Genome-Wide Characterization of the MDS Gene Family in Gossypium Reveals GhMDS11 as a Key Mediator of Cold Stress Response
Abstract
1. Introduction
2. Results
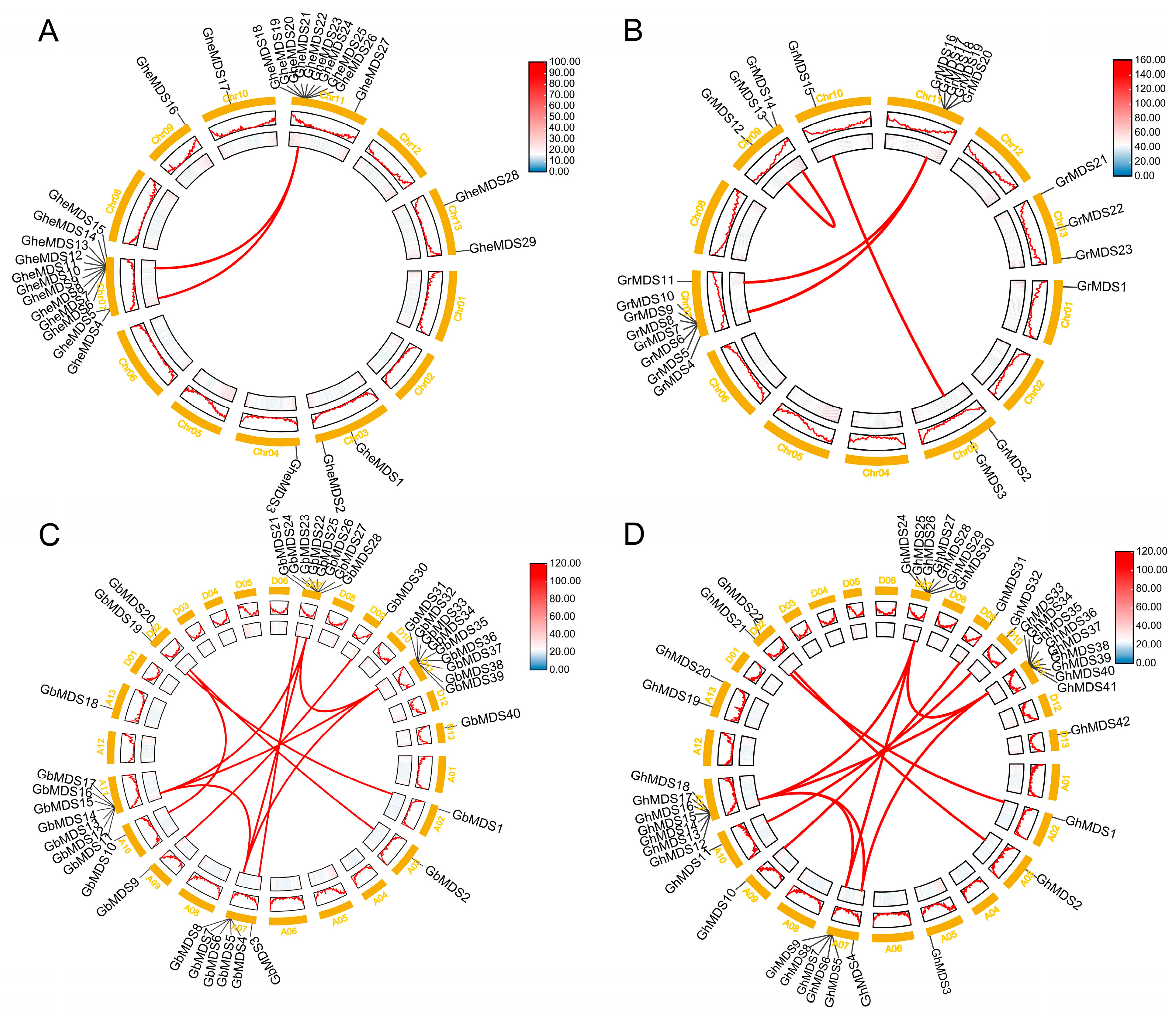
2.1. Identification, Chromosomal Localization, and Physicochemical Property Analysis of the MDS Gene Family
2.2. Analysis of Gene Duplication and Evolutionary Selection Patterns in the MDS Gene Family
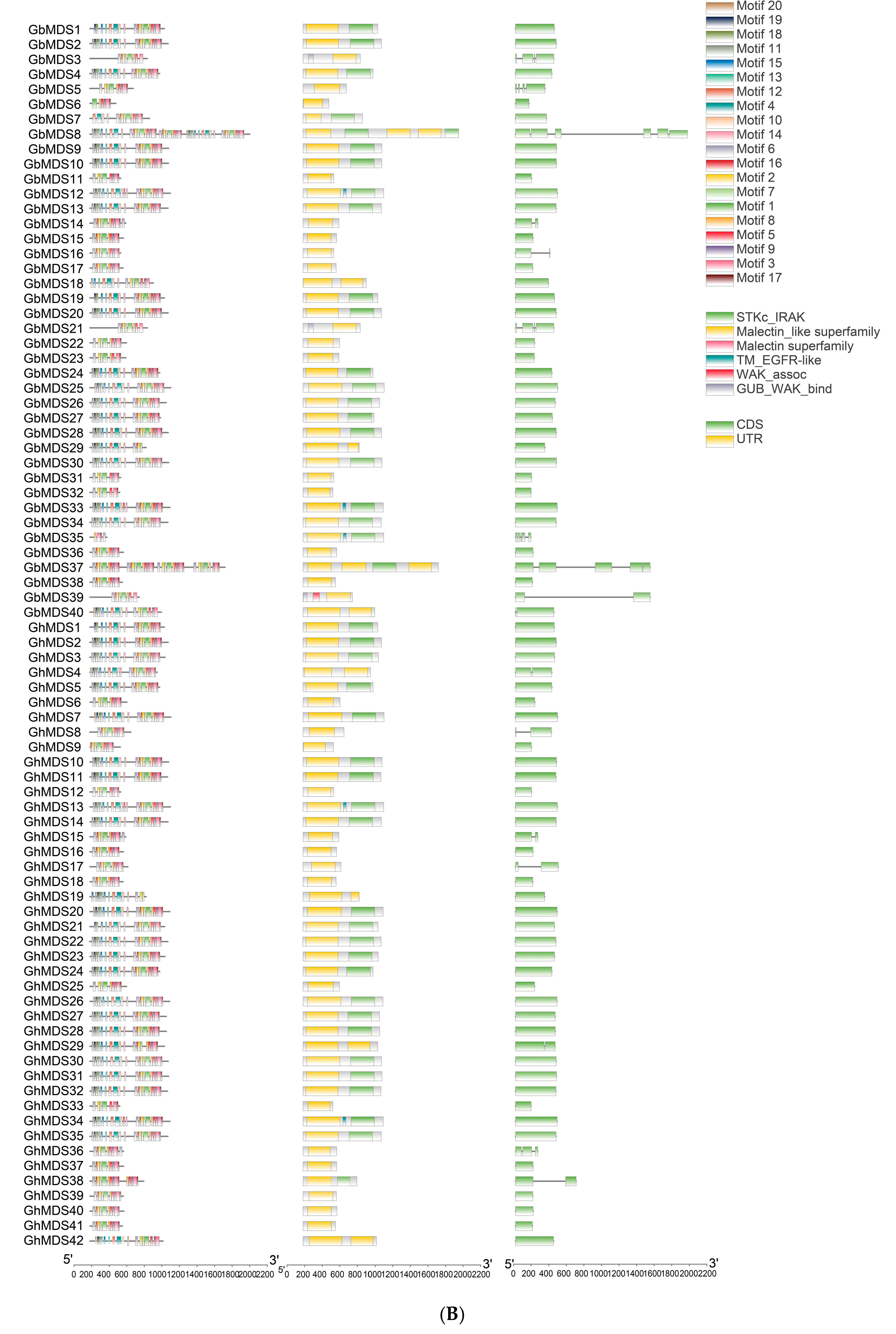
2.3. Gene Structure Analysis of the MDS Gene Family
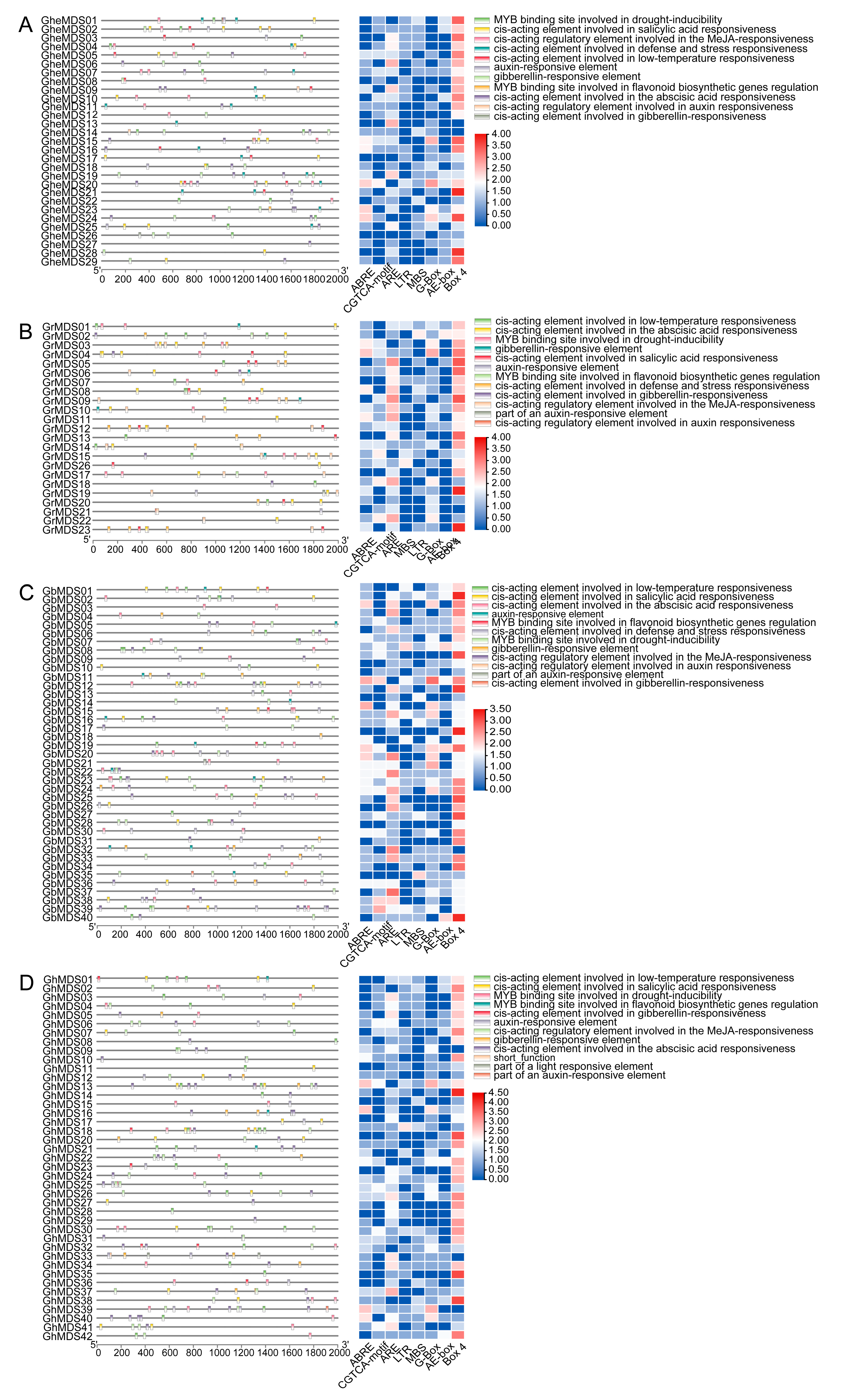
2.4. Prediction and Analysis of Promoter Function in the MDS Gene Family
2.5. Expression Profiles of GhMDS Genes in Different Tissues and Under Various Stress Conditions
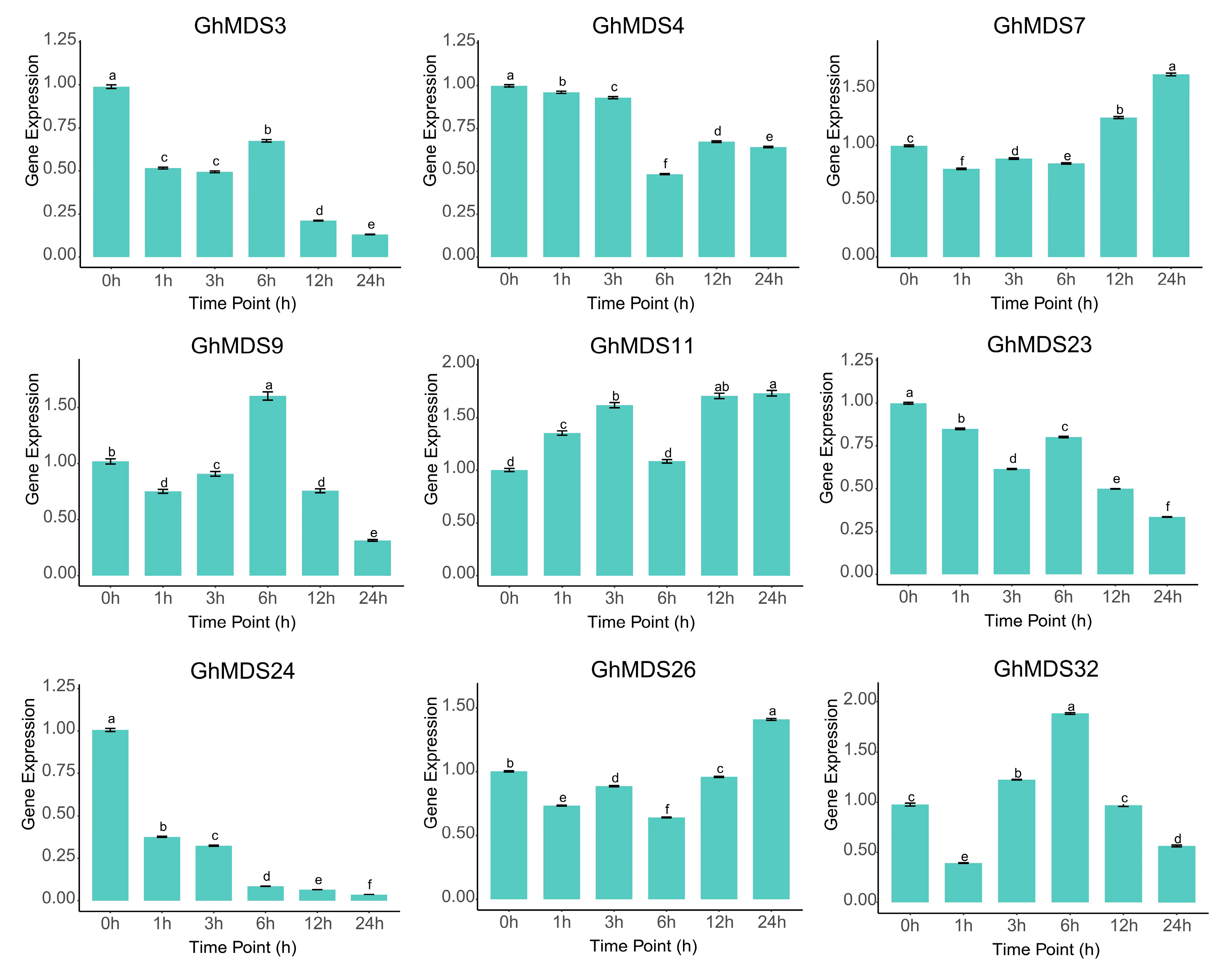
2.6. Functional Validation Analysis of GhMDS Gene Transcriptomes
2.7. GhMDS11-Silenced Cotton Plants Exhibit Increased Sensitivity to Cold Stress
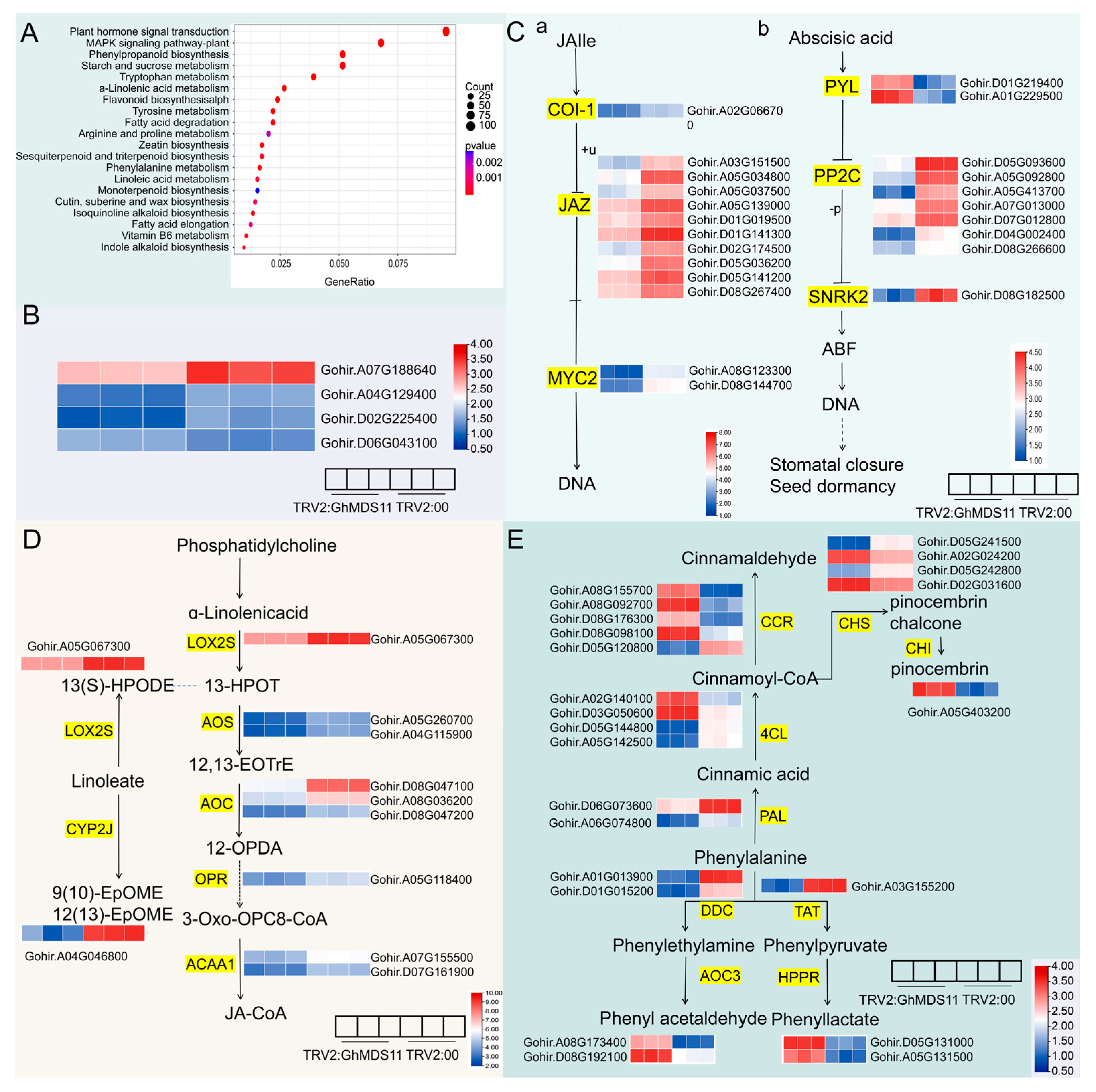
2.8. Comparative Transcriptome Analysis of GhMDS11-Silenced Cotton Plants Through GO Enrichment Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification of MDS Gene Family Members
4.3. Phylogenetic Analysis of MDS Gene Family
4.4. Chromosomal Localization and Synteny Analysis of MDS Gene Family
4.5. Analysis of Cis-Acting Elements in MDS Gene Family Promoters
4.6. Expression Analysis of GhMDS Genes
4.7. Virus-Induced Gene Silencing (VIGS) Assay
4.8. Data Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smith, C.W.; Cothren, J.T. Cotton: Origin, History, Technology, and Production; Wiley: Hoboken, NJ, USA, 1999; Volume 40, p. 1492a. [Google Scholar]
