Integrative Single-Cell and Bulk RNA Sequencing Identifies a Macrophage-Related Prognostic Signature for Predicting Prognosis and Therapy Responses in Colorectal Cancer
Abstract
1. Introduction
2. Results
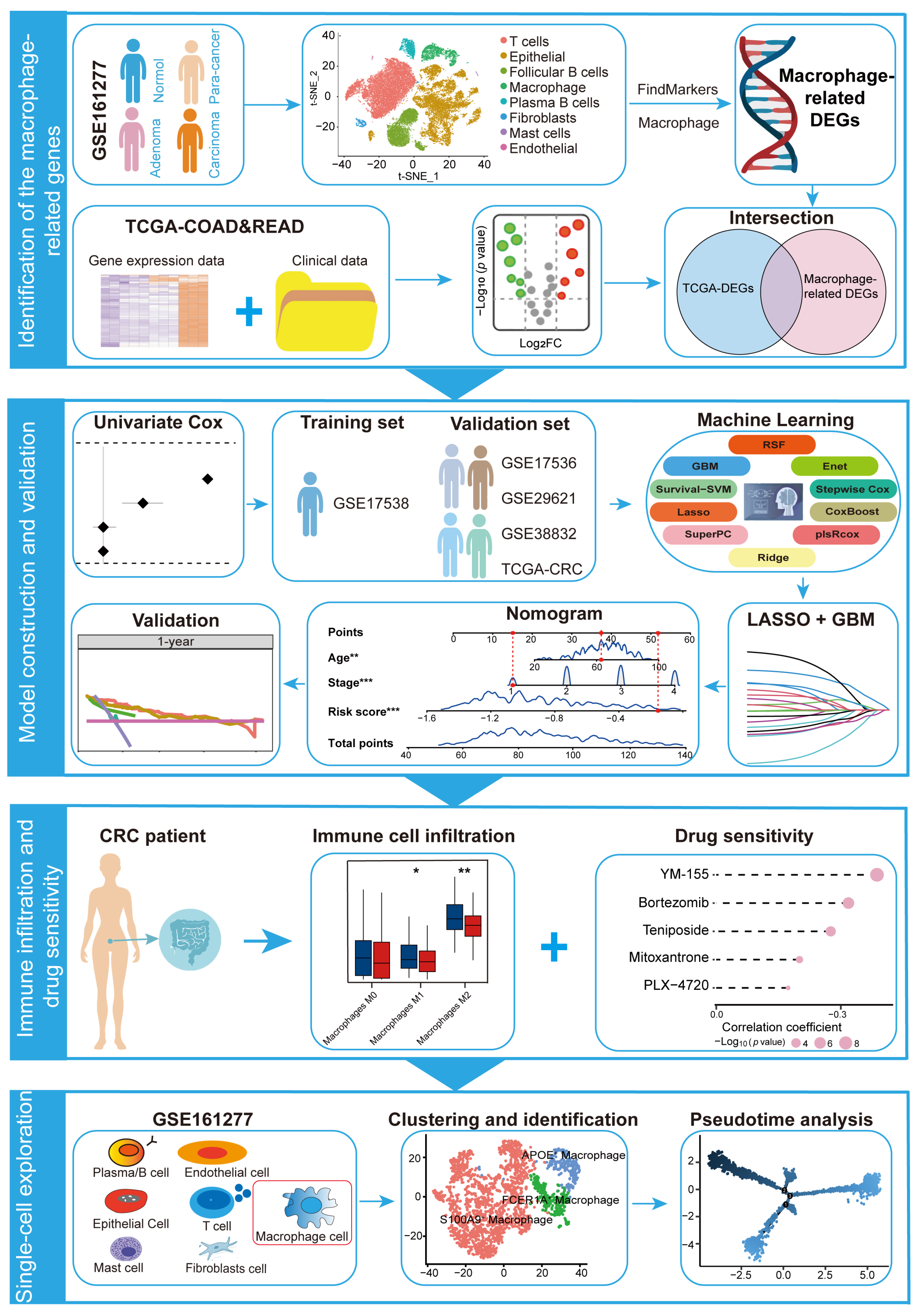
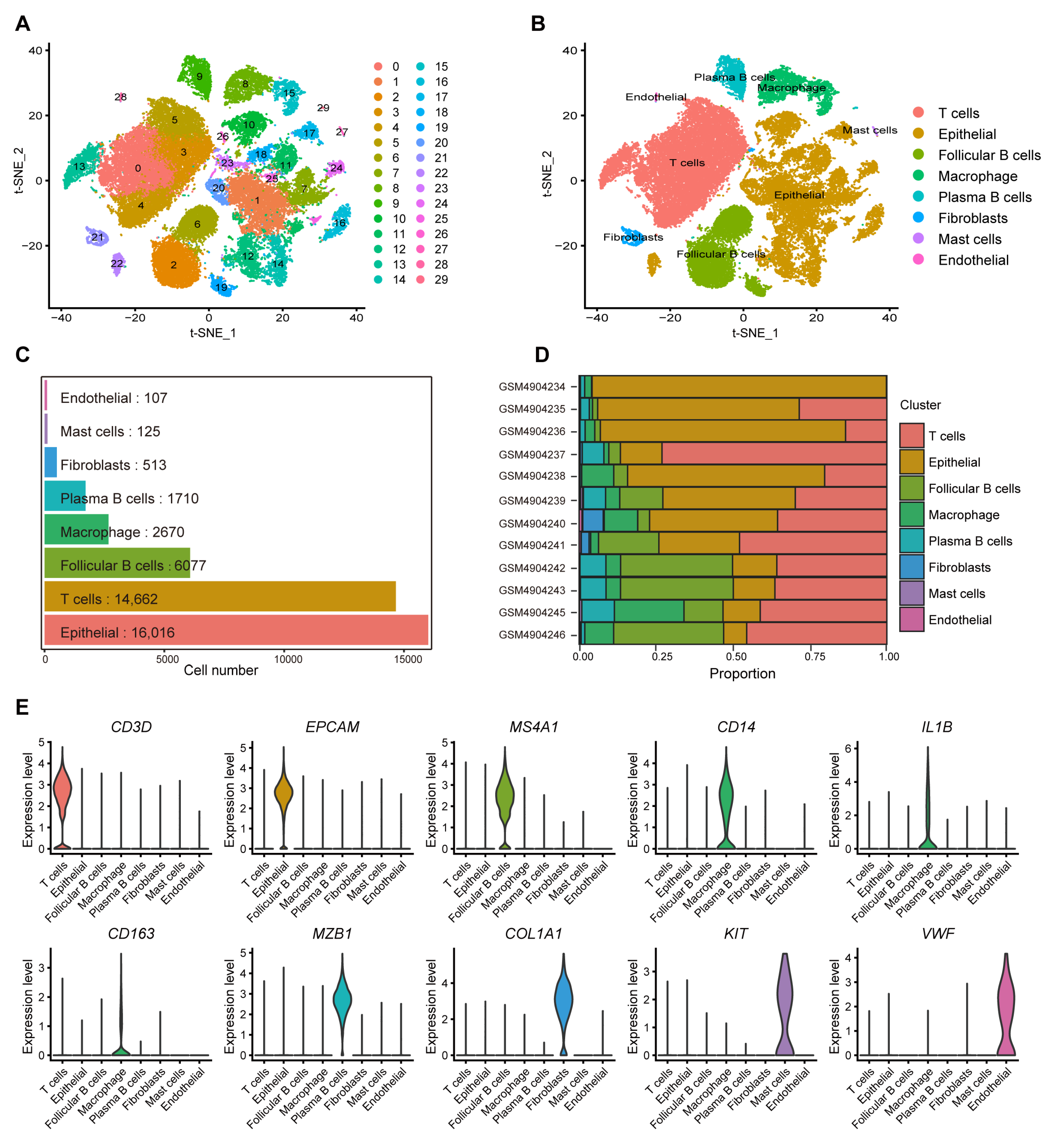
2.1. Identification of Macrophage-Related Genes in CRC Through scRNA-Seq Analysis
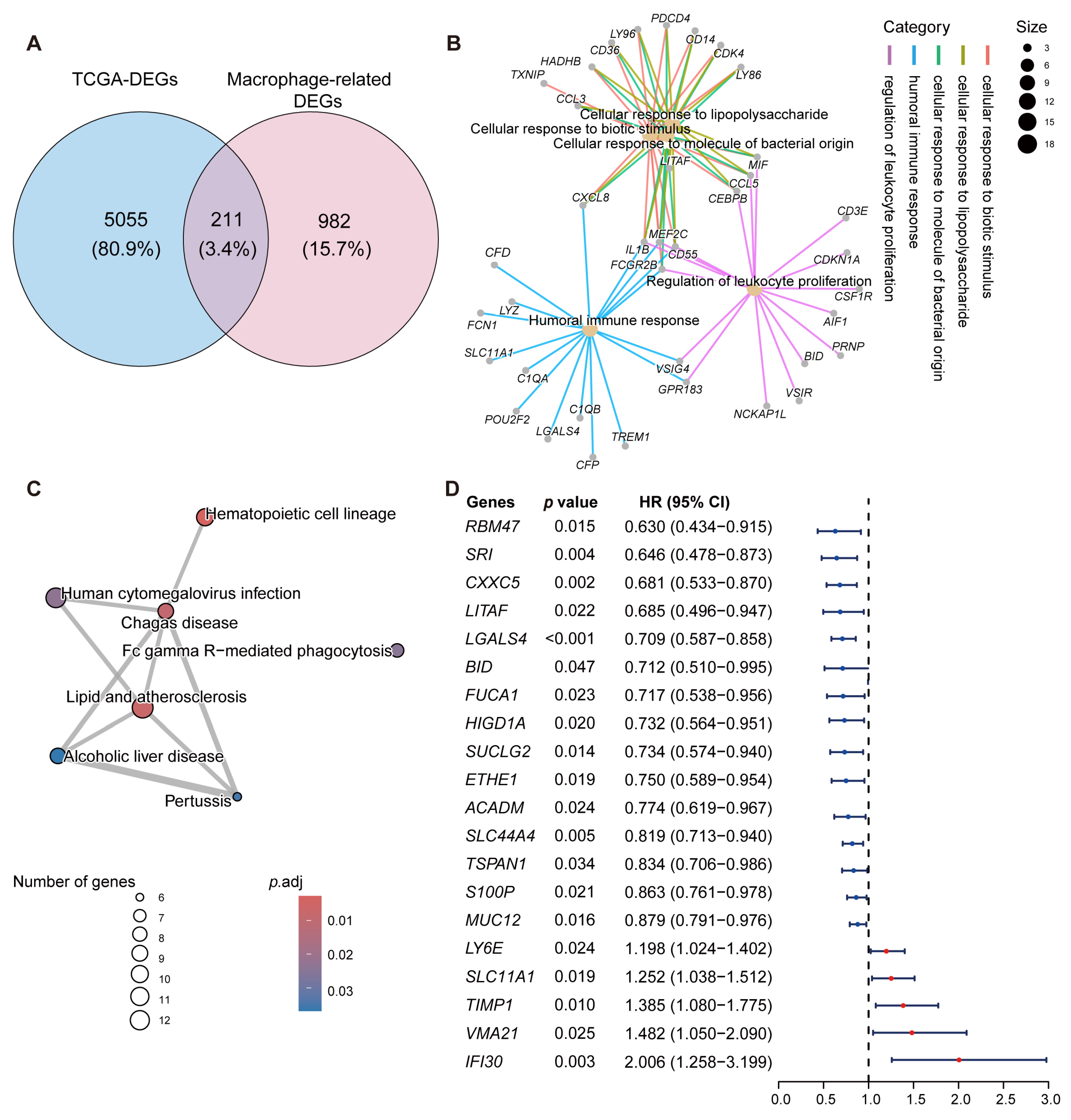
2.2. Identification of Macrophage-Related Genes with Prognostic Significance
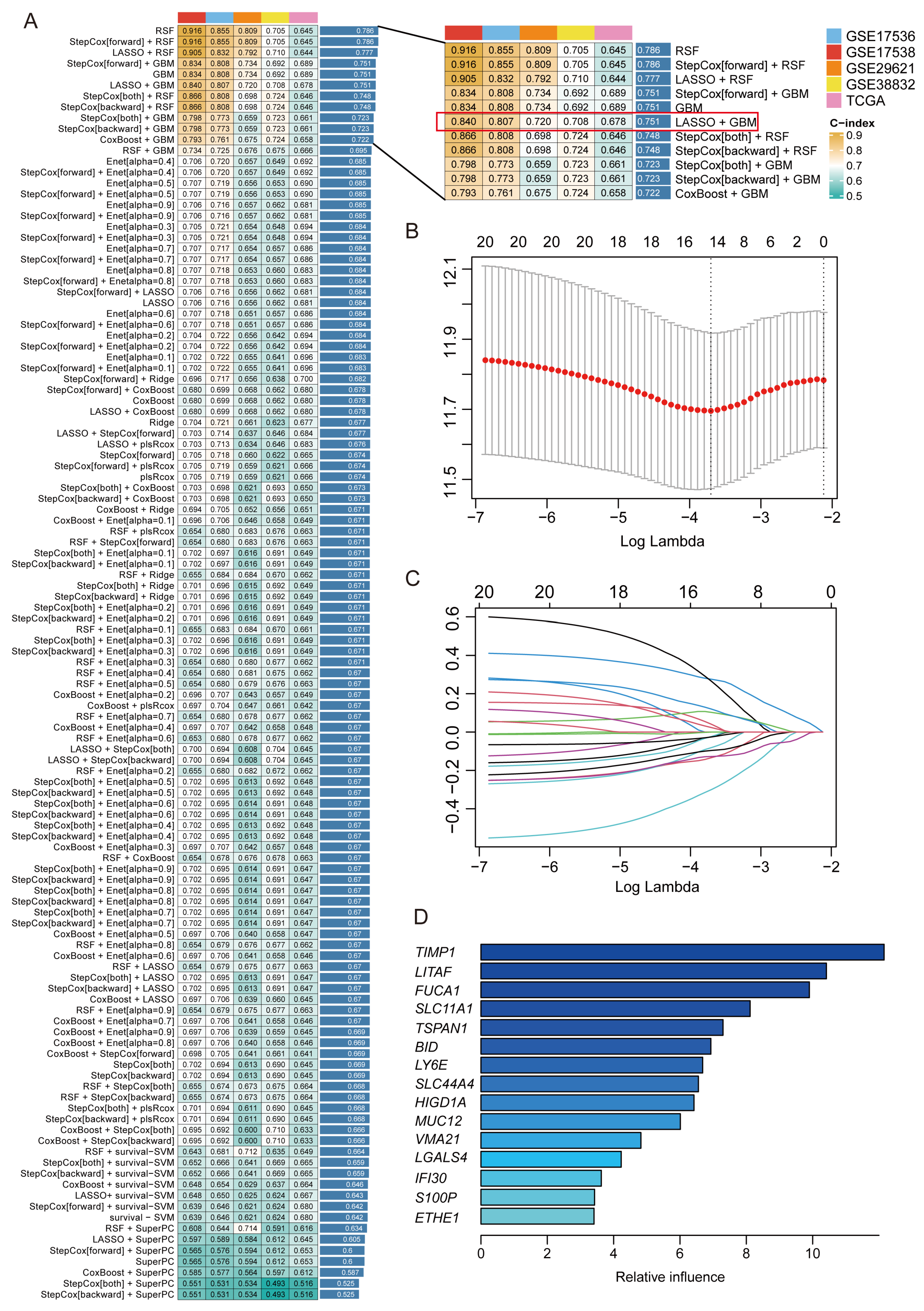
2.3. Construction of 15-Gene MRPS Based on Machine Learning
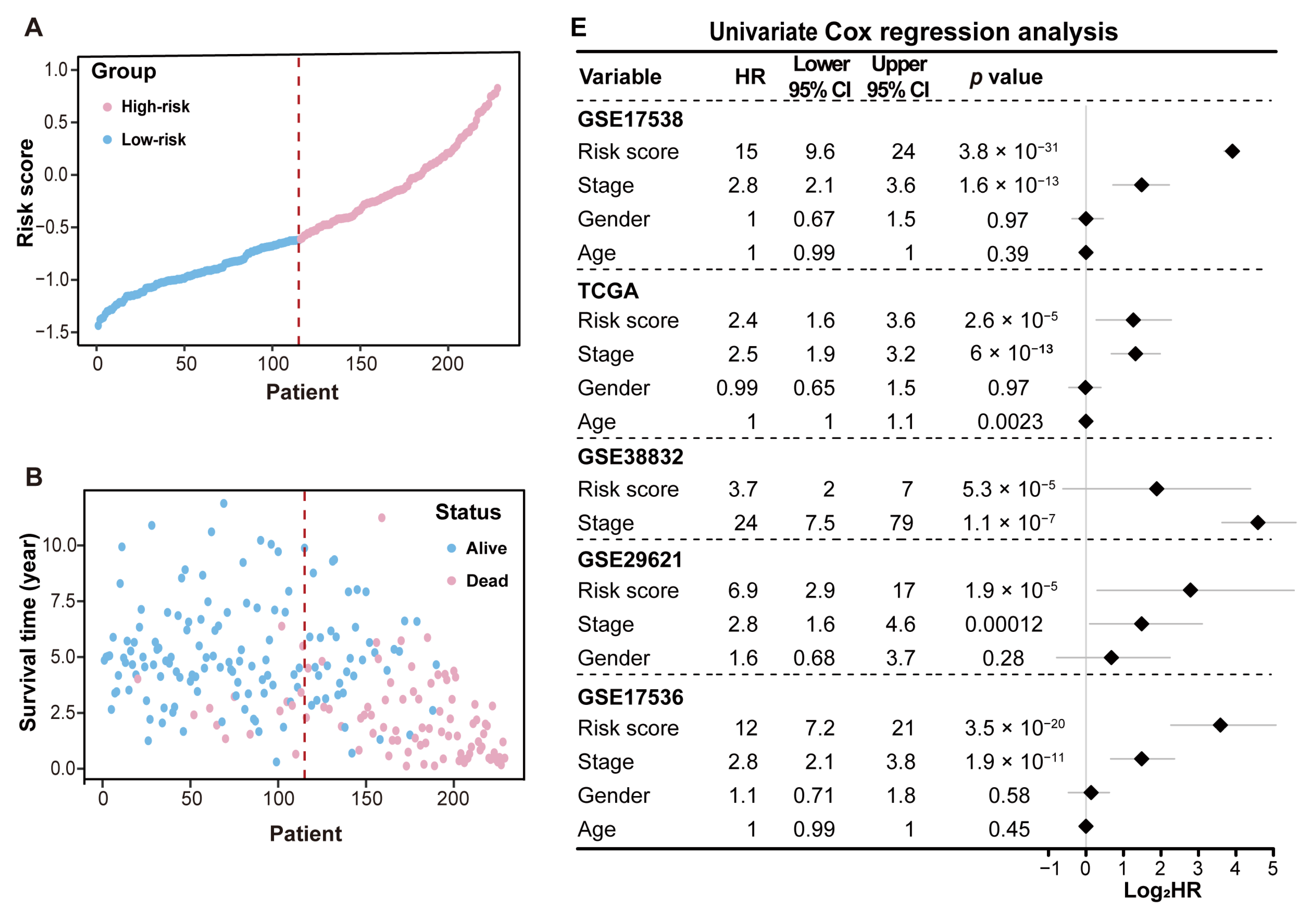
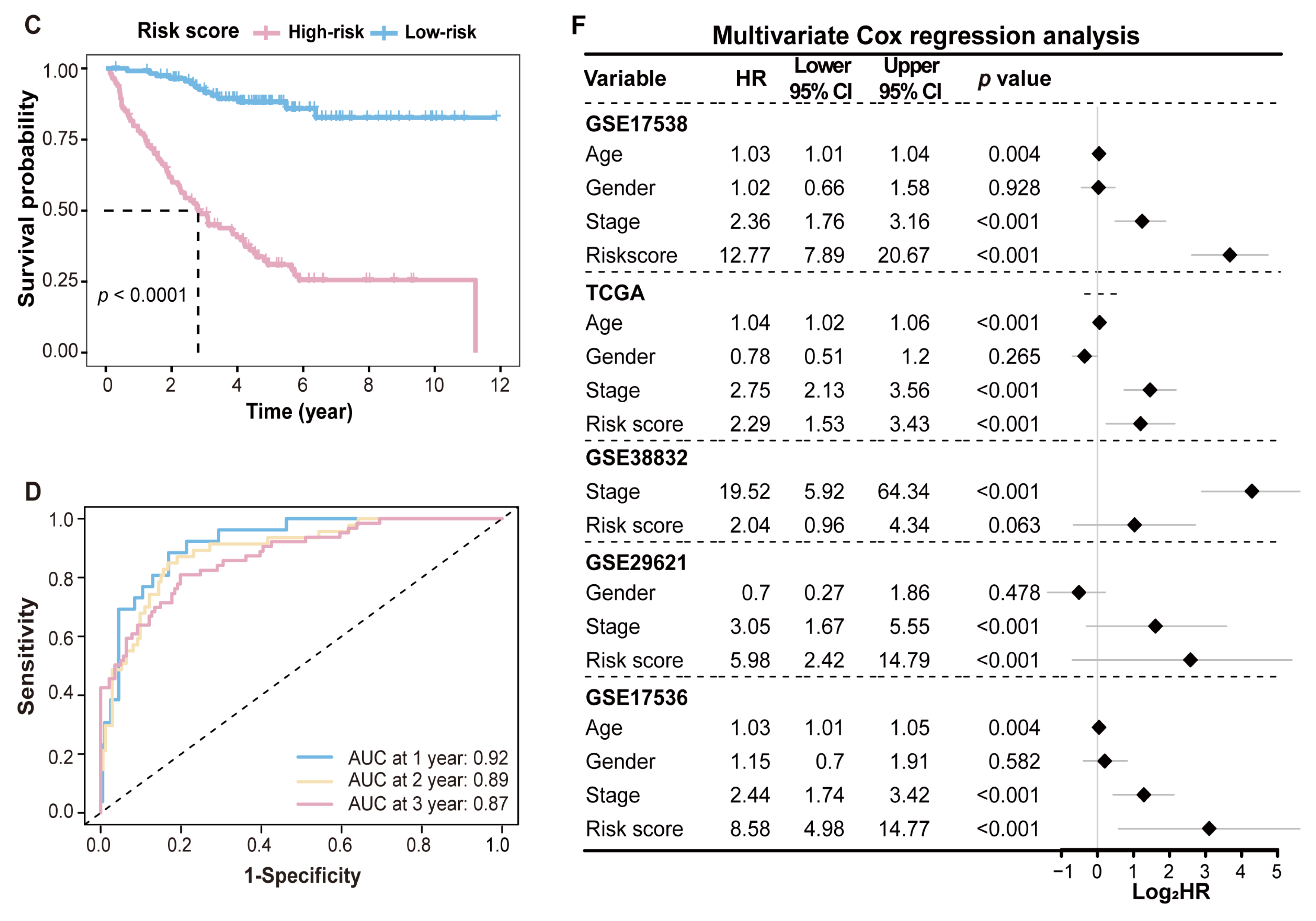
2.4. Performance Evaluation and Validation of MRPS in CRC
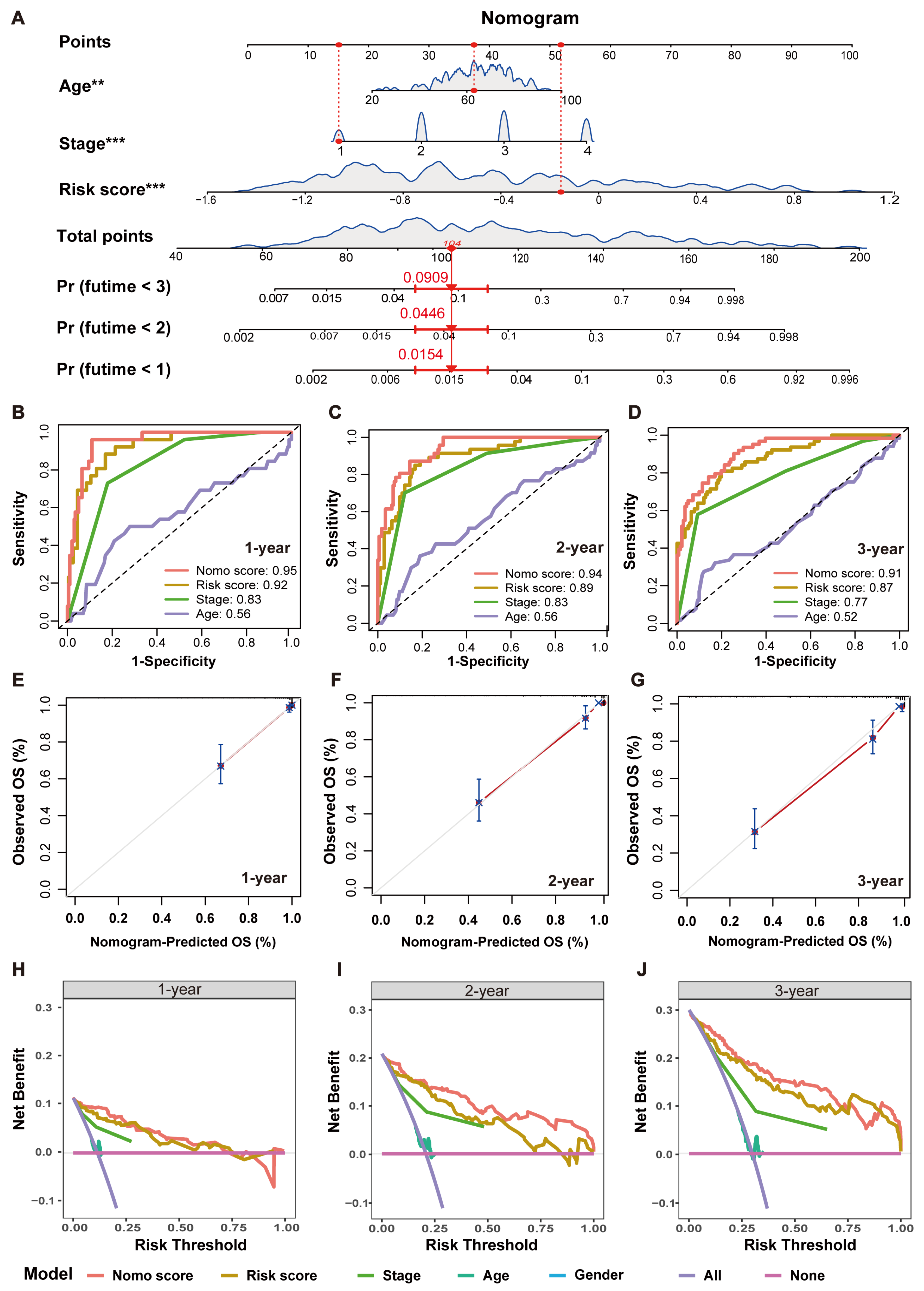
2.5. Establishment and Validation of an MRPS-Based Nomogram for CRC
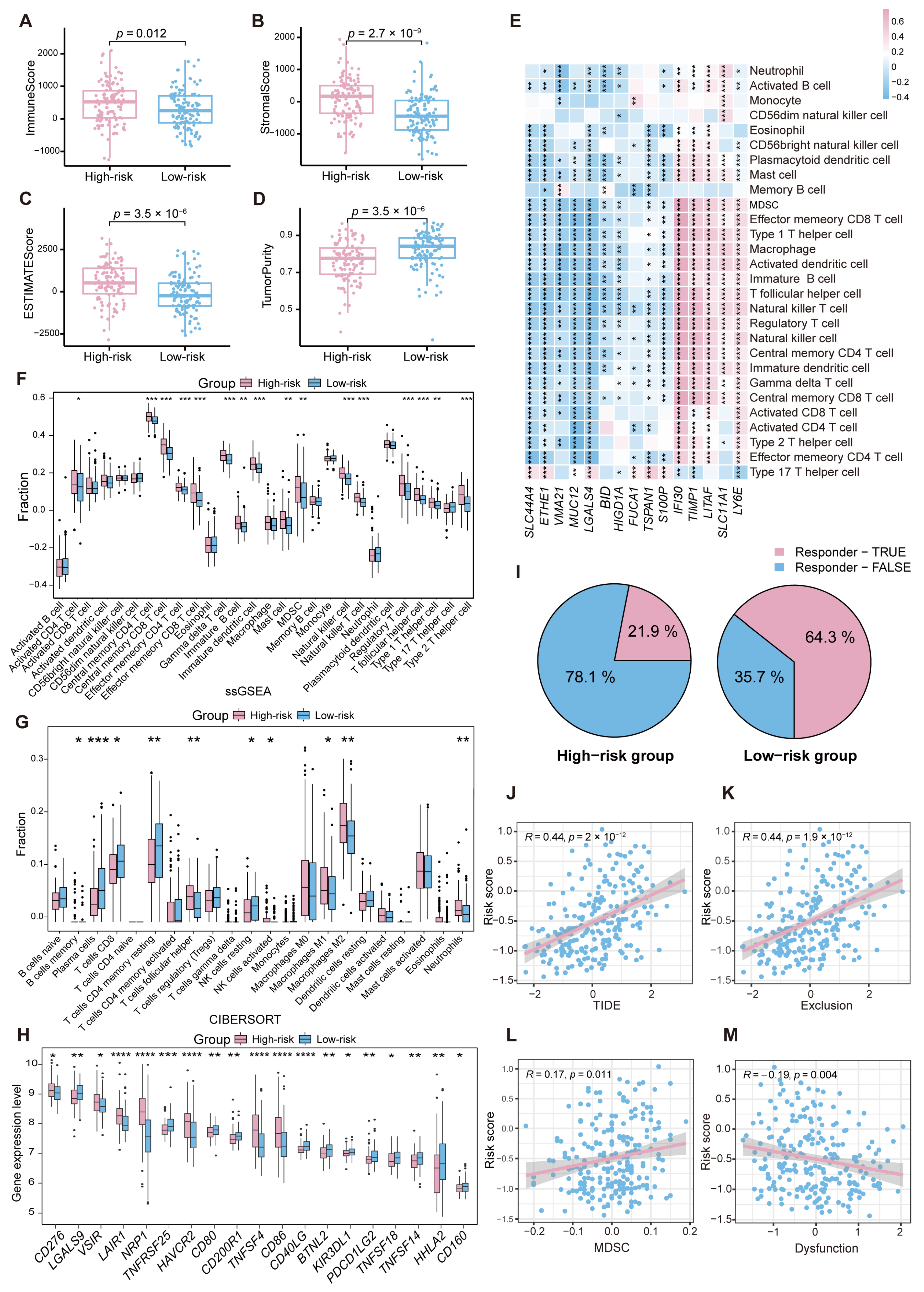
2.6. Immune Landscape Variations Between High- and Low-Risk Groups in CRC
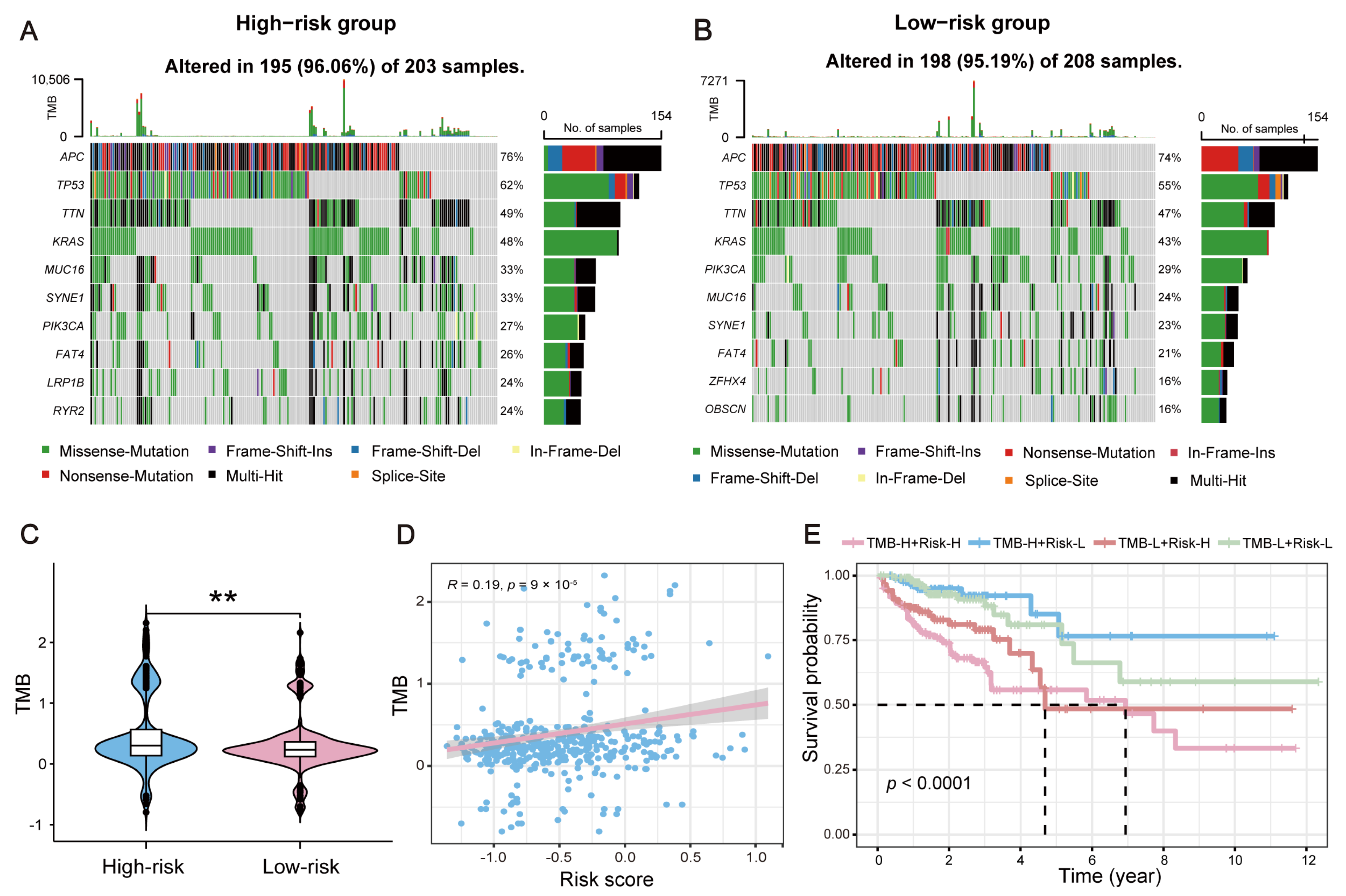
2.7. Impact of TMB on the Survival of CRC Patients with Distinct Risks
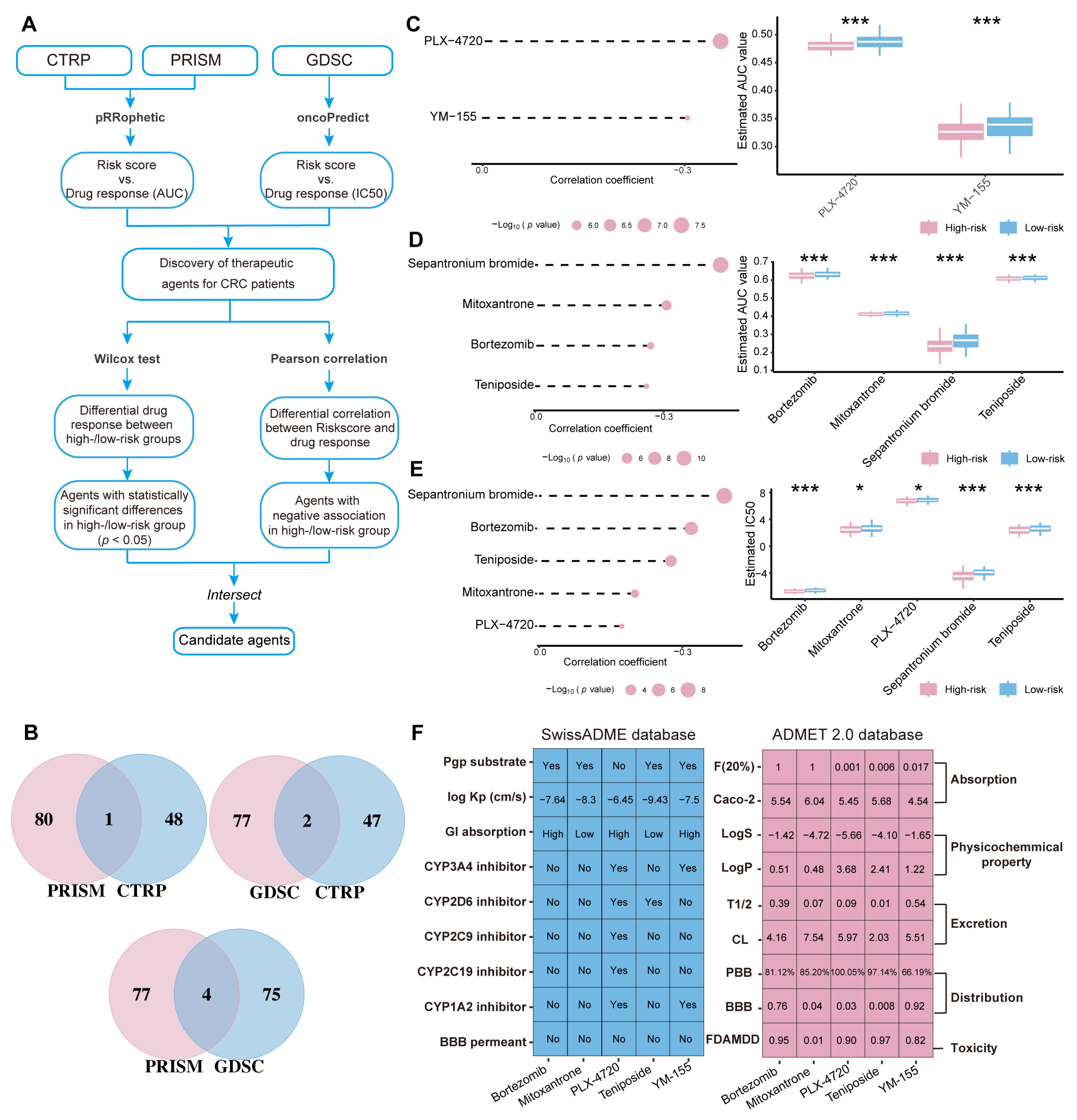
2.8. Screening of Potential Drugs for CRC Patients
