Confounder-Adjusted Differentiation of Colorectal Cancer via Dynamic Propagation of Pathway Influence
Abstract
1. Introduction
2. Results
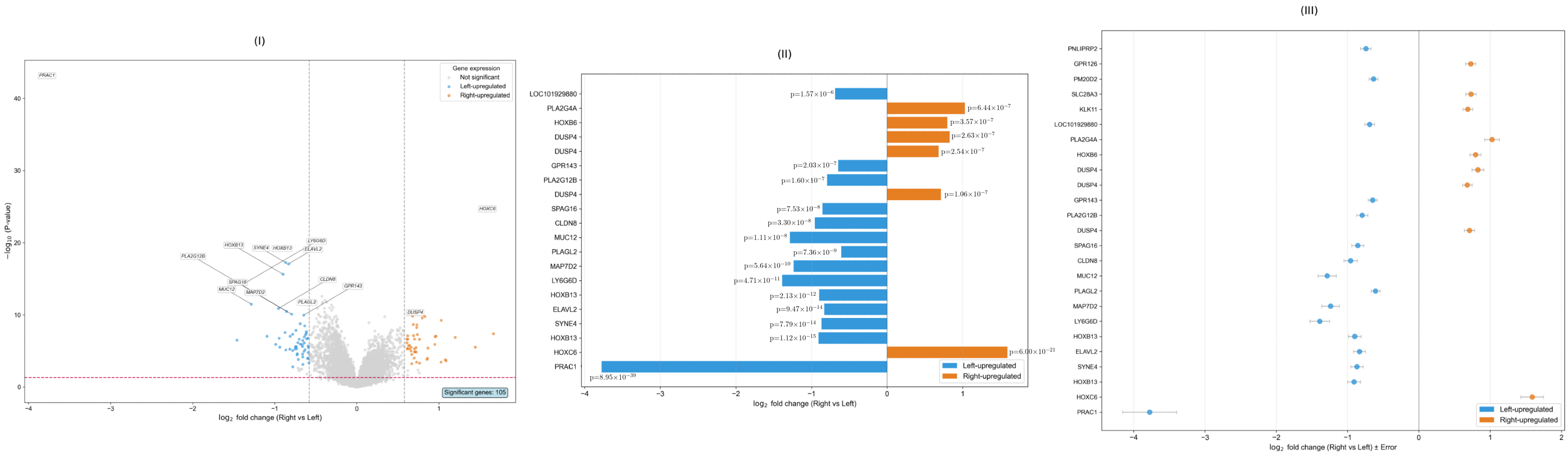
2.1. Distinct Functional Signatures in Left Versus Right-Sided CRC
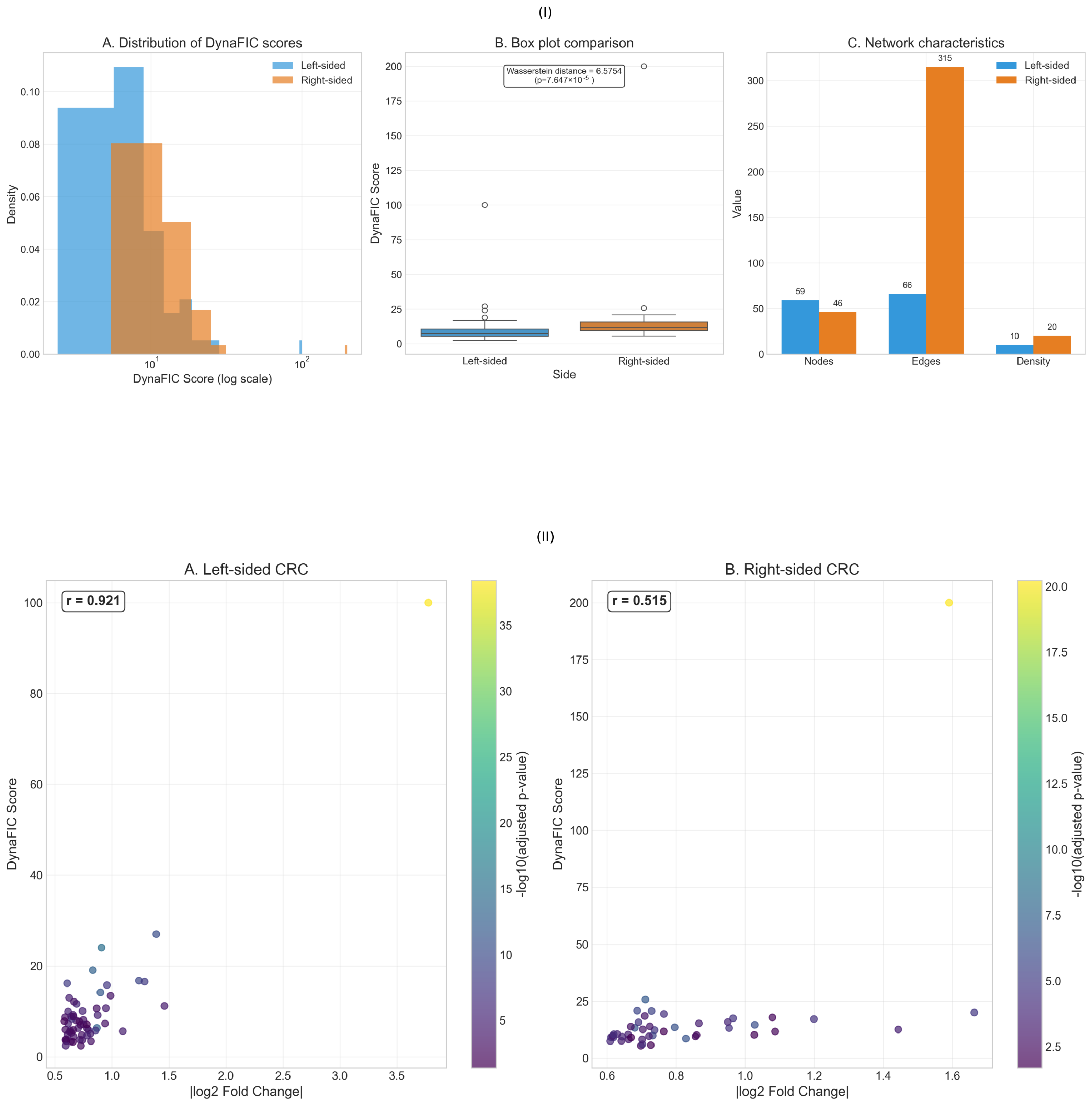
2.2. Network Influence Heterogeneity Between Tumor Sides
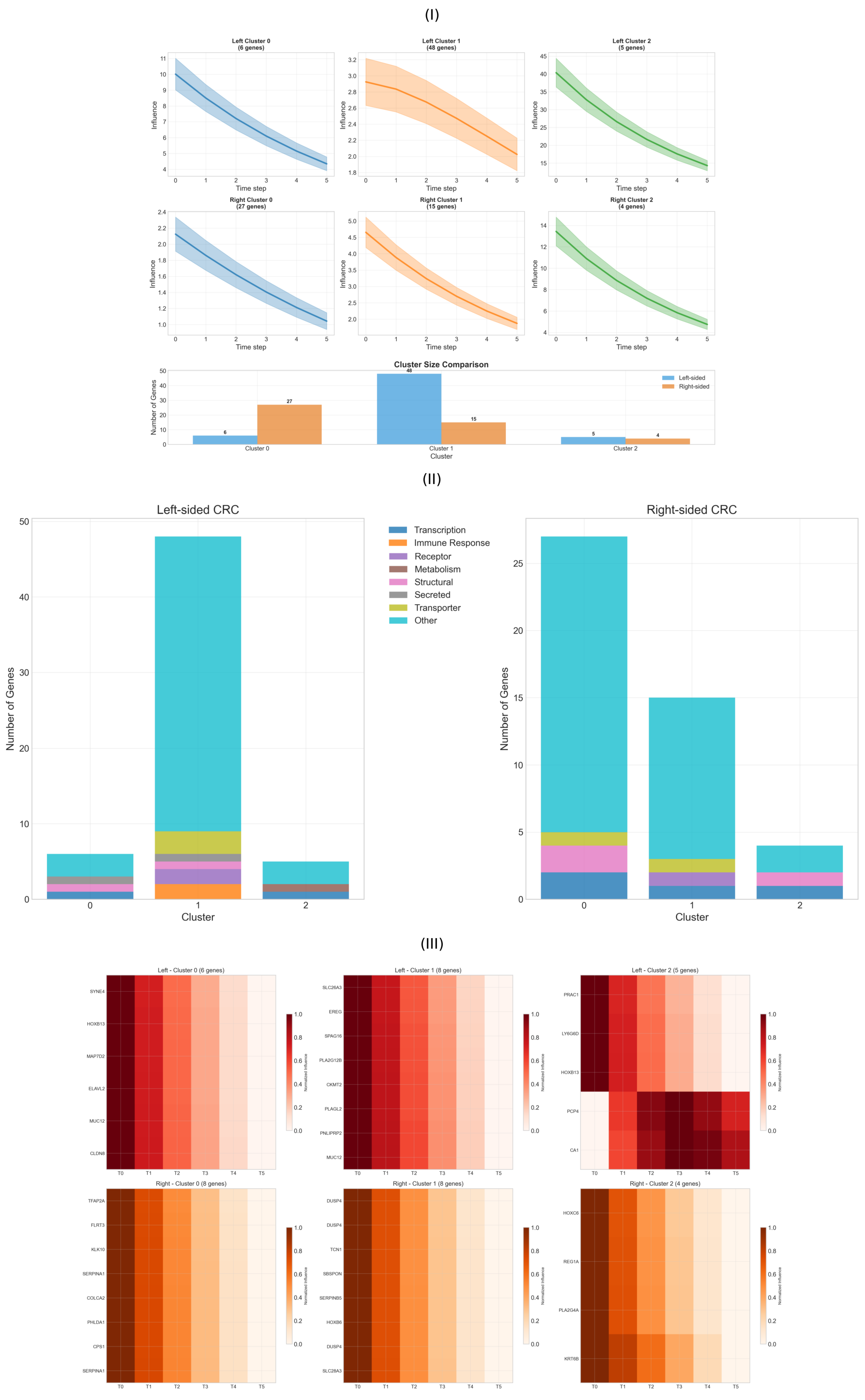
2.3. Temporal Propagation Patterns
2.4. Functional Heterogeneity Analysis
2.5. Dynamic Influence Flow and Cross-Module Communication
2.6. Multi-Scale Network Validation and Performance Assessment
3. Discussions
3.1. Network Architecture and Refined Topological Organization
3.2. Molecular Signatures and Functional Specialization
3.3. Expression-Influence Relationships and Signal Processing
3.4. Temporal Dynamics and Therapeutic Windows
3.5. Functional Heterogeneity and Microenvironmental Integration
3.6. Stability Volatility and Network Resilience
3.7. Context-Dependent Regulation and Therapeutic Resistance
3.8. Network Validation and Biological Authenticity
3.9. Translational Implications and Precision Medicine
4. Methodology
4.1. Data Acquisition and Preprocessing
4.2. DynaFIC Framework Overview
4.3. Comparative Analysis and Statistical Validation
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Complete DynaFIC Results
| Rank | Gene Symbol | DynaFIC Score | log2FC |
|---|---|---|---|
| 1 | PRAC1 | 100.0000 | −3.7759 |
| 2 | LY6G6D | 27.0247 | −1.3891 |
| 3 | HOXB13 | 24.0226 | −0.9086 |
| 4 | ELAVL2 | 19.0742 | −0.8324 |
| 5 | MAP7D2 | 16.7755 | −1.2369 |
| 6 | MUC12 | 16.5606 | −1.2863 |
| 7 | PLAGL2 | 16.1849 | −0.6073 |
| 8 | CLDN8 | 15.7750 | −0.9600 |
| 9 | HOXB13 | 14.1672 | −0.8990 |
| 10 | GPR15LG | 13.4444 | −0.9900 |
| 11 | AK4 | 12.9826 | −0.6249 |
| 12 | CYP2B6 | 12.1070 | −0.6700 |
| 13 | LOC101929880 | 11.6324 | −0.6901 |
| 14 | SLC26A3 | 11.1726 | −1.4600 |
| 15 | C10orf99 | 10.6950 | −0.9500 |
| 16 | PCK1 | 10.6500 | −0.8700 |
| 17 | PNLIPRP2 | 10.0700 | −0.7400 |
| 18 | AMACR | 9.9439 | −0.6155 |
| 19 | SATB2 | 9.2300 | −0.6600 |
| 20 | CKMT2 | 9.1700 | −0.8700 |
| 21 | QPRT | 8.8974 | −0.6418 |
| 22 | GPR143 | 8.3565 | −0.6476 |
| 23 | KIAA0226L | 7.9698 | −0.6874 |
| 24 | RUBCNL | 7.0913 | −0.7517 |
| 25 | SYNE4 | 6.3342 | −0.8680 |
| 26 | PLA2G12B | 5.8670 | −0.6262 |
| 27 | LPAR1 | 5.5446 | −0.6187 |
| 28 | OSBPL3 | 5.3892 | −0.6839 |
| 29 | BEST2 | 5.3421 | −0.7321 |
| 30 | PLAC9 | 5.1952 | −0.6420 |
| 31 | ATP2A3 | 4.9747 | −0.6078 |
| 32 | PRSS8 | 4.9652 | −0.7154 |
| 33 | MS4A12 | 4.8906 | −0.6853 |