- Thorp, K.R.; Ale, S.; Bange, M.P.; Barnes, E.M.; Hoogenboom, G.; Lascano, R.J.; McCarthy, A.C.; Nair, S.; Paz, J.O.; Rajan, N.; et al. Development and Application of Process-based Simulation Models for Cotton Production: A Review of Past, Present, and Future Directions. J. Cotton Sci. 2014, 18, 10–47. [Google Scholar] [CrossRef]
- Wei, T.; Zheng, J.; Hou, Y.; Xu, Y.; Aziz, K.; Lu, P.; Wang, Y.; Wang, K.; Liu, F.; Cai, X.; et al. GhGTG1 enhances cold stress tolerance by improving sensitivity to ABA in cotton and Arabidopsis. Environ. Exp. Bot. 2023, 208, 105256. [Google Scholar] [CrossRef]
- Ijaz, A.; Anwar, Z.; Ali, A.; Ditta, A.; Shani, M.Y.; Haidar, S.; Wang, B.; Fang, L.; Khan, S.M.U.D.; Khan, M.K.R. Unraveling the genetic and molecular basis of heat stress in cotton. Front. Genet. 2024, 15, 1296622. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Sarwar, R.; Zhang, W.; Geng, R.; Zhu, K.M.; Tan, X.L. Research progress on the physiological response and molecular mechanism of cold response in plants. Front. Plant Sci. 2024, 15, 1334913. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, Y.; Feng, S.; Zhao, T.; Wang, L.; Zheng, Z.; Ai, N.; Guan, X. The impact of temperature on cotton yield and production in Xinjiang, China. npj Sustain. Agric. 2024, 2, 33. [Google Scholar] [CrossRef]
- Adnan, A.; Ali, R.M.; Yue, Z.; Lizhen, Z.; Xuejiao, W.; Mukhtar, A.; Muhammad, H. Impact of climate warming on cotton growth and yields in China and pakistan: A regional perspective. Agriculture 2021, 11, 97. [Google Scholar] [CrossRef]