2.9. Exploring the Role of MRPS-Related Genes in Macrophage Development via Pseudotime Trajectory Analysis
3. Discussion
4. Materials and Methods
4.1. Data Collection and Processing
4.2. Single-Cell RNA-Seq Analysis
4.3. Construction of a Prognostic Signature Through an Integrative Machine Learning Framework
4.4. Functional Enrichment Analysis
4.5. Predictive Performance Evaluation of the MRPS
4.6. Independence Assessment of the MRPS Ability
4.7. Performance Comparison of the MRPS with Established Models in CRC
4.8. Construction and Validation of an MRPS-Based Nomogram for CRC
4.9. Comprehensive Analysis of Immune Cell Infiltration in High- and Low-Risk Groups
4.10. Correlation Analysis of the Prognostic Model and Response to Immune Checkpoint Inhibitor Therapy
4.11. Screening Potential Drugs for CRC Treatment Based on the GDSC, CTRP, and PRISM Databases
4.12. Analysis of Tumor Mutation Burden Across Different Risk Groups
4.13. Pseudo-Temporal Analysis of Macrophage Subpopulations in CRC
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cao, J.H.; Cao, C.H.; Lin, J.L.; Li, S.Y.; He, L.J.; Han, K.; Chen, J.W.; Li, S.; Wang, X.; Xie, D.; et al. NEIL1 drives the initiation of colorectal cancer through transcriptional regulation of COL17A1. Cell Rep. 2024, 43, 113654. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- White, M.T.; Sears, C.L. The microbial landscape of colorectal cancer. Nat. Rev. Microbiol. 2024, 22, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Desai, S.A.; Patel, V.P.; Bhosle, K.P.; Nagare, S.D.; Thombare, K.C. The tumor microenvironment: Shaping cancer progression and treatment response. J. Chemother. 2024, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Zhu, W.; Feng, S.; Yan, P.; Qin, S. The role of cancer-associated fibroblasts in the invasion and metastasis of colorectal cancer. Front. Cell Dev. Biol. 2024, 12, 1375543. [Google Scholar] [CrossRef]
- Gao, J.; Liang, Y.; Wang, L. Shaping Polarization Of Tumor-Associated Macrophages In Cancer Immunotherapy. Front. Immunol. 2022, 13, 888713. [Google Scholar] [CrossRef]
- de Visser, K.E.; Joyce, J.A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef]
