Genome-Wide Characterization of Kiwifruit Invertase Gene Family Reveals Roles of AcCWINV4 in Sugar Accumulation and Cold Tolerance
Abstract
1. Introduction
2. Results
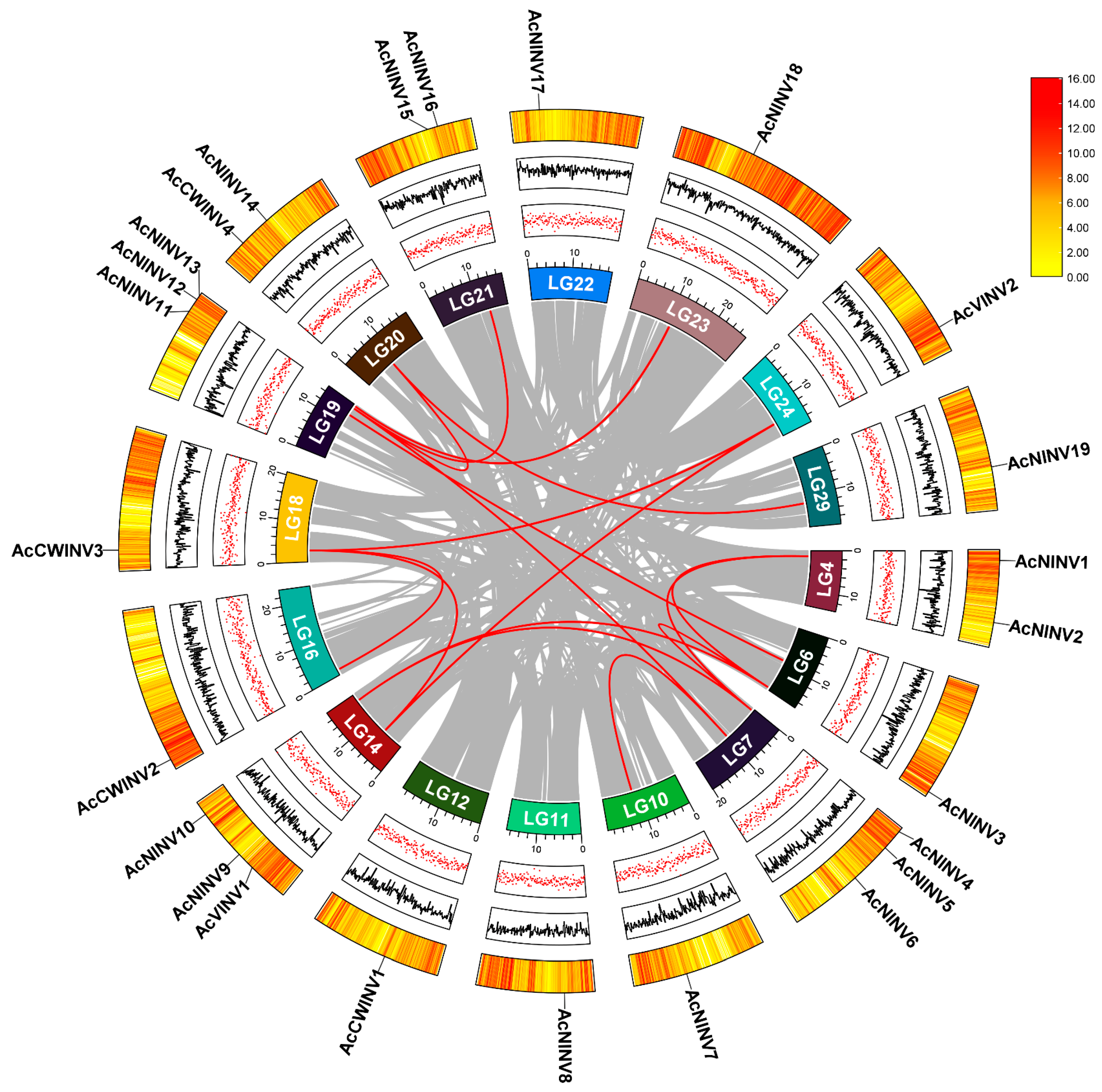
2.1. Identification and Characterization Analyses of the INV Gene Family in Kiwifruit
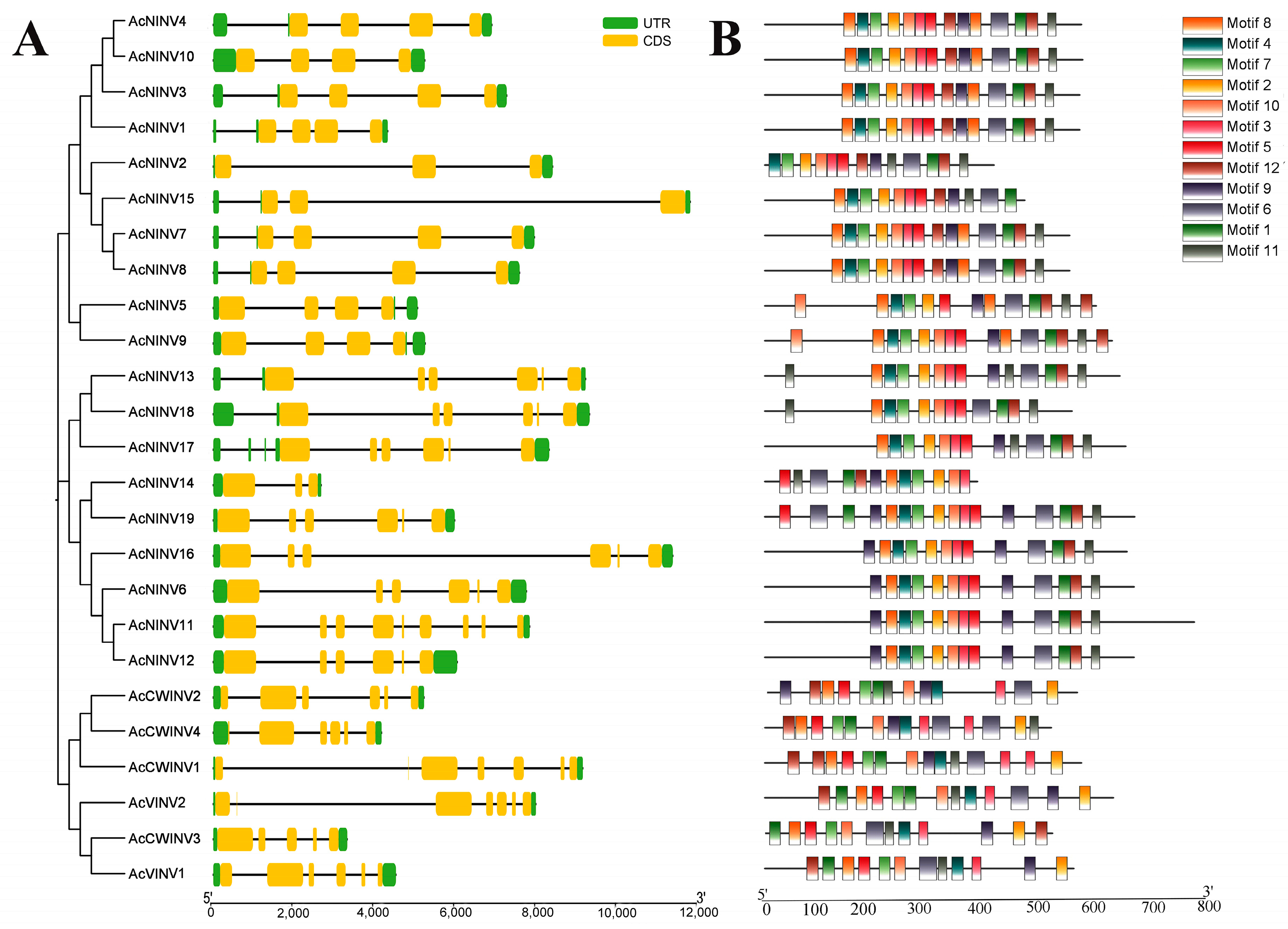
2.2. Structure and Selection Pressure Analyses of AcINVs
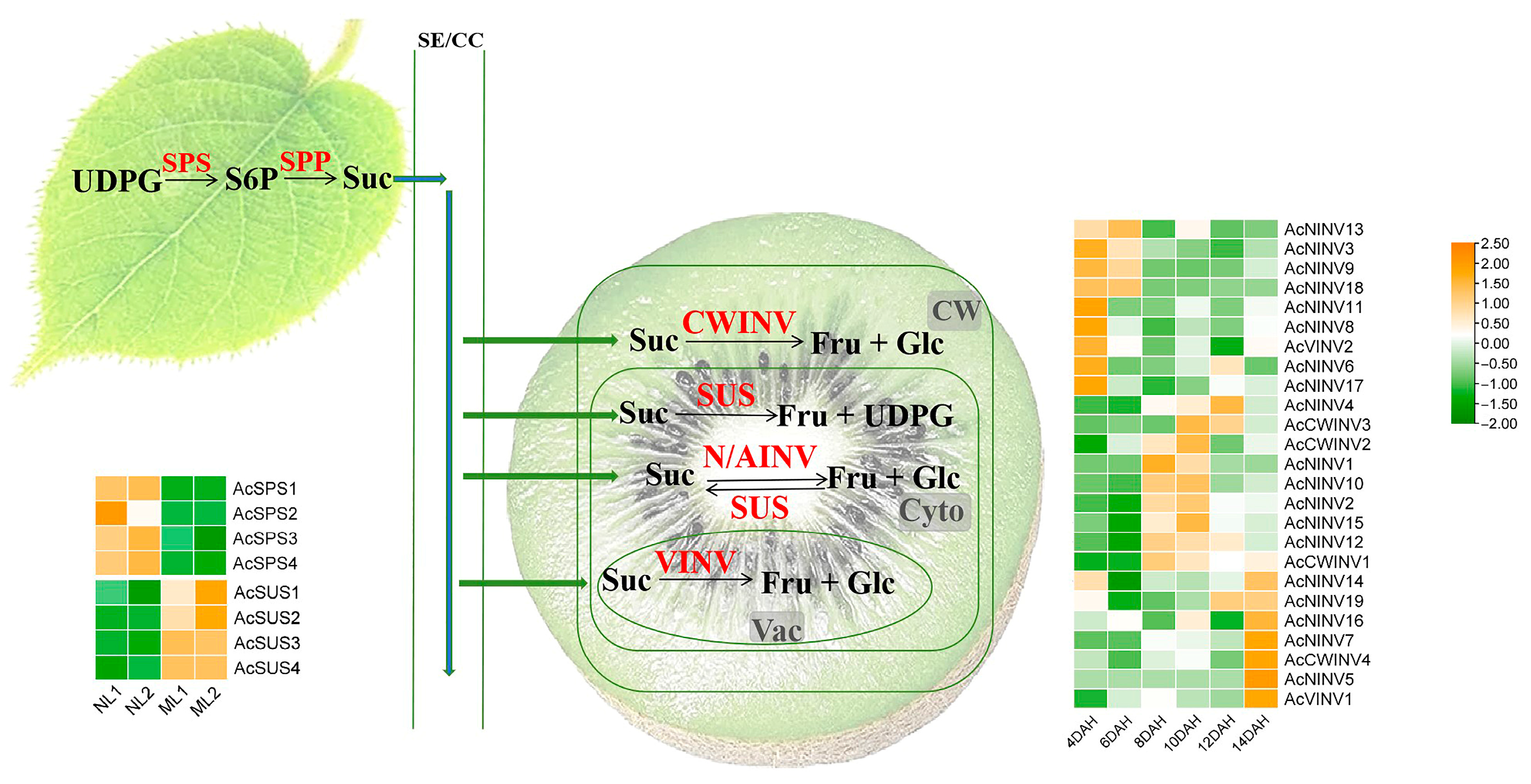
2.3. Expression Patterns of AcINV Family Gene in Source and Sink Tissues
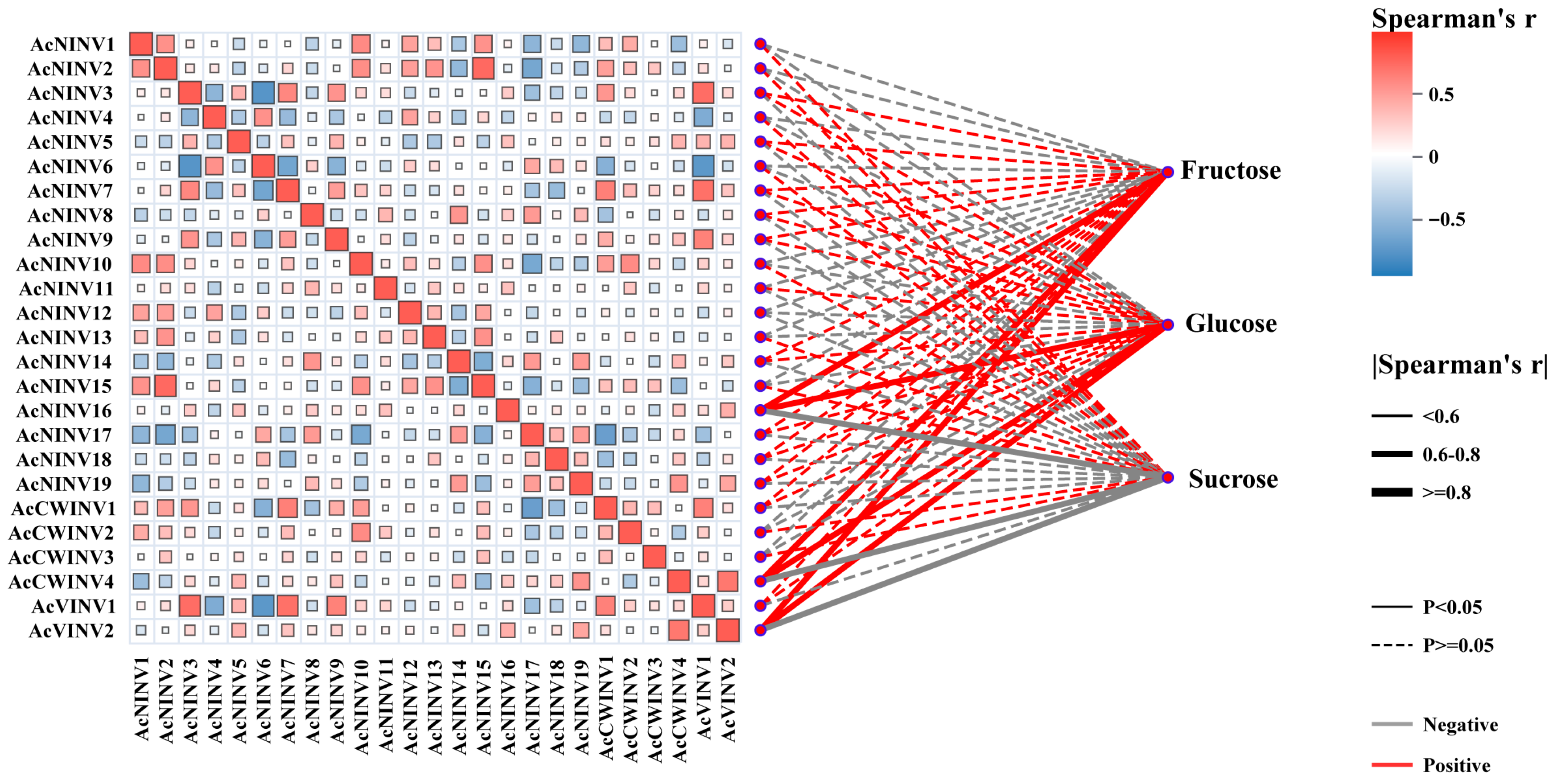
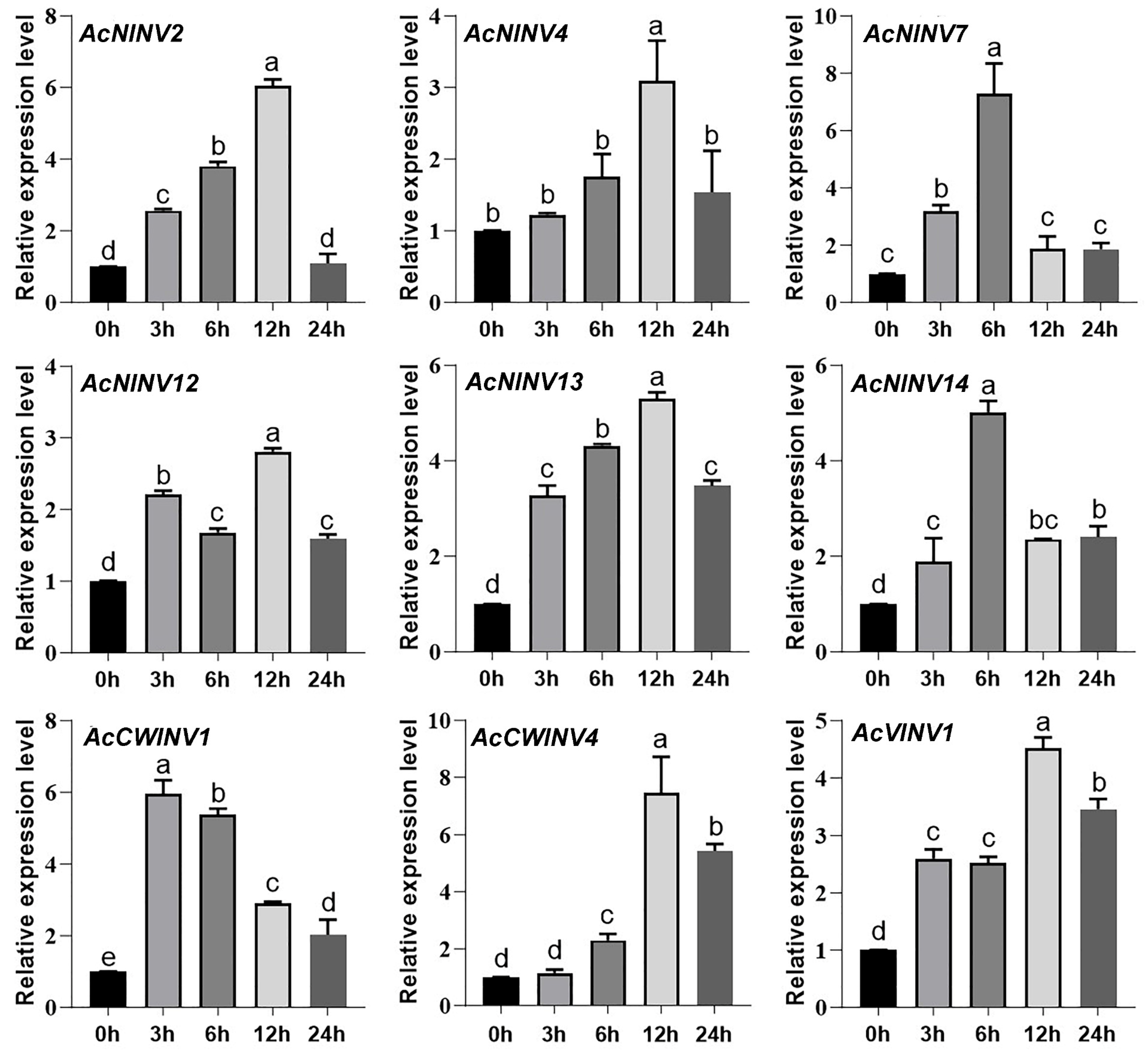
2.4. Correlation Analysis Between INV Expression and Sugar Content
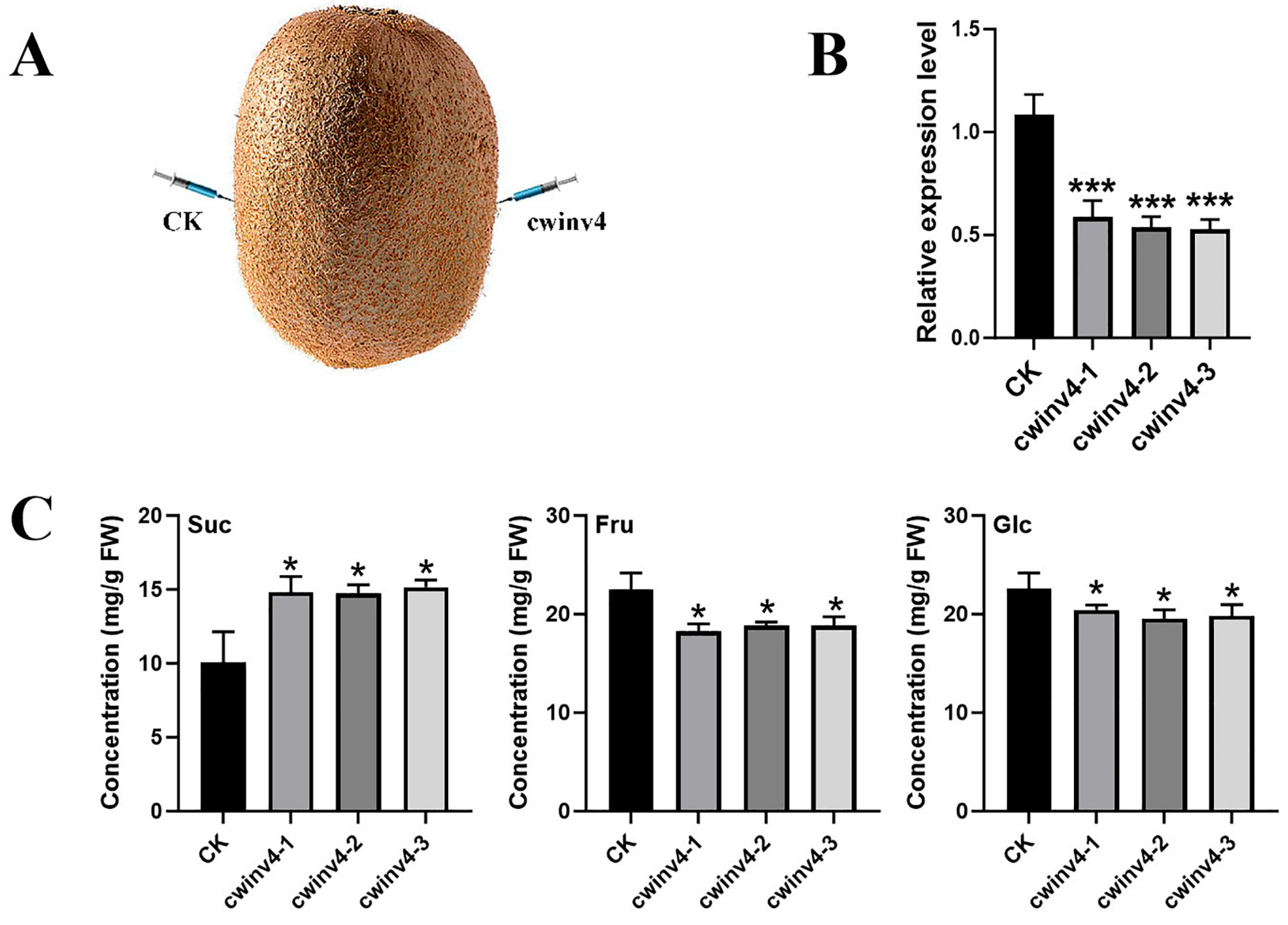
2.5. Functional Analysis of AcCWINV4 Regarding Sugar Contents
2.6. The Expression of AcCWINV4 Is Induced by Low Temperature
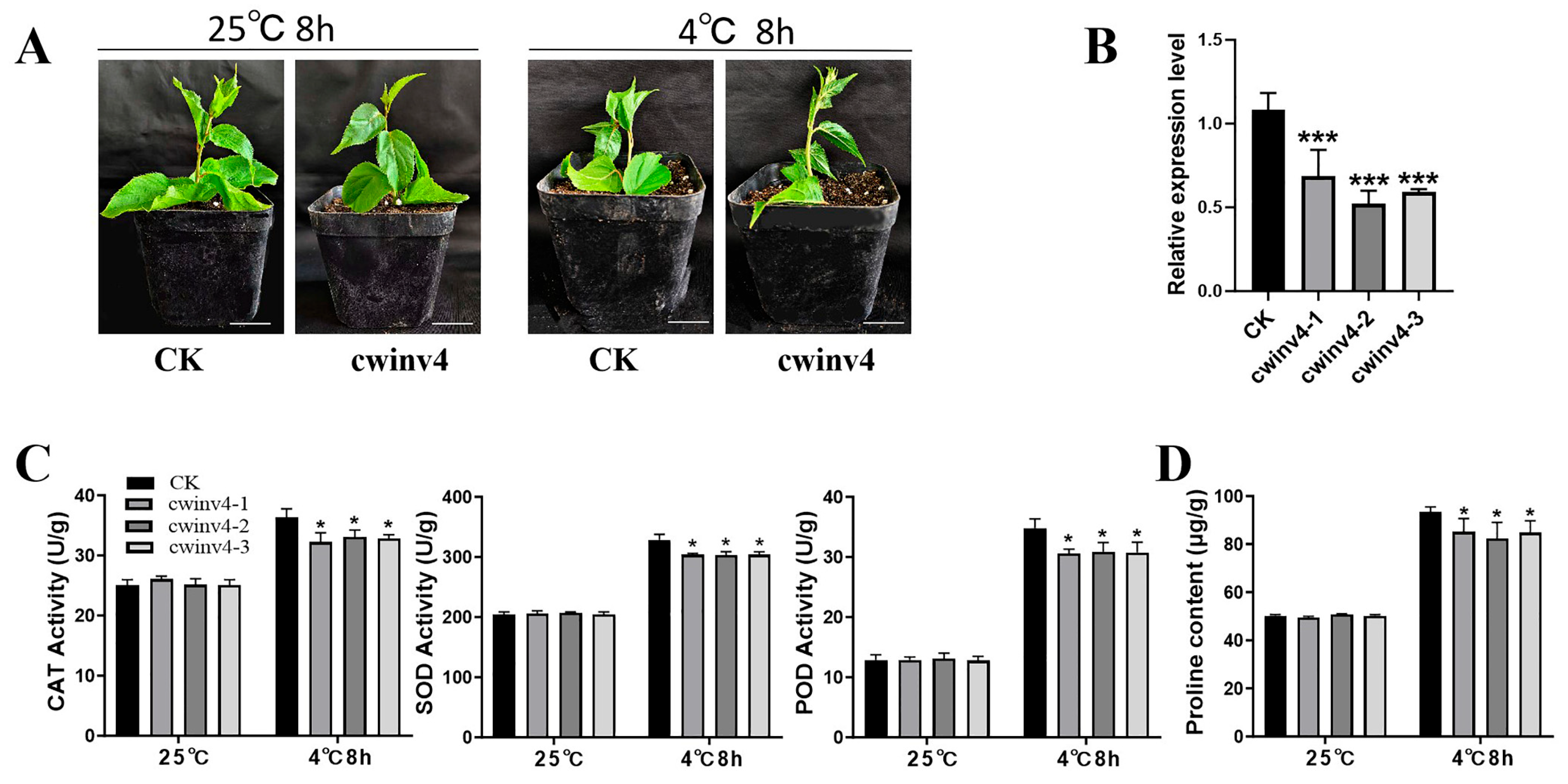
2.7. Functional Analysis of AcCWINV4 Regarding Cold Tolerance
3. Discussion
4. Materials and Methods
4.1. Plant Material and Processing Methods
4.2. Identification of Kiwifruit INV Gene Families
4.3. Bioinformatic Analysis of the INV Gene Family in Kiwifruit
4.4. Gene Silencing of AcCWINV4
4.5. Determination of Sugar Content
4.6. RNA Extraction and qRT-PCR Analysis
4.7. Measurement of Cold Resistance
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Persia, D.; Cai, G.; Del Casino, C.; Faleri, C.; Willemse, M.T.; Cresti, M. Sucrose synthase is associated with the cell wall of tobacco pollen tubes. Plant Physiol. 2008, 147, 1603–1618. [Google Scholar] [CrossRef]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, Y.; Lv, L. Genome-wide identification and function analysis of the sucrose phosphate synthase MdSPS gene family in apple. J. Integ. Agric. 2023, 22, 2080–2093. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, C.; Jia, R.; Yang, N.; Jin, L.; Zhu, L.; Ma, B.; Yao, Y.; Ma, F.; Li, M. Malate metabolism mediated by the cytoplasmic malate dehydrogenase gene MdcyMDH affects sucrose synthesis in apple fruit. Hortic. Res. 2022, 9, uhac194. [Google Scholar] [CrossRef]