- Liu, Z.; Ji, M.; He, R.; Dai, Y.; Liu, Y.; Mou, N.; Du, J.; Zhang, X.; Chen, D.; Chen, Y. Effect of low temperature on insecticidal protein contents of cotton (Gossypium herbaceum L.) in the boll shell and its physiological mechanism. Plants 2023, 12, 1767. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, J.; Xu, J.; Zhang, X.; Xie, Z.; Li, Z. Effect of cold stress on photosynthetic physiological characteristics and molecular mechanism analysis in cold-resistant cotton (ZM36) seedlings. Front. Plant Sci. 2024, 15, 1396666. [Google Scholar] [CrossRef] [PubMed]
- Abro, A.A.; Qasim, M.; Abbas, M.; Muhammad, N.; Ali, I.; Khalid, S.; Ahmed, J.; Waqas, M.; Ercisli, S.; Iqbal, R.; et al. Integrating physiological and molecular insights in cotton under cold stress conditions. Genet. Resour. Crop Evol. 2024, 72, 2561–2591. [Google Scholar] [CrossRef]
- Lorençone, J.A.; Lorençone, P.A.; Aparecido, L.E.d.O.; Torsoni, G.B.; Rolim, G.d.S.; Macedo, F.G. The future of cotton in brazil: Agroclimatic suitability and climate change impacts. AgriEngineering 2025, 7, 198. [Google Scholar] [CrossRef]
- Cabusora, C.C. Developing climate-resilient crops: Adaptation to abiotic stress-affected areas. Technol. Agron. 2024, 4, e005. [Google Scholar] [CrossRef]
- Abdelraheem, A.; Adams, N.; Zhang, J. Effects of drought on agronomic and fiber quality in an introgressed backcross inbred line population of Upland cotton under field conditions. Field Crops Res. 2020, 254, 107850. [Google Scholar] [CrossRef]
- Huang, G.; Huang, J.Q.; Chen, X.Y.; Zhu, Y.X. Recent Advances and Future Perspectives in Cotton Research. Annu. Rev. Plant Biol. 2021, 72, 437–462. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Lichen, W. Mechanism of cotton resistance to abiotic stress, and recent research advances in the osmoregulation related genes. Front. Plant Sci. 2022, 13, 972635. [Google Scholar] [CrossRef] [PubMed]