- Jing, L.; An, Y.; Cai, T.; Xiang, J.; Li, B.; Guo, J.; Ma, X.; Wei, L.; Tian, Y.; Cheng, X.; et al. A subpopulation of CD146(+) macrophages enhances antitumor immunity by activating the NLRP3 inflammasome. Cell. Mol. Immunol. 2023, 20, 908–923. [Google Scholar] [CrossRef]
- Popena, I.; Abols, A.; Saulite, L.; Pleiko, K.; Zandberga, E.; Jekabsons, K.; Endzelins, E.; Llorente, A.; Line, A.; Riekstina, U. Effect of colorectal cancer-derived extracellular vesicles on the immunophenotype and cytokine secretion profile of monocytes and macrophages. Cell Commun. Signal. 2018, 16, 17. [Google Scholar] [CrossRef]
- Mola, S.; Pandolfo, C.; Sica, A.; Porta, C. The Macrophages-Microbiota Interplay in Colorectal Cancer (CRC)-Related Inflammation: Prognostic and Therapeutic Significance. Int. J. Mol. Sci. 2020, 21, 6866. [Google Scholar] [CrossRef]
- Papalexi, E.; Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018, 18, 35–45. [Google Scholar] [CrossRef]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef]
- Solinas, G.; Germano, G.; Mantovani, A.; Allavena, P. Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J. Leukoc. Biol. 2009, 86, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.S.; Wahyuningtyas, R.; Aui, S.P.; Chang, K.T. Autocrine VEGF signalling on M2 macrophages regulates PD-L1 expression for immunomodulation of T cells. J. Cell. Mol. Med. 2019, 23, 1257–1267. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wei, F.; Wang, J.; Yu, W.; Shen, M.; Liu, T.; Zhang, D.; Wang, Y.; Ren, X.; Sun, Q. Myeloid-derived suppressor cells regulate the immunosuppressive functions of PD-1(-)PD-L1(+) Bregs through PD-L1/PI3K/AKT/NF-kappaB axis in breast cancer. Cell Death Dis. 2021, 12, 465. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Sharabi, A. Targeting myeloid-derived suppressor cells to enhance natural killer cell-based immunotherapy. Pharmacol. Ther. 2022, 235, 108114. [Google Scholar] [CrossRef] [PubMed]
- Bos, R.; Sherman, L.A. CD4+ T-cell help in the tumor milieu is required for recruitment and cytolytic function of CD8+ T lymphocytes. Cancer Res. 2010, 70, 8368–8377. [Google Scholar] [CrossRef]
- Zander, R.; Schauder, D.; Xin, G.; Nguyen, C.; Wu, X.; Zajac, A.; Cui, W. CD4(+) T Cell Help Is Required for the Formation of a Cytolytic CD8(+) T Cell Subset that Protects against Chronic Infection and Cancer. Immunity 2019, 51, 1028–1042.e1024. [Google Scholar] [CrossRef] [PubMed]
- Seung, E.; Xing, Z.; Wu, L.; Rao, E.; Cortez-Retamozo, V.; Ospina, B.; Chen, L.; Beil, C.; Song, Z.; Zhang, B.; et al. A trispecific antibody targeting HER2 and T cells inhibits breast cancer growth via CD4 cells. Nature 2022, 603, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Bruno, T.C.; Ebner, P.J.; Moore, B.L.; Squalls, O.G.; Waugh, K.A.; Eruslanov, E.B.; Singhal, S.; Mitchell, J.D.; Franklin, W.A.; Merrick, D.T.; et al. Antigen-Presenting Intratumoral B Cells Affect CD4(+) TIL Phenotypes in Non-Small Cell Lung Cancer Patients. Cancer Immunol. Res. 2017, 5, 898–907. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Mao, J.; Leng, X.; Zhu, L.; Rui, X.; Jin, Z.; Jiang, H.; Liu, H.; Zhang, F.; Bi, X.; et al. Co-delivery of proanthocyanidin and mitoxantrone induces synergistic immunogenic cell death to potentiate cancer immunotherapy. Biomater. Sci. 2022, 10, 4549–4560. [Google Scholar] [CrossRef]
- Hong, Y.S.; Hong, S.W.; Kim, S.M.; Jin, D.H.; Shin, J.S.; Yoon, D.H.; Kim, K.P.; Lee, J.L.; Heo, D.S.; Lee, J.S.; et al. Bortezomib induces G2-M arrest in human colon cancer cells through ROS-inducible phosphorylation of ATM-CHK1. Int. J. Oncol. 