- Li, M.; Li, D.; Feng, F.; Zhang, S.; Ma, F.; Cheng, L. Proteomic analysis reveals dynamic regulation of fruit development and sugar and acid accumulation in apple. J. Exp. Bot. 2016, 67, 5145–5157. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, Y.; Luo, Z.; Lv, L.; Wang, C.; Zhu, L.; Ma, F.; Li, M.; Han, D. Overexpression of a transcription factor MdWRKY126 altered soluble sugar accumulation in apple and tomato fruit. Hortic. Plant J. 2025, 11, 989–998. [Google Scholar] [CrossRef]
- Sturm, A. Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999, 121, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L. CWIN-sugar transporter nexus is a key component for reproductive success. J. Plant Physiol. 2022, 268, 153572. [Google Scholar] [CrossRef]
- Roitsch, T.; González, M.C. Function and regulation of plant invertases: Sweet sensations. Trends Plant Sci. 2004, 9, 606–613. [Google Scholar] [CrossRef]
- Lemoine, R.; Camera, S.L.; Atanassova, R.; Dédaldéchamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.; Laloi, L.; Thévenot, P.; Maurousset, L.; et al. Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Wu, X.; Li, Y.; Liu, G.; Cui, Z.; Jiang, T.; Ma, Q.; Luo, L.; Zhang, P. Cell wall invertase 3 affects cassava productivity via regulating sugar allocation from source to sink. Front. Plant Sci. 2019, 10, 541. [Google Scholar] [CrossRef]
- Jin, Y.; Ni, D.A.; Ruan, Y.L. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell 2009, 21, 2072–2089. [Google Scholar] [CrossRef]
- Wang, Z.; Wei, X.; Yang, J.; Li, H.; Ma, B.; Zhang, K.; Zhang, Y.; Cheng, L.; Ma, F.; Li, M. Heterologous expression of the apple hexose transporter MdHT 2.2 altered sugar concentration with increasing cell wall invertase activity in tomato fruit. Plant Biotechnol. J. 2020, 18, 540–552. [Google Scholar]