- Soni, M.; Sheshukov, A.Y.; Aguilar, J. The critical role of temperature in determining optimal planting schedule for cotton: A review. Agric. For. Meteorol. 2025, 373, 110741. [Google Scholar] [CrossRef]
- Demeke, B.W.; Rathore, L.S.; Mekonnen, M.M.; Liu, W. Spatiotemporal dynamics of the water footprint and virtual water trade in global cotton production and trade. Clean. Prod. Lett. 2024, 7, 100074. [Google Scholar] [CrossRef]
- Liu, J.; Magwanga, R.O.; Xu, Y.; Wei, T.; Kirungu, J.N.; Zheng, J.; Hou, Y.; Wang, Y.; Agong, S.G.; Okuto, E. Functional characterization of cotton C-repeat binding factor genes reveal their potential role in cold stress tolerance. Front. Plant Sci. 2021, 12, 766130. [Google Scholar] [CrossRef]
- Liu, Y.; Zhong, X.; Zhang, Z.; Lan, J.; Huang, X.; Tian, H.; Li, X.; Zhang, Y. Receptor-like kinases MDS1 and MDS2 promote SUMM2-mediated immunity. J. Integr. Plant Biol. 2021, 63, 277–282. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, J. Multiple functions and regulatory networks of WRKY33 and its orthologs. Gene 2024, 931, 148899. [Google Scholar] [CrossRef]
- Ma, Z.; Hu, L. WRKY transcription factor responses and tolerance to abiotic stresses in plants. Int. J. Mol. Sci. 2024, 25, 6845. [Google Scholar] [CrossRef]
- Yu, M.; Luobu, Z.; Zhuoga, D.; Wei, X.; Tang, Y. Advances in plant response to low-temperature stress. Plant Growth Regul. 2024, 105, 167–185. [Google Scholar] [CrossRef]
- Qian, Z.; He, L.; Li, F. Understanding cold stress response mechanisms in plants: An overview. Front. Plant Sci. 2024, 15, 1443317. [Google Scholar] [CrossRef] [PubMed]
- Julia, R.; Matthew, W.J.; Peter, S.; Monika, B.; Jana, N.; Matthias, B.; Peggy, S.-B.; Vera, S.; Marie-Theres, H. Multiplex mutagenesis of four clustered CrRLK1L with CRISPR/Cas9 exposes their growth regulatory roles in response to metal ions. Sci. Rep. 2018, 8, 12182. [Google Scholar]
- Li, J. Functional Analysis of MDS1/MDS3 in Arabidopsis thaliana. Master’s Thesis, Shandong Agricultural University, Tai’an, China, 2022. [Google Scholar]
- Ge, Z.; Yun, S.; Qianhua, W.; Dongxia, Y.; Dongliang, L.; Wenqiang, Q.; Xiaoyang, G.; Zuoren, Y.; Wenying, X.; Zhen, S.; et al. Gossypium hirsutum Salt Tolerance Is Enhanced by Overexpression of G. arboreum JAZ1. Front. Bioeng. Biotechnol. 2020, 8, 157. [Google Scholar] [CrossRef]
- Clark, M.K.; Behmer, S.T.; Sword, G.A. Selection of stable reference genes for accurate reverse-transcription quantitative PCR in cotton-herbivore studies using virus-induced gene silencing. Sci. Rep. 2025, 15, 24482. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Shuang, J.; Li, Z.; Xiao, H.; Liu, Y.; Wang, T.; Wei, Y.; Hu, S.; Wan, S.; Peng, R. Identification of the Golden-2-like transcription factors gene family in Gossypium hirsutum. PeerJ 2021, 9, e12484. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, J.; Huang, Y.; Wang, S.; Wei, L.; Liu, D.; Weng, Y.; Xiang, J.; Zhu, Q.; Yang, Z. CottonMD: A multi-omics database for cotton biological study. Nucleic Acids Res. 2023, 51, D1446–D1456. [Google Scholar] [CrossRef]
- Paul, S.K.; Islam, M.S.U.; Akter, N.; Zohra, F.T.; Rashid, S.B.; Ahmed, M.S.; Rahman, S.M.; Sarkar, M.A.R. Genome-wide identification and characterization of FORMIN gene family in cotton (Gossypium hirsutum L.) and their expression profiles in response to multiple abiotic stress treatments. PLoS ONE 2025, 20, e0319176. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R. Interactive Data Analyses Using TBtools. In Integrative Bioinformatics: History and Future; Chen, M., Hofestädt, R., Eds.; Springer: Singapore, 2022; pp. 343–375. [Google Scholar]
- Chen, C.; Wu, Y.; Xia, R. A painless way to customize Circos plot: From data preparation to visualization using TBtools. iMeta 2022, 1, e35. [Google Scholar]
- Abro, A.A.; Sun, C.; Abbas, M.; Liu, Q.; Jie, Z.; Xu, Y.; Hou, Y.; Zhou, Z.; Iqbal, R.; Liu, F. Comprehensive profiling of Bcl-2-associated athanogene (BAG) genes and their genetic potential role under cold stress in Cotton. Funct. Integr. Genom. 2025, 25, 104. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Li, H.; Li, K.; Guo, Y.; Guo, J.; Miao, K.; Botella, J.R.; Song, C.-P.; Miao, Y. A transient transformation system for gene characterization in upland cotton (Gossypium hirsutum). Plant Methods 2018, 14, 50. [Google Scholar] [CrossRef] [PubMed]
- Maren, N.A.; Duduit, J.R.; Huang, D.; Zhao, F.; Ranney, T.G.; Liu, W. Stepwise optimization of real-time RT-PCR analysis. Methods Mol. Biol. 2023, 2653, 317–332. [Google Scholar] [PubMed]
- Purkayastha, A.; Dasgupta, I. Virus-induced gene silencing: A versatile tool for discovery of gene functions in plants. Plant Physiol. Biochem. 2009, 47, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Broderick, S.R.; Jones, M.L. An optimized protocol to increase virus-induced gene silencing efficiency and minimize viral symptoms in petunia. Plant Mol. Biol. Rep. 2014, 32, 219–233. [Google Scholar] [CrossRef]
- Mardini, M.; Kazancev, M.; Ivoilova, E.; Utkina, V.; Vlasova, A.; Demurin, Y.; Soloviev, A.; Kirov, I. Advancing virus-induced gene silencing in sunflower: Key factors of VIGS spreading and a novel simple protocol. Plant Methods 2024, 20, 122. [Google Scholar] [CrossRef]
- Sung, Y.C.; Lin, C.P.; Chen, J.C. Optimization of virus-induced gene silencing in Catharanthus roseus. Plant Pathol. 2014, 63, 1159–1167. [Google Scholar] [CrossRef]
- Su, Y.; Wang, G.; Huang, Z.; Hu, L.; Fu, T.; Wang, X. Silencing GhIAA43, a member of cotton AUX/IAA genes, enhances wilt resistance via activation of salicylic acid-mediated defenses. Plant Sci. 2022, 314, 111126. [Google Scholar] [CrossRef]
- Yang, X.; Li, H.; Niyitanga, S.; Zhang, L.; Li, X.; Qi, J.; Xu, J.; Tao, A.; Fang, P.; Zhang, L. Establishment of TRV-mediated Gene Silencing and Application for Elucidating Functions of Anthocyanidin Reductase Gene HcANR in Kenaf (Hibiscus cannabinus L.). Trop. Plant Biol. 2023, 16, 146–155. [Google Scholar] [CrossRef]
- Ge, X.; Wu, J.; Zhang, C.; Wang, Q.; Hou, Y.; Yang, Z.; Yang, Z.; Xu, Z.; Wang, Y.; Lu, L.; et al. Prediction of VIGS efficiency by the Sfold program and its reliability analysis in Gossypium hirsutum. Sci. Bull. 2016, 61, 543–551. [Google Scholar] [CrossRef]
- Gao, X.; Shan, L. Functional genomic analysis of cotton genes with agrobacterium-mediated virus-induced gene silencing. In Virus-Induced Gene Silencing: Methods and Protocols; Becker, A., Ed.; Humana Press: Totowa, NJ, USA, 2013; pp. 157–165. [Google Scholar]
- Gao, X.; Britt, R.C., Jr.; Shan, L.; He, P. Agrobacterium-mediated virus-induced gene silencing assay in cotton. J. Vis. Exp. 2011, 54, e2938. [Google Scholar] [CrossRef]
- Wasserstein, R.L.; Schirm, A.L.; Lazar, N.A. Moving to a World Beyond “p < 0.05”. Am. Stat. 2019, 73, 1–19. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Weissgerber, T.L.; Milic, N.M.; Winham, S.J.; Garovic, V.D. Beyond bar and line graphs: Time for a new data presentation paradigm. PLoS Biol. 2015, 13, e1002128. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Khan, A.H.; Liu, Y.; Madad, A.; Zhu, F.; Wang, J.; Zhang, G.; Wang, F.; Li, Z.; Shi, S.; et al. Genome-Wide Characterization of the MDS Gene Family in Gossypium Reveals GhMDS11 as a Key Mediator of Cold Stress Response. Int. J. Mol. Sci. 2025, 26, 10144. https://doi.org/10.3390/ijms262010144
Zhu X, Khan AH, Liu Y, Madad A, Zhu F, Wang J, Zhang G, Wang F, Li Z, Shi S, et al. Genome-Wide Characterization of the MDS Gene Family in Gossypium Reveals GhMDS11 as a Key Mediator of Cold Stress Response. International Journal of Molecular Sciences. 2025; 26(20):10144. https://doi.org/10.3390/ijms262010144
Chicago/Turabian StyleZhu, Xuehan, Ahmad Haris Khan, Yihao Liu, Allah Madad, Faren Zhu, Junwei Wang, Ganggang Zhang, Fei Wang, Zihan Li, Shandang Shi, and et al. 2025. "Genome-Wide Characterization of the MDS Gene Family in Gossypium Reveals GhMDS11 as a Key Mediator of Cold Stress Response" International Journal of Molecular Sciences 26, no. 20: 10144. https://doi.org/10.3390/ijms262010144
APA StyleZhu, X., Khan, A. H., Liu, Y., Madad, A., Zhu, F., Wang, J., Zhang, G., Wang, F., Li, Z., Shi, S., & Li, H. (2025). Genome-Wide Characterization of the MDS Gene Family in Gossypium Reveals GhMDS11 as a Key Mediator of Cold Stress Response. International Journal of Molecular Sciences, 26(20), 10144. https://doi.org/10.3390/ijms262010144