2012, 41, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Liao, A.; Chen, J.; Wang, L.; Yang, L.; Li, Y.; Zou, Y.; Shi, J.; Yu, S.; Yu, Z.; Guo, J. cGAS-STING activation by nanodelivery of teniposide achieves colorectal cancer chemo-immunotherapy. Eur. Polym. J. 2024, 219, 113379. [Google Scholar] [CrossRef]
- Zhan, T.; Faehling, V.; Rauscher, B.; Betge, J.; Ebert, M.P.; Boutros, M. Multi-omics integration identifies a selective vulnerability of colorectal cancer subtypes to YM155. Int. J. Cancer 2021, 148, 1948–1963. [Google Scholar] [CrossRef] [PubMed]
- Koumaki, K.; Kontogianni, G.; Kosmidou, V.; Pahitsa, F.; Kritsi, E.; Zervou, M.; Chatziioannou, A.; Souliotis, V.L.; Papadodima, O.; Pintzas, A. BRAF paradox breakers PLX8394, PLX7904 are more effective against BRAFV600Epsilon CRC cells compared with the BRAF inhibitor PLX4720 and shown by detailed pathway analysis. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166061. [Google Scholar] [CrossRef] [PubMed]
- Ji, T.; Fu, H.; Wang, L.; Chen, J.; Tian, S.; Wang, G.; Wang, L.; Wang, Z. Single-cell RNA profiling reveals classification and characteristics of mononuclear phagocytes in colorectal cancer. PLoS Genet. 2024, 20, e1011176. [Google Scholar] [CrossRef]
- Bao, X.; Li, Q.; Chen, D.; Dai, X.; Liu, C.; Tian, W.; Zhang, H.; Jin, Y.; Wang, Y.; Cheng, J.; et al. A multiomics analysis-assisted deep learning model identifies a macrophage-oriented module as a potential therapeutic target in colorectal cancer. Cell Rep. Med. 2024, 5, 101399. [Google Scholar] [CrossRef] [PubMed]
- Wyllie, S.; Seu, P.; Goss, J.A. The natural resistance-associated macrophage protein 1 Slc11a1 (formerly Nramp1) and iron metabolism in macrophages. Microbes Infect. 2002, 4, 351–359. [Google Scholar] [CrossRef]
- Xu, L.; Li, Z.; Song, S.; Chen, Q.; Mo, L.; Wang, C.; Fan, W.; Yan, Y.; Tong, X.; Yan, H. Downregulation of alpha-l-fucosidase 1 suppresses glioma progression by enhancing autophagy and inhibiting macrophage infiltration. Cancer Sci. 2020, 111, 2284–2296. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Guo, Q.; Feng, Y.; Cheng, P.; Wu, A. Dual role of fucosidase in cancers and its clinical potential. J. Cancer 2022, 13, 3121–3132. [Google Scholