato, N. Is monoglucosyldiacylglycerol a precursor to monogalactosyldiacylglycerol in all cyanobacteria? Plant Cell Physiol. 2015, 56, 1890–1899. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Popova, A.V.; Velitchkova, M.; Zanev, Y. Effect of membrane fluidity on photosynthetic oxygen production reactions. Z. Naturforschung C 2007, 62, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Hölzl, G.; Witt, S.; Gaude, N.; Melzer, M.; Schöttler, M.A.; Dörmann, P. The role of diglycosyl lipids in photosynthesis and membrane lipid homeostasis in Arabidopsis. Plant Physiol. 2009, 150, 1147–1159. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, A.; Nagata, N.; Wada, H.; Kobayashi, K. Plastid anionic lipids are essential for the development of both photosynthetic and non-photosynthetic organs in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 4860. [Google Scholar] [CrossRef]
- Ostermeier, M.; Garibay-Hernández, A.; Holzer, V.J.C.; Schroda, M.; Nickelsen, J. Structure, biogenesis, and evolution of thylakoid membranes. Plant Cell 2024, 36, 4014–4035. [Google Scholar] [CrossRef]
- Zhukov, A.V. On Qualitative Composition of Membrane Lipids in Plant Cells. Russ. J. Plant Physiol. 2021, 68, 367–383. [Google Scholar] [CrossRef]
- Gidden, J.; Denson, J.; Liyanage, R.; Ivey, D.M.; Lay, J.O. Lipid Compositions in Escherichia coli and Bacillus subtilis During Growth as Determined by MALDI-TOF and TOF/TOF Mass Spectrometry. Int. J. Mass Spectrom. 2009, 283, 178–184. [Google Scholar] [CrossRef]
- Santos, T.C.; Futerman, A.H. The fats of the matter: Lipids in prebiotic chemistry and in origin of life studies. Prog. Lipid Res. 2023, 92, 101253. [Google Scholar] [CrossRef]
- Yoon, H.S.; Hackett, J.D.; Ciniglia, C.; Pinto, G.; Bhattacharya, D. A Molecular Timeline for the Origin of Photosynthetic Eukaryotes. Mol. Biol. Evol. 2004, 21, 809–818. [Google Scholar] [CrossRef]
- Demé, B.; Cataye, C.; Block, M.A.; Maréchal, E.; Jouhet, J. Contribution of galactoglycerolipids to the 3-dimensional architecture of thylakoids. FASEB J. 2014, 28, 3373–3383. [Google Scholar] [CrossRef]
- Frentzen, M. Phosphatidylglycerol and sulfoquinovosyldiacylglycerol: Anionic membrane lipids and phosphate regulation. Curr. Opin. Plant Biol. 2004, 7, 270–276. [Google Scholar] [CrossRef]
- Gombos, Z.; Várkonyi, Z.; Hagio, M.; Iwaki, M.; Kovács, L.; Masamoto, K.; Itoh, S.; Wada, H. Phosphatidylglycerol requirement for the function of electron acceptor plastoquinone QB in the photosystem II reaction center. Biochemistry 2002, 41, 3796–3802. [Google Scholar] [CrossRef]
- Endo, K.; Kobayashi, K.; Wada, H. Sulfoquinovosyldiacylglycerol has an essential role in Thermosynechococcus elongatus BP-1 under phosphate-deficient conditions. Plant Cell Physiol. 2016, 57, 2461–2471. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Fujii, S.; Sato, M.; Toyooka, K.; Wada, H. Specific role of phosphatidylglycerol and functional overlaps with other thylakoid lipids in Arabidopsis chloroplast biogenesis. Plant Cell Rep. 2015, 34, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Magyar, M.; Akhtar, P.; Sipka, G.; Domonkos, I.; Han, W.; Li, X.; Han, G.; Shen, J.R.; Lambrev, P.H.; Garab, G. Effects of lipids on the rate-limiting steps in the dark-to-light transition of Photosystem II core complex of Thermostichus vulcanus. Front. Plant Sci. 2024, 15, 1381040. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Jimbo, H.; Nakamura, Y.; Wada, H. Biosynthesis of phosphatidylglycerol in photosynthetic organisms. Prog. Lipid Res. 2024, 93, 101266. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Endo, K.; Wada, H. Roles of Lipids in Photosynthesis. In Lipids in Plant and Algae Development; Nakamura, Y., Li-Beisson, Y., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 21–49. [Google Scholar] [CrossRef]