- Huang, D.; Wu, B.; Chen, G.; Xing, W.; Xu, Y.; Ma, F.; Li, H.; Hu, W.; Huang, H.; Yang, L.; et al. Genome-wide analysis of the passion fruit invertase gene family reveals involvement of PeCWINV5 in hexose accumulation. BMC Plant Biol. 2024, 24, 836. [Google Scholar] [CrossRef]
- Khan, M.A.; Zaman, F.; Liu, Y.Z.; Alam, S.M.; Han, H.; Luo, Y.; Ateeq, M. CsMYB1-CwINV6 Module Involves in the Promotion of Soluble Sugar Accumulation in Citrus Fruits Under Drought Stress. Plant Cell Environ. 2025, 48, 5572–5583. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, H.; Wu, M.; Zhou, Y.; Yu, C.; Yang, Q.; Rolland, F.; Poel, B.; Bouzayen, M.; Hu, N.; et al. A vacuolar invertase gene SlVI modulates sugar metabolism and postharvest fruit quality and stress resistance in tomato. Hortic. Res. 2025, 12, uhae283. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zheng, F.; Feng, Z.; Li, Y.; Ma, M.; Wang, G.; Zhao, H. A vacuolar invertase CsVI2 regulates sucrose metabolism and increases drought tolerance in Cucumis sativus L. Int. J. Mol. Sci. 2021, 23, 176. [Google Scholar]
- Vargas, W.A.; Salerno, G.L. The Cinderella story of sucrose hydrolysis: Alkaline/neutral invertases, from cyanobacteria to unforeseen roles in plant cytosol and organelles. Plant Sci. 2010, 178, 1–8. [Google Scholar] [CrossRef]
- Peng, Y.; Zhu, L.; Tian, R.; Wang, L.; Su, J.; Yuan, Y.; Ma, F.; Li, M.; Ma, B. Genome-wide identification, characterization and evolutionary dynamic of invertase gene family in apple, and revealing its roles in cold tolerance. Int. J. Biol. Macromol. 2023, 229, 766–777. [Google Scholar] [CrossRef] [PubMed]
- Dahro, B.; Wang, Y.; Alhag, A.; Li, C.; Guo, D.; Liu, J.H. Genome-wide identification and expression profiling of invertase gene family for abiotic stresses tolerance in Poncirus trifoliata. BMC Plant Biol. 2021, 21, 559. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Shi, Y.; Yu, Z.; Liu, W.; Lv, B.; Wang, B.; Yang, G. Isolation and functional analysis of MdCS1: A gene encoding a citrate synthase in Malus domestica (L.). Plant Growth Regul. 2015, 75, 209–218. [Google Scholar] [CrossRef]
- He, X.; Wei, Y.; Kou, J.; Xu, F.; Chen, Z.; Shao, X. PpVIN2, an acid invertase gene family member, is sensitive to chilling temperature and affects sucrose metabolism in postharvest peach fruit. Plant Growth Regul. 2018, 86, 169–180. [Google Scholar] [CrossRef]
- Wan, H.; Wu, L.; Yang, Y.; Zhou, G.; Ruan, Y. Evolution of sucrose metabolism: The dichotomy of invertases and beyond. Trends Plant Sci. 2018, 23, 163–177. [Google Scholar] [CrossRef]
- Qian, W.; Xiao, B.; Wang, L.; Hao, X.; Yue, C.; Cao, H.; Wang, Y.; Li, N.; Yu, Y.; Zeng, J. CsINV5, a tea vacuolar invertase gene enhances cold tolerance in transgenic Arabidopsis. BMC Plant Biol. 2018, 18, 228. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, M.; Wang, M.; Zhang, W.; Qiao, C.; Luo, Q.; Lu, X. Freshwater cyanobacterium Synechococcus elongatus PCC 7942 adapts to an environment with salt stress via ion-induced enzymatic balance of compatible solutes. Appl. Environ. Microbiol. 2020, 86, e02904-19. [Google Scholar] [CrossRef]
- Dahro, B.; Wang, Y.; Khan, M.; Zhang, Y.; Fang, T.; Ming, R.; Li, C.; Liu, J.H. Two AT-Hook proteins regulate A/NINV7 expression to modulate sucrose catabolism for cold tolerance in Poncirus trifoliata. New Phytol. 2022, 235, 2331–2349. [Google Scholar] [CrossRef]
- Lunn, J.E. Evolution of sucrose synthesis. Plant Physiol. 2002, 128, 1490–1500. [Google Scholar] [CrossRef]
- Ma, T.; Lan, T.; Ju, Y.; Cheng, G.; Que, Z.; Geng, T.; Fang, Y.; Sun, X. Comparison of the nutritional properties and biological activities of kiwifruit (Actinidia) and their different forms of products: Towards making kiwifruit more nutritious and functional. Food Funct. 2019, 10, 1317–1329. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; An, B.; Zhang, Y.; Yang, J.; Li, S.; Sun, T.; Li, Y. Sugar transporter proteins (STPs) in Gramineae crops: Comparative analysis, phylogeny, evolution, and expression profiling. Cells 2019, 8, 560. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.S.; Raes, J. Duplication and divergence: The evolution of new genes and old ideas. Annu. Rev. Genet. 2004, 38, 615–643. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Xu, H.; Zou, Q.; Zhang, J.; Jiang, S.; Fang, H.; Wang, Y.; Su, M.; Wang, N.; Chen, X. The vacuolar membrane sucrose transporter MdSWEET16 plays essential roles in the cold tolerance of apple. Plant Cell Tissue Organ Cult. (PCTOC) 2020, 140, 129–142. [Google Scholar] [CrossRef]
- Zhou, H.; Hou, Y.; Tan, L.; Li, Q.; Li, W.; Kafle, S.; Xu, M.; Kiselev, K.V.; Meng, L.; Xin, H. VaEIN3.1-VaERF057-VaFBA1 Module Positively Regulates Cold Tolerance by Accumulating Soluble Sugar in Grapevine. Plant Cell Environ. 2025, 48, 5429–5447. [Google Scholar] [CrossRef]
- Li, X.; Xu, Y.; Liu, K.; Li, Z.; Cao, Y.; Zhang, L. VcTT2 enhances drought tolerance in blueberry by regulating ROS scavenging. Fruit Res. 2025, 5, e029. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, Y.; Lv, L.; Wang, T.; Liu, W.; Li, X.; Li, W.; Huo, J.; Han, D. MbbHLH93, a transcription factor associated with cold and drought tolerance in Malus baccata. Fruit Res. 2024, 4, e038. [Google Scholar] [CrossRef]
- Ahiakpa, J.K.; Magdy, M.; Karikari, B.; Munir, S.; Mumtaz, M.A.; Tamim, S.A.; Mahmood, S.; Liu, G.; Chen, W.; Wang, Y. Genome-wide identification and expression profiling of tomato invertase genes indicate their response to stress and phytohormones. J. Plant Growth Regul. 2022, 41, 1481–1498. [Google Scholar] [CrossRef]
- Ji, X.; Van den Ende, W.; Van Laere, A.; Cheng, S.; Bennett, J. Structure, evolution, and expression of the two invertase gene families of rice. J. Mol. Evol. 2005, 60, 615–634. [Google Scholar] [CrossRef]
- Yuan, H.; Pang, F.; Cai, W.; Chen, X.; Zhao, M.; Yu, H. Genome-wide analysis of the invertase genes in strawberry (Fragaria × ananassa). Integr. Agric. 2021, 20, 2652–2665. [Google Scholar] [CrossRef]
- Shen, L.; Yao, Y.; He, H.; Qin, Y.; Liu, Z.; Liu, W.; Qi, Z.; Yang, L.; Cao, Z.; Yang, Y. Genome-wide identification, expression, and functional analysis of the alkaline/neutral invertase gene family in pepper. Int. J. Mol. Sci. 2018, 19, 224. [Google Scholar] [CrossRef]
- Yao, Y.; Geng, M.T.; Wu, X.; Liu, J.; Li, R.; Hu, X.; Guo, J. Genome-wide identification, expression, and activity analysis of alkaline/neutral invertase gene family from cassava (Manihot esculenta Crantz). Plant Mol. Biol. Rep. 2015, 33, 304–315. [Google Scholar] [CrossRef]
- Xu, X.; Yang, Y.; Liu, C.; Sun, Y.; Zhang, T.; Hou, M.; Huang, S.; Yuan, H. The evolutionary history of the sucrose synthase gene family in higher plants. BMC Plant Biol. 2019, 19, 566. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Cai, C.; Lu, Y.; Li, Y.; Xie, Z. Identification and expression analysis of invertase family genes during grape (Vitis vinifera L.) berry development under CPPU and GA treatment. Mol. Genet. Genomics. 2023, 298, 777–789. [Google Scholar]
- Braun, D.; Wang, L.; Ruan, Y. Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J. Exp. Bot. 2014, 65, 1713–1735. [Google Scholar] [CrossRef]
- Tao, H.; Sun, H.; Wang, Y.; Wang, X.; Guo, Y. Effects of water stress on quality and sugar metabolism in ‘Gala’apple fruit. Hortic. Plant J. 2023, 9, 60–72. [Google Scholar] [CrossRef]
- Wang, R.; Shu, P.; Zhang, C.; Zhang, J.; Chen, Y.; Zhang, Y.; Du, K.; Xie, Y.; Li, M.; Ma, T. Integrative analyses of metabolome and genome-wide transcriptome reveal the regulatory network governing flavor formation in kiwifruit (Actinidia chinensis). Biol. Plant. 2022, 233, 373–389. [Google Scholar] [CrossRef]
- Kumar, S.; Sharma, D.R. Review Article In vitro propagation of kiwifruit. J. Hortic. Sci. Biotechnol. 2002, 77, 503–508. [Google Scholar]
- Feng, K.; Yu, J.; Cheng, Y.; Ruan, M.; Wang, R.; Ye, Q.; Zhou, G.; Li, Z.; Yao, Z.; Yang, Y.; et al. The SOD gene family in tomato: Identification, phylogenetic relationships, and expression patterns. Front. Plant Sci. 2016, 7, 1279. [Google Scholar] [CrossRef]
- Xu, Z.; Dai, J.; Su, W.; Wu, H.; Shah, K.; Xing, L.; Ma, J.; Zhang, D.; Zhao, C. Selection and validation of reliable reference genes for gene expression studies in different genotypes and TRV-infected fruits of peach (Prunus persica L. Batsch) during Ripening. Genes 2022, 13, 160. [Google Scholar] [CrossRef] [PubMed]
- Han, D.G.; Wang, Y.; Zhang, L.; Ma, L.; Zhang, X.Z.; Xu, X.F.; Han, Z.H. Isolation and functional characterization of MxCS1: A gene encoding a citrate synthase in Malus xiaojinensis. Biol. Plant. 2012, 56, 50–56. [Google Scholar] [CrossRef]
- Han, D.; Wang, L.; Wang, Y.; Yang, G.; Gao, C.; Yu, Z.; Li, T.; Zhang, X.; Ma, L.; Xu, X.; et al. Overexpression of Malus xiaojinensis CS1 gene in tobacco affects plant development and increases iron stress tolerance. Sci. Hortic. 2013, 150, 65–72. [Google Scholar] [CrossRef]
- Bai, C.; Wu, C.; Ma, L.; Fu, A.; Zheng, Y.; Han, J.; Li, C.; Yuan, S.; Zheng, S.; Gao, L. Transcriptomics and metabolomics analyses provide insights into postharvest ripening and senescence of tomato fruit under low temperature. Hortic. Plant J. 2023, 9, 109–121. [Google Scholar] [CrossRef]
- Xu, G.; Li, L.; Zhou, J.; Lyu, D.; Zhao, D.; Qin, S. Comparison of transcriptome and metabolome analysis revealed differences in cold resistant metabolic pathways in different apple cultivars under low temperature stress. Hortic. Plant J. 2023, 9, 183–198. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, Q.; Gichuki, D.K.; Hou, Y.; Liu, Y.; Zhou, H.; Xu, C.; Fang, L.; Gong, L.; Zheng, B. Genome-wide profiling of histone H3 lysine 27 trimethylation and its modification in response to chilling stress in grapevine leaves. Hortic. Plant J. 2023, 9, 496–508. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, A.; Yang, X.; Liu, D.; Luo, Z.; Zhang, Z.; Huo, J.; Han, D.; Zhang, Y.; Zhang, L. Genome-Wide Characterization of Kiwifruit Invertase Gene Family Reveals Roles of AcCWINV4 in Sugar Accumulation and Cold Tolerance. Int. J. Mol. Sci. 2025, 26, 10089. https://doi.org/10.3390/ijms262010089
Zhang A, Yang X, Liu D, Luo Z, Zhang Z, Huo J, Han D, Zhang Y, Zhang L. Genome-Wide Characterization of Kiwifruit Invertase Gene Family Reveals Roles of AcCWINV4 in Sugar Accumulation and Cold Tolerance. International Journal of Molecular Sciences. 2025; 26(20):10089. https://doi.org/10.3390/ijms262010089
Chicago/Turabian StyleZhang, Aoning, Xiaomeng Yang, Deshuai Liu, Zhexing Luo, Zhihao Zhang, Junwei Huo, Deguo Han, Yan Zhang, and Lihua Zhang. 2025. "Genome-Wide Characterization of Kiwifruit Invertase Gene Family Reveals Roles of AcCWINV4 in Sugar Accumulation and Cold Tolerance" International Journal of Molecular Sciences 26, no. 20: 10089. https://doi.org/10.3390/ijms262010089
APA StyleZhang, A., Yang, X., Liu, D., Luo, Z., Zhang, Z., Huo, J., Han, D., Zhang, Y., & Zhang, L. (2025). Genome-Wide Characterization of Kiwifruit Invertase Gene Family Reveals Roles of AcCWINV4 in Sugar Accumulation and Cold Tolerance. International Journal of Molecular Sciences, 26(20), 10089. https://doi.org/10.3390/ijms262010089