| 34 | SLC26A2 | 4.8298 | −0.6513 |
| 35 | GPR35 | 4.7890 | −0.6011 |
| 36 | CAPN9 | 4.7105 | −0.6284 |
| 37 | CA2 | 4.6712 | −0.6405 |
| 38 | CA4 | 4.6288 | −0.6938 |
| 39 | CLCA1 | 4.5754 | −0.6724 |
| 40 | BTNL8 | 4.5032 | −0.7223 |
| 41 | ALDH1A2 | 4.4891 | −0.6115 |
| 42 | MGAM | 4.4258 | −0.6447 |
| 43 | SLC9A2 | 4.3896 | −0.6758 |
| 44 | CEACAM7 | 4.2967 | −0.6932 |
| 45 | PADI2 | 4.2045 | −0.6584 |
| 46 | SCNN1B | 4.1567 | −0.6291 |
| 47 | ANPEP | 4.0934 | −0.6178 |
| 48 | ABCG2 | 4.0423 | −0.6235 |
| 49 | EPHX2 | 3.9845 | −0.6067 |
| 50 | CYP2C18 | 3.9234 | −0.6389 |
| 51 | CDH3 | 3.8679 | −0.6521 |
| 52 | FCGBP | 3.7892 | −0.6734 |
| 53 | GUCA2B | 3.7234 | −0.6845 |
| 54 | VAV3 | 3.3435 | −0.6402 |
| 55 | ADRA2A | 3.2891 | −0.6157 |
| 56 | TSPAN8 | 3.1245 | −0.6623 |
| 57 | MYOC | 2.9876 | −0.6089 |
| 58 | GPR110 | 2.7234 | −0.6345 |
| 59 | SLC39A5 | 2.4037 | −0.6012 |
| Rank | Gene Symbol | DynaFIC Score | log2FC |
|---|---|---|---|
| 1 | HOXC6 | 200.0000 | 1.5906 |
| 2 | DUSP4 | 25.7421 | 0.7107 |
| 3 | KLK11 | 20.8406 | 0.6869 |
| 4 | GPR126 | 20.6906 | 0.7285 |
| 5 | REG1A | 20.0147 | 1.6637 |
| 6 | SBSPON | 19.4070 | 0.7600 |
| 7 | CPS1 | 18.5440 | 0.7100 |
| 8 | REG4 | 17.8925 | 1.0785 |
| 9 | SBSPON | 17.5200 | 0.9600 |
| 10 | TCN1 | 17.1700 | 1.2000 |
| 11 | L1TD1 | 15.8730 | 0.9500 |
| 12 | SCG5 | 15.7690 | 0.6900 |
| 13 | KLK10 | 15.2746 | 0.8658 |
| 14 | PLA2G4A | 14.6221 | 1.0274 |
| 15 | CPS1 | 13.9430 | 0.7200 |
| 16 | HOXA10-HOXA9 | 13.8400 | 0.6700 |
| 17 | HOXB6 | 13.5208 | 0.7956 |
| 18 | DUSP4 | 13.2994 | 0.6800 |
| 19 | SERPINB5 | 13.1948 | 0.9532 |
| 20 | CLDN2 | 12.6400 | 0.7000 |
| 21 | RAB27B | 12.3224 | 0.7372 |
| 22 | REG4 | 11.7007 | 1.0866 |
| 23 | SLC28A3 | 9.9127 | 0.7317 |
| 24 | ANO1 | 9.8775 | 0.6161 |
| 25 | TFAP2A | 9.1400 | 0.6185 |
| 26 | DUSP4 | 8.5720 | 0.8280 |
| 27 | CLDN18 | 8.0000 | 0.7500 |
| 28 | HOXB5 | 7.8900 | 0.8200 |
| 29 | HOXC10 | 7.6700 | 1.1200 |
| 30 | HOXC9 | 7.4500 | 1.0800 |
| 31 | HOXC4 | 7.2300 | 0.9500 |
| 32 | GPR137B | 7.0100 | 0.7800 |
| 33 | ZBED2 | 6.8900 | 0.8100 |
| 34 | ASCL2 | 6.7600 | 0.9300 |
| 35 | GREM1 | 6.6400 | 0.7600 |
| 36 | VSTM2A | 6.5200 | 0.8400 |
| 37 | CDX1 | 6.4000 | 0.9100 |
| 38 | AXIN2 | 6.2800 | 0.7900 |
| 39 | EPHB3 | 6.1600 | 0.8200 |
| 40 | MSX1 | 6.0400 | 0.8700 |
| 41 | BMP7 | 5.9200 | 0.7300 |
| 42 | INHBA | 5.8000 | 0.8600 |
| 43 | SFRP2 | 5.6800 | 0.7400 |
| 44 | WNT3 | 5.5600 | 0.8900 |
| 45 | CTSE | 5.4103 | 0.6975 |
| 46 | FGF20 | 5.3000 | 0.8000 |
Appendix B. Detailed Mathematical Formulations