- Guskov, A.; Kern, J.; Gabdulkhakov, A.; Broser, M.; Zouni, A.; Saenger, W. Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 2009, 16, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Rajasekharan, A.; Gummadi, S.N. Flip-flop of phospholipids in proteoliposomes reconstituted from detergent extract of chloroplast membranes: Kinetics and phospholipid specificity. PLoS ONE 2011, 6, e28401. [Google Scholar] [CrossRef]
- Alfonso, M.; Luján, M.A.; Picorel, R. Role of Lipids and Fatty Acids in the Maintenance of Photosynthesis and the Assembly of Photosynthetic Complexes During Photosystem II Turnover. In Photosynthesis: Molecular Approaches to Solar Energy Conversion; Shen, J.-R., Satoh, K., Allakhverdiev, S.I., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 395–427. [Google Scholar] [CrossRef]
- Takahashi, T.; Inoue-Kashino, N.; Ozawa, S.-I.; Takahashi, Y.; Kashino, Y.; Satoh, K. Photosystem II complex in vivo is a monomer. J. Biol. Chem. 2009, 284, 15598–15606. [Google Scholar] [CrossRef]
- Itoh, S.; Kozuki, T.; Nishida, K.; Fukushima, Y.; Yamakawa, H.; Domonkos, I.; Laczkó-Dobos, H.; Kis, M.; Ughy, B.; Gombos, Z. Two functional sites of phosphatidylglycerol for regulation of reaction of plastoquinone Q(B) in photosystem II. Biochimica Biophysica Acta BBA Bioenerg. 2012, 1817, 287–297. [Google Scholar] [CrossRef]
- Yao, H.; Shi, Y.; Gao, R.; Zhang, G.; Zhang, R.; Zheng, C.; Xu, B. Isolation of lipids from photosystem I complex and its characterization with high performance liquid chromatography/electrospray ionization mass spectrometry. J. Chromatogr. B 2006, 837, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Malone, L.A.; Qian, P.; Mayneord, G.E.; Hitchcock, A.; Farmer, D.A.; Thompson, R.F.; Swainsbury, D.J.K.; Ranson, N.A.; Hunter, C.N.; Johnson, M.P. Cryo-EM structure of the spinach cytochrome b6f complex at 3.6 Å. resolution. Nature 2019, 575, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Cao, D.; Xie, F.; Xu, F.; Su, X.; Mi, H.; Zhang, X.; Li, M. Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase. Nat. Commun. 2020, 11, 610. [Google Scholar] [CrossRef] [PubMed]
- Latowski, D.; Åkerlund, H.-E.; Strzałka, K. Violaxanthin de-epoxidase, the xanthophyll cycle enzyme, requires lipid inverted hexagonal structures for its activity. Biochemistry 2024, 43, 4417–4420. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, Y.; Allakhverdiev, S.I.; Murata, N. Protein synthesis is the primary target of reactive oxygen species in the photoinhibition of photosystem II. Physiol. Plant. 2011, 142, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Sato, N. Roles of the acidic lipids sulfoquinovosyl diacylglycerol and phosphatidylglycerol in photosynthesis: Their specificity and evolution. J. Plant Res. 2004, 117, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Allakhverdiev, S.I.; Los, D.A.; Murata, N. Regulatory Roles in Photosynthesis of Unsaturated Fatty Acids in Membrane Lipids. In Lipids in Photosynthesis: Essential and Regulatory Functions; Wada, H., Murata, N., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 373–388. [Google Scholar] [CrossRef]
- Kim, E.-H.; Razeghifard, R.; Anderson, J.M.; Chow, W.S. Multiple sites of retardation of electron transfer in Photosystem II after hydrolysis of phosphatidylglycerol. Photosynth. Res. 2007, 93, 149–158. [Google Scholar] [CrossRef]
- Sakurai, I.; Mizusawa, N.; Ohashi, S.; Kobayashi, M.; Wada, H. Effects of the lack of phosphatidylglycerol on the donor side of photosystem II. Plant Physiol. 2007, 144, 1336–1346. [Google Scholar] [CrossRef]
- Domonkos, I.; Malec, P.; Sallai, A.; Kovács, L.; Itoh, K.; Shen, G.; Ughy, B.; Bogos, B.; Sakurai, I.; Kis, M.; et al. Phosphatidylglycerol is essential for oligomerization of photosystem I reaction center. Plant Physiol. 2004, 134, 1471–1478. [Google Scholar] [CrossRef]
- Endo, K.; Kobayashi, K.; Wang, H.-T.; Chu, H.-A.; Shen, J.-R.; Wada, H. Site-directed mutagenesis of two amino acid residues in cytochrome b 559 α subunit that interact with a phosphatidylglycerol molecule (PG772) induces quinone-dependent inhibition of photosystem II activity. Photosynth. Res. 2019, 139, 267–279. [Google Scholar] [CrossRef]
- Pan, X.; Liu, Z.; Li, M.; Chang, W. Architecture and function of plant light-harvesting complexes II. Curr. Opin. Struct. Biol. 2013, 23, 515–525. [Google Scholar] [CrossRef]
- Páli, T.; Garab, G.; Horváth, L.I.; Kóta, Z. Functional significance of the lipid–protein interface in photosynthetic membranes. Cell. Mol. Life Sci. 2003, 60, 1591–1606. [Google Scholar] [CrossRef] [PubMed]
- Grotjohann, I.; Fromme, P. Structure of cyanobacterial photosystem I. Photosynth. Res. 2005, 85, 51–72. [Google Scholar] [CrossRef] [PubMed]
- Jimbo, H.; Takagi, K.; Hirashima, T.; Nishiyama, Y.; Wada, H. Long-Chain Saturated Fatty Acids, Palmitic and Stearic Acids, Enhance the Repair of Photosystem II. Int. J. Mol. Sci. 2020, 21, 7509. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhang, Z.; Bi, Y.; Yang, W.; Xu, Y.; Zhang, L. Decreased stability of photosystem I in dgd1 mutant of Arabidopsis thaliana. FEBS Lett. 2005, 579, 3619–3624. [Google Scholar] [CrossRef] [PubMed]
- Malavath, T.; Caspy, I.; Netzer-El, S.Y.; Klaiman, D.; Nelson, N. Structure and function of wild-type and subunit-depleted photosystem I in Synechocystis. Biochim. Biophys. Acta BBA Bioenerg. 2018, 1859, 645–654. [Google Scholar] [CrossRef]
- Dobson, Z.; Ahad, S.; Vanlandingham, J.; Toporik, H.; Vaughn, N.; Vaughn, M.; Williams, D.; Reppert, M.; Fromme, P.; Mazor, Y. The structure of photosystem I from a high-light tolerant cyanobacteria. eLife 2021, 10, e67518. [Google Scholar] [CrossRef]
- Qin, X.; Suga, M.; Kuang, T.; Shen, J.R. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 2015, 348, 989–995. [Google Scholar] [CrossRef]
- Kurisu, G.; Zhang, H.; Smith, J.L.; Cramer, W.A. Structure of the cytochrome b6f complex of oxygenic photosynthesis: Tuning the cavity. Science 2003, 302, 1009–1014. [Google Scholar] [CrossRef]
- Baniulis, D.; Yamashita, E.; Whitelegge, J.P.; Zatsman, A.I.; Hendrich, M.P.; Hasan, S.S.; Ryan, C.M.; Cramer, W.A. Structure–function, stability, and chemical modification of the cyanobacterial cytochrome b6f complex from Nostoc sp. PCC 7120. J. Biol. Chem. 2009, 284, 9861–9869. [Google Scholar] [CrossRef]
- Cramer, W.A. Structure function of the cytochrome b6f lipoprotein complex: A scientific odyssey and personal perspective. Photosynth. Res. 2019, 139, 53–65. [Google Scholar] [CrossRef] [PubMed]
- White, S.H. Membrane Proteins of Known 3D Structure. 2018. Available online: https://blanco.biomol.uci.edu/mpstruc/ (accessed on 15 August 2025).
- Hasan, S.S.; Yamashita, E.; Ryan, C.M.; Whitelegge, J.P.; Cramer, W.A. Conservation of lipid functions in cytochrome bc complexes. J. Mol. Biol. 2011, 414, 145–162. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Ping, W.; Wu, H.; Li, M.; Gu, D.; Xu, Y. Monogalactosyldiacylglycerol deficiency in tobacco inhibits the cytochrome b6f-mediated intersystem electron transport process and affects the photostability of the photosystem II apparatus. Biochim. Biophys. Acta BBA Bioenerg. 2013, 1827, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Stroebel, D.; Choquet, Y.; Popot, J.-L.; Picot, D. An atypical haem in the cytochrome b6f complex. Nature 2003, 426, 413–418. [Google Scholar] [CrossRef]
- Kern, J.; Zouni, A.; Guskov, A.; Krauß, N. Lipids in the Structure of Photosystem I, Photosystem II and the Cytochrome b6f Complex. In Lipids in Photosynthesis: Essential and Regulatory Functions; Wada, H., Murata, N., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 203–242. [Google Scholar] [CrossRef]
- Peltier, G.; Aro, E.-M.; Shikanai, T. NDH-1 and NDH-2 plastoquinone reductases in oxygenic photosynthesis. Annu. Rev. Plant Biol. 2016, 67, 55–80. [Google Scholar] [CrossRef]
- Schuller, J.M.; Saura, P.; Thiemann, J.; Schuller, S.K.; Gamiz-Hernandez, A.P.; Kurisu, G.; Nowaczyk, M.M.; Kaila, V.R. Redox-coupled proton pumping drives carbon concentration in the photosynthetic complex I. Nat. Commun. 2020, 11, 494. [Google Scholar] [CrossRef]
- Shen, L.; Tang, K.; Wang, W.; Wang, C.; Wu, H.; Mao, Z.; An, S.; Chang, S.; Kuang, T.; Shen, J.-R.; et al. Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare. Nature 2022, 601, 649–654. [Google Scholar] [CrossRef]
- Su, X.; Cao, D.; Pan, X.; Shi, L.; Liu, Z.; Dall’oSto, L.; Bassi, R.; Zhang, X.; Li, M. Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana. Mol. Plant 2022, 15, 454–467. [Google Scholar] [CrossRef]
- Introini, B.; Hahn, A.; Kühlbrandt, W. Cryo-EM structure of the NDH-PSI-LHCI supercomplex from Spinacia oleracea. Nat. Struct. Mol. Biol. 2025, 32, 968–978. [Google Scholar] [CrossRef]
- Nguyen, H.C.; Melo, A.A.; Kruk, J.; Frost, A.; Gabruk, M. Photocatalytic LPOR forms helical lattices that shape membranes for chlorophyll synthesis. Nat. Plants 2021, 7, 437–444. [Google Scholar] [CrossRef]
- Williams, W.P. The role of lipids in the structure and function of photosynthetic membranes. Prog. Lipid Res. 1994, 33, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Suga, M.; Shen, J.-R. Structural variations of photosystem I-antenna supercomplex in response to adaptations to different light environments. Curr. Opin. Struct. Biol. 2020, 63, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Wickramanayake, J.S.; Goss, J.A.; Zou, M.; Goggin, F.L. Loss of Function of Fatty Acid Desaturase 7 in Tomato Enhances Photosynthetic Carbon Fixation Efficiency. Front. Plant Sci. 2020, 11, 932. [Google Scholar] [CrossRef] [PubMed]
- Vereshchagin, A.G.; Novitskaya, G.V. Anomalous Ionization of Methyl Linolenate by Metastable Argon Atoms: Possible Linolenic Acid Participation in Photosynthetic Reactions. Nature 1964, 203, 1384–1385. [Google Scholar] [CrossRef]
- Klyachko-Gurvich, G.L.; Tsoglin, L.N.; Doucha, J.; Kopetskii, J.; Shebalina, I.B.; Semenenko, V.E. Desaturation of fatty acids as an adaptive response to shifts in light intensity 1. Physiol. Plant. 2002, 107, 240–249. [Google Scholar] [CrossRef]
- Zhukov, A.V. Palmitic Acid and Its Role in the Structure and Functions of Plant Cell Membranes. Russ. J. Plant Physiol. 2015, 62, 206–213. [Google Scholar] [CrossRef]
- Chan, T.; Shimizu, Y.; Pospíšil, P.; Nijo, N.; Fujiwara, A.; Taninaka, Y.; Ishikawa, T.; Hori, H.; Nanba, D.; Imai, A.; et al. Quality control of photosystem II: Lipid peroxidation accelerates photoinhibition under excessive illumination. PLoS ONE 2012, 7, e52100, Correction in PLoS ONE 2013, 8, 10–1371. [Google Scholar] [CrossRef]
- Ivanov, A.; Allakhverdiev, S.; Huner, N.; Murata, N. Genetic decrease in fatty acid unsaturation of phosphatidylglycerol increased photoinhibition of photosystem I at low temperature in tobacco leaves. Biochim. Biophys. Acta BBA Bioenerg. 2012, 1817, 1374–1379. [Google Scholar] [CrossRef]
- LaBrant, E.; Barnes, A.C.; Roston, R.L. Lipid transport required to make lipids of photosynthetic membranes. Photosynth. Res. 2018, 138, 345–360. [Google Scholar] [CrossRef]
- Pospíšil, P.; Yamamoto, Y. Damage to photosystem II by lipid peroxidation products. Biochim. Biophys. Acta BBA Gen. Subj. 2017, 1861, 457–466. [Google Scholar] [CrossRef]
- Pospíšil, P.; Kumar, A.; Prasad, A. Reactive oxygen species in photosystem II: Relevance for oxidative signaling. Photosynth. Res. 2022, 152, 245–260. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhukov, A.; Volkov, V. The Photosynthetic Complexes of Thylakoid Membranes of Photoautotrophs and a Quartet of Their Polar Lipids. Int. J. Mol. Sci. 2025, 26, 9869. https://doi.org/10.3390/ijms26209869
Zhukov A, Volkov V. The Photosynthetic Complexes of Thylakoid Membranes of Photoautotrophs and a Quartet of Their Polar Lipids. International Journal of Molecular Sciences. 2025; 26(20):9869. https://doi.org/10.3390/ijms26209869
Chicago/Turabian StyleZhukov, Anatoly, and Vadim Volkov. 2025. "The Photosynthetic Complexes of Thylakoid Membranes of Photoautotrophs and a Quartet of Their Polar Lipids" International Journal of Molecular Sciences 26, no. 20: 9869. https://doi.org/10.3390/ijms26209869
APA StyleZhukov, A., & Volkov, V. (2025). The Photosynthetic Complexes of Thylakoid Membranes of Photoautotrophs and a Quartet of Their Polar Lipids. International Journal of Molecular Sciences, 26(20), 9869. https://doi.org/10.3390/ijms26209869