Appendix B.1. Multi-Layer Network Construction
Appendix B.1.1. Protein–Protein Interaction Network
Appendix B.1.2. Transcriptional Regulatory Network
Appendix B.1.3. Functional Similarity Network
Appendix B.1.4. Tissue-Specific Weight Computation
Appendix B.2. Network Diffusion Dynamics
Appendix B.2.1. Initial Influence Vector
Appendix B.2.2. Weighted Adjacency Matrix
Appendix B.2.3. Regulatory Amplification Matrix
Appendix B.3. Functional Hierarchy Metrics
Appendix B.3.1. Cancer GO Enrichment Score
Appendix B.3.2. GO Depth Score
Appendix B.3.3. Context Modulation Index
Appendix B.4. DynaFIC Score Computation
Appendix B.4.1. Network Centrality
Appendix B.4.2. Final Score Integration
Appendix B.4.3. Score Normalization
Appendix B.5. Analytical Methods
Appendix B.5.1. Dynamic Signature Clustering Analysis
Appendix B.5.2. Hierarchical Disruption Testing
Appendix B.5.3. Stability Volatility Index
Appendix B.5.4. Multi-Scale Network Propagation Validation
Appendix B.5.5. Tissue-Specific Context Modulation Validation
References
- Biller, L.H.; Schrag, D. Diagnosis and treatment of metastatic colorectal cancer: A review. JAMA 2021, 325, 669–685. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Abdel Hamid, M.; Pammer, L.M.; Oberparleiter, S.; Günther, M.; Amann, A.; Gruber, R.A.; Mair, A.; Nocera, F.I.; Ormanns, S.; Zimmer, K.; et al. Multidimensional differences of right-and left-sided colorectal cancer and their impact on targeted therapies. NPJ Precis. Oncol. 2025, 9, 116. [Google Scholar] [CrossRef] [PubMed]
- Tejpar, S.; Stintzing, S.; Ciardiello, F.; Tabernero, J.; Van Cutsem, E.; Beier, F.; Esser, R.; Lenz, H.J.; Heinemann, V. Prognostic and predictive relevance of primary tumor location in patients with RAS wild-type metastatic colorectal cancer: Retrospective analyses of the CRYSTAL and FIRE-3 trials. JAMA Oncol. 2017, 3, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, J.; Muro, K.; Shitara, K.; Yamazaki, K.; Shiozawa, M.; Ohori, H.; Takashima, A.; Yokota, M.; Makiyama, A.; Akazawa, N.; et al. Panitumumab vs bevacizumab added to standard first-line chemotherapy and overall survival among patients with RAS wild-type, left-sided metastatic colorectal cancer: A randomized clinical trial. JAMA 2023, 329, 1271–1282. [Google Scholar] [CrossRef]
- Salem, M.E.; Weinberg, B.A.; Xiu, J.; El-Deiry, W.S.; Hwang, J.J.; Gatalica, Z.; Philip, P.A.; Shields, A.F.; Lenz, H.J.; Marshall, J.L. Comparative molecular analyses of left-sided colon, right-sided colon, and rectal cancers. Oncotarget 2017, 8, 86356. [Google Scholar] [CrossRef]
- Gavrić, I.; Hodžić, E.; Sarajlić, L.; Salibašić, M.; Bajramagić, S.; Dizdarević, A.; Kulović, E. Analysis of TP53, APC, KRAS, and MMR Genetic mutations in colorectal cancer: A review article. Sanamed 2024, 19, 333–341. [Google Scholar] [CrossRef]
- Oh, S.; Song, S.; Dasgupta, N.; Grabowski, G. The analytical landscape of static and temporal dynamics in transcriptome data. Front. Genet. 2014, 5, 35. [Google Scholar] [CrossRef]
- Thakur, S.; Ghosh, S.; Dasgupta, M. Implementation of WGCNA for Identifying Regulatory Modules in Biological Networks. In Next-Generation Sequencing; CRC Press: Boca Raton, FL, USA, 2025; pp. 80–97. [Google Scholar]
- Carels, N.; Sgariglia, D.; Junior, M.G.V.; Lima, C.R.; Carneiro, F.R.G.; Silva, G.F.d.; Silva, F.A.B.d.; Scardini, R.; Tuszynski, J.A.; Andrade, C.V.d.; et al. A strategy utilizing protein–protein interaction hubs for the treatment of cancer diseases. Int. J. Mol. Sci. 2023, 24, 16098. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Dao, T.K.; Pham, D.T.; Duong, T.H. Exploring the molecular terrain: A survey of analytical methods for biological network analysis. Symmetry 2024, 16, 462. [Google Scholar] [CrossRef]
- Wang, S.; Wu, R.; Lu, J.; Jiang, Y.; Huang, T.; Cai, Y.D. Protein-protein interaction networks as miners of biological discovery. Proteomics 2022, 22, 2100190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Kong, F.; Deng, D.; Tang, X.; Wu, X.; Xu, C.; Zhu, L.; Liu, J.; Ai, B.; Han, Z.; et al. Moving Target Defense Meets Artificial Intelligence-Driven Network: A Comprehensive Survey. IEEE Internet Things J. 2025, 12, 13384–13397. [Google Scholar] [CrossRef]
- Naarala, J.; Kolehmainen, M.; Juutilainen, J. Electromagnetic fields, genomic instability and cancer: A systems biological view. Genes 2019, 10, 479. [Google Scholar] [CrossRef]
- Prokop, A. Towards the first principles in biology and cancer: New vistas in computational systems biology of cancer. Life 2021, 12, 21. [Google Scholar] [CrossRef]
- Jacquemin, V.; Antoine, M.; Dom, G.; Detours, V.; Maenhaut, C.; Dumont, J.E. Dynamic cancer cell heterogeneity: Diagnostic and therapeutic implications. Cancers 2022, 14, 280. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Giuliani, A.; Yoshikawa, K.; Falk, M.; Hildenbrand, G.; Salmina, K.; Freivalds, T.; Vainshelbaum, N.; Weidner, J.; Sievers, A.; et al. Spatial-temporal genome regulation in stress-response and cell-fate change. Int. J. Mol. Sci. 2023, 24, 2658. [Google Scholar] [CrossRef]
- Li, S.; Pan, T.; Xu, G.; Gao, Y.; Zhang, Y.; Xu, Q.; Pan, J.; Zhou, W.; Xu, J.; Li, Q.; et al. Deep immunophenotyping reveals clinically distinct cellular states and ecosystems in large-scale colorectal cancer. Commun. Biol. 2023, 6, 785. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, J.; Shi, Z.; Franklin, J.L.; Deane, N.G.; Coffey, R.J.; Beauchamp, R.D.; Zhang, B. Deciphering genomic alterations in colorectal cancer through transcriptional subtype-based network analysis. PLoS ONE 2013, 8, e79282. [Google Scholar] [CrossRef]
- Yan, W.; Xue, W.; Chen, J.; Hu, G. Biological networks for cancer candidate biomarkers discovery. Cancer Inform. 2016, 15, CIN-S39458. [Google Scholar] [CrossRef] [PubMed]
- Mukund, K.; Syulyukina, N.; Ramamoorthy, S.; Subramaniam, S. Right and left-sided colon cancers-specificity of molecular mechanisms in tumorigenesis and progression. BMC Cancer 2020, 20, 317. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Chen, J.; Zhou, B.; Xu, K.; Wang, K.; Fang, Z.; Shao, Y.; Yuan, Y.; Zheng, S.; Hu, W. HomeoboxC6 promotes metastasis by orchestrating the DKK1/Wnt/β-catenin axis in right-sided colon cancer. Cell Death Dis. 2021, 12, 337. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- André, T.; Shiu, K.K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab in microsatellite-instability–high advanced colorectal cancer. N. Engl. J. Med. 2020, 383, 2207–2218. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, D.; Li, Y.; Qi, L.; Si, W.; Bo, Y.; Chen, X.; Ye, Z.; Fan, H.; Liu, B.; et al. Spatiotemporal single-cell analysis decodes cellular dynamics underlying different responses to immunotherapy in colorectal cancer. Cancer Cell 2024, 42, 1268–1285. [Google Scholar] [CrossRef]
- Jin, M.; Wu, J.; Shi, L.; Zhou, B.; Shang, F.; Chang, X.; Dong, X.; Deng, S.; Liu, L.; Cai, K.; et al. Gut microbiota distinct between colorectal cancers with deficient and proficient mismatch repair: A study of 230 CRC patients. Front. Microbiol. 2022, 13, 993285. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Guo, Y.; Liu, J.; Guo, F.; Du, L.; Yang, Y.; Li, X.; Ma, Y. Depicting the landscape of gut microbial-metabolic interaction and microbial-host immune heterogeneity in deficient and proficient DNA mismatch repair colorectal cancers. J. Immunother. Cancer 2023, 11, e007420. [Google Scholar] [CrossRef] [PubMed]
- Amodio, V.; Vitiello, P.; Bardelli, A.; Germano, G. DNA repair-dependent immunogenic liabilities in colorectal cancer: Opportunities from errors. Br. J. Cancer 2024, 131, 1576–1590. [Google Scholar] [CrossRef]
- Gharib, E.; Robichaud, G.A. From crypts to cancer: A holistic perspective on colorectal carcinogenesis and therapeutic strategies. Int. J. Mol. Sci. 2024, 25, 9463. [Google Scholar] [CrossRef]
- Kasi, P.M.; Afable, M.G.; Herting, C.; Lukanowski, M.; Jin, Z. Anti-EGFR antibodies in the management of advanced colorectal cancer. Oncologist 2023, 28, 1034–1048. [Google Scholar] [CrossRef]
- Kang, J.; Guo, Z.; Zhang, H.; Guo, R.; Zhu, X.; Guo, X. Dual inhibition of EGFR and IGF-1R signaling leads to enhanced antitumor efficacy against esophageal squamous cancer. Int. J. Mol. Sci. 2022, 23, 10382. [Google Scholar] [CrossRef] [PubMed]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017, 18, 1182–1191, Correction in Lancet Oncol. 2017, 18, 510. [Google Scholar] [CrossRef]
- Lenz, H.J.; Lonardi, S.; Elez, E.; Van Cutsem, E.; Jensen, L.H.; Bennouna, J.; Mendez, G.; Schenker, M.; De La Fouchardiere, C.; Limon, M.L.; et al. Nivolumab (NIVO) plus ipilimumab (IPI) vs chemotherapy (chemo) as first-line (1L) treatment for microsatellite instability-high/mismatch repair-deficient (MSI-H/dMMR) metastatic colorectal cancer (mCRC): Expanded efficacy analysis from CheckMate 8HW. J. Clin. Oncol. 2024, 42, 3503. [Google Scholar] [CrossRef]
- Ros, J.; Balconi, F.; Baraibar, I.; Saoudi Gonzalez, N.; Salva, F.; Tabernero, J.; Elez, E. Advances in immune checkpoint inhibitor combination strategies for microsatellite stable colorectal cancer. Front. Oncol. 2023, 13, 1112276. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Ooft, S.N.; Weeber, F.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van Werkhoven, E.; Schipper, L.; Hoes, L.; Vis, D.J.; van de Haar, J.; et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019, 11, eaay2574. [Google Scholar] [CrossRef]
- Smabers, L.P.; Wensink, E.; Verissimo, C.S.; Koedoot, E.; Pitsa, K.C.; Huismans, M.A.; Higuera Barón, C.; Doorn, M.; Valkenburg-van Iersel, L.B.; Cirkel, G.A.; et al. Organoids as a biomarker for personalized treatment in metastatic colorectal cancer: Drug screen optimization and correlation with patient response. J. Exp. Clin. Cancer Res. 2024, 43, 61. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Jiang, Y.; Zhang, L.; Li, Y.; Hu, X.; Hua, G.; Cai, S.; Mo, S.; Peng, J. Patient-derived organoids as a platform for drug screening in metastatic colorectal cancer. Front. Bioeng. Biotechnol. 2023, 11, 1190637. [Google Scholar] [CrossRef]
- Jensen, L.H.; Rogatto, S.R.; Lindebjerg, J.; Havelund, B.; Abildgaard, C.; do Canto, L.M.; Vagn-Hansen, C.; Dam, C.; Rafaelsen, S.; Hansen, T.F. Precision medicine applied to metastatic colorectal cancer using tumor-derived organoids and in-vitro sensitivity testing: A phase 2, single-center, open-label, and non-comparative study. J. Exp. Clin. Cancer Res. 2023, 42, 115. [Google Scholar] [CrossRef]
- Abdolahi, S.; Ghazvinian, Z.; Muhammadnejad, S.; Saleh, M.; Asadzadeh Aghdaei, H.; Baghaei, K. Patient-derived xenograft (PDX) models, applications and challenges in cancer research. J. Transl. Med. 2022, 20, 206. [Google Scholar] [CrossRef]
- Liu, X.; Xin, Z.; Wang, K. Patient-derived xenograft model in colorectal cancer basic and translational research. Anim. Model. Exp. Med. 2023, 6, 26–40. [Google Scholar] [CrossRef]
- Yoshida, G.J. Applications of patient-derived tumor xenograft models and tumor organoids. J. Hematol. Oncol. 2020, 13, 4. [Google Scholar] [CrossRef]
- Marisa, L.; de Reyniès, A.; Duval, A.; Selves, J.; Gaub, M.P.; Vescovo, L.; Etienne-Grimaldi, M.C.; Schiappa, R.; Guenot, D.; Ayadi, M.; et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013, 10, e1001453. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Mering, C.v.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Han, H.; Cho, J.W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, X.F.; Ou-Yang, L. Inferring cancer common and specific gene networks via multi-layer joint graphical model. Comput. Struct. Biotechnol. J. 2023, 21, 974–990. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Ding, X.; Zhao, Z. Multi-omics integration with weighted affinity and self-diffusion applied for cancer subtypes identification. J. Transl. Med. 2024, 22, 79. [Google Scholar] [CrossRef]
- Picart-Armada, S.; Barrett, S.J.; Willé, D.R.; Perera-Lluna, A.; Gutteridge, A.; Dessailly, B.H. Benchmarking network propagation methods for disease gene identification. PLoS Comput. Biol. 2019, 15, e1007276. [Google Scholar] [CrossRef]
- Visonà, G.; Bouzigon, E.; Demenais, F.; Schweikert, G. Network propagation for GWAS analysis: A practical guide to leveraging molecular networks for disease gene discovery. Brief. Bioinform. 2024, 25, bbae014. [Google Scholar] [CrossRef] [PubMed]
- Cowen, L.; Ideker, T.; Raphael, B.J.; Sharan, R. Network propagation: A universal amplifier of genetic associations. Nat. Rev. Genet. 2017, 18, 551–562. [Google Scholar] [CrossRef] [PubMed]
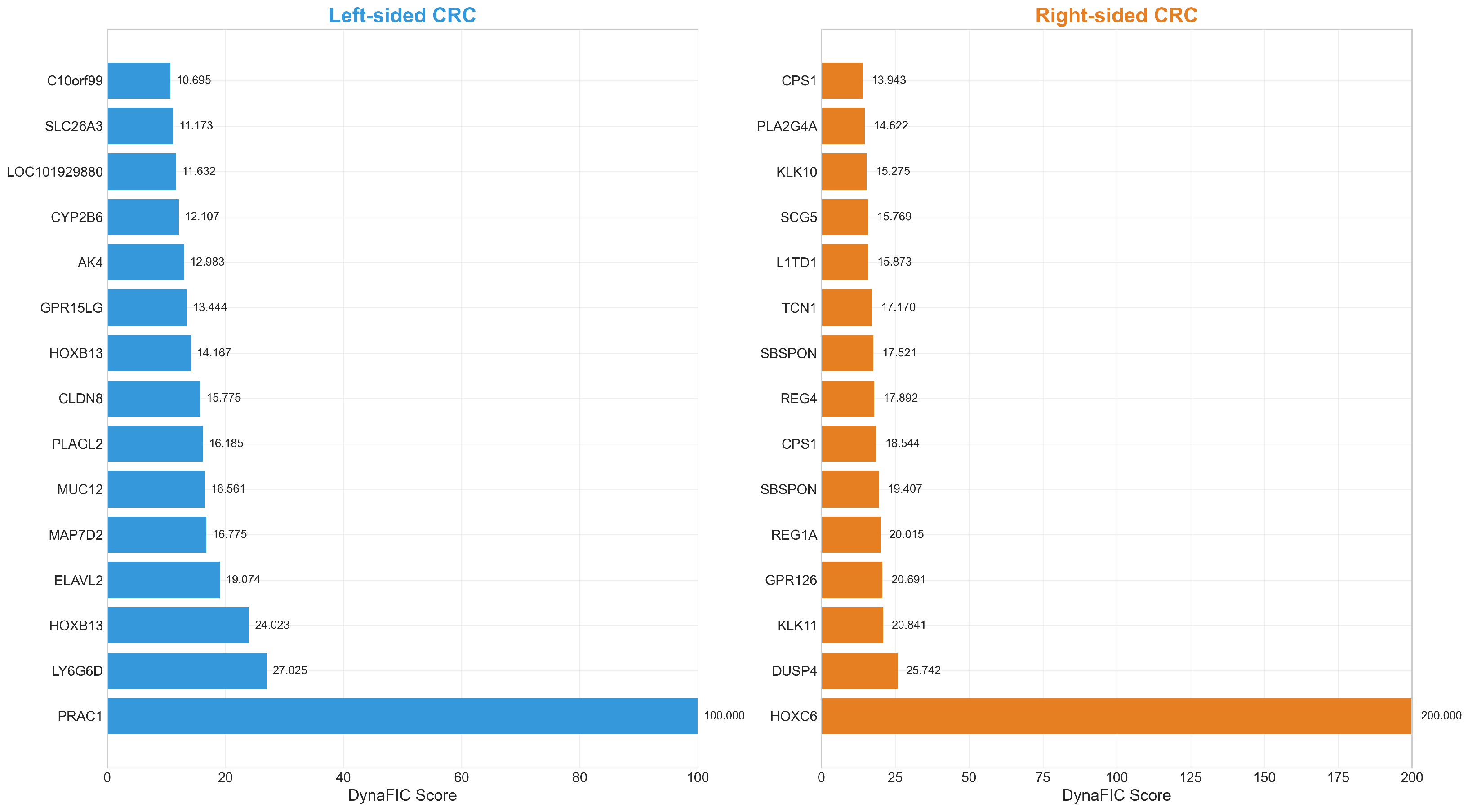



| Left-Sided Colorectal Cancer | Right-Sided Colorectal Cancer | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Rank | Gene | Score | log2FC | Rank | Gene | Score | log2FC | ||
| 1 | PRAC1 | 100.00 | −3.78 | 1 | HOXC6 | 200.00 | 1.59 | ||
| 2 | LY6G6D | 27.02 | −1.39 | 2 | DUSP4 | 25.74 | 0.71 | ||
| 3 | HOXB13 | 24.02 | −0.91 | 3 | KLK11 | 20.84 | 0.69 | ||
| 4 | ELAVL2 | 19.07 | −0.83 | 4 | GPR126 | 20.69 | 0.73 | ||
| 5 | MAP7D2 | 16.78 | −1.24 | 5 | REG1A | 20.01 | 1.66 | ||
| 6 | MUC12 | 16.56 | −1.29 | 6 | SBSPON | 19.41 | 0.76 | ||
| 7 | PLAGL2 | 16.18 | −0.61 | 7 | CPS1 | 18.54 | 0.71 | ||
| 8 | CLDN8 | 15.77 | −0.96 | 8 | REG4 | 17.89 | 1.08 | ||
| 9 | HOXB13 | 14.17 | −0.90 | 9 | SBSPON | 17.52 | 0.96 | ||
| 10 | GPR15LG | 13.44 | −0.99 | 10 | TCN1 | 17.17 | 1.20 | ||
| 11 | AK4 | 12.98 | −0.62 | 11 | L1TD1 | 15.87 | 0.95 | ||
| 12 | CYP2B6 | 12.11 | −0.67 | 12 | SCG5 | 15.77 | 0.69 | ||
| 13 | LOC101929880 | 11.63 | −0.69 | 13 | KLK10 | 15.27 | 0.87 | ||
| 14 | SLC26A3 | 11.17 | −1.46 | 14 | PLA2G4A | 14.62 | 1.03 | ||
| 15 | C10orf99 | 10.69 | −0.95 | 15 | CPS1 | 13.94 | 0.72 | ||
| 16 | PCK1 | 10.65 | −0.87 | 16 | HOXA10-HOXA9 | 13.84 | 0.67 | ||
| 17 | PNLIPRP2 | 10.07 | −0.74 | 17 | HOXB6 | 13.52 | 0.80 | ||
| 18 | AMACR | 9.94 | −0.62 | 18 | DUSP4 | 13.30 | 0.68 | ||
| 19 | SATB2 | 9.23 | −0.66 | 19 | SERPINB5 | 13.19 | 0.95 | ||
| 20 | CKMT2 | 9.170 | −0.87 | 20 | CLDN2 | 12.64 | 0.70 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batrancea, L.M.; Akgüller, Ö.; Balcı, M.A.; Çalıbaşı Koçal, G.; Gaban, L. Confounder-Adjusted Differentiation of Colorectal Cancer via Dynamic Propagation of Pathway Influence. Int. J. Mol. Sci. 2025, 26, 10023. https://doi.org/10.3390/ijms262010023
Batrancea LM, Akgüller Ö, Balcı MA, Çalıbaşı Koçal G, Gaban L. Confounder-Adjusted Differentiation of Colorectal Cancer via Dynamic Propagation of Pathway Influence. International Journal of Molecular Sciences. 2025; 26(20):10023. https://doi.org/10.3390/ijms262010023
Chicago/Turabian StyleBatrancea, Larissa Margareta, Ömer Akgüller, Mehmet Ali Balcı, Gizem Çalıbaşı Koçal, and Lucian Gaban. 2025. "Confounder-Adjusted Differentiation of Colorectal Cancer via Dynamic Propagation of Pathway Influence" International Journal of Molecular Sciences 26, no. 20: 10023. https://doi.org/10.3390/ijms262010023
APA StyleBatrancea, L. M., Akgüller, Ö., Balcı, M. A., Çalıbaşı Koçal, G., & Gaban, L. (2025). Confounder-Adjusted Differentiation of Colorectal Cancer via Dynamic Propagation of Pathway Influence. International Journal of Molecular Sciences, 26(20), 10023. https://doi.org/10.3390/ijms262010023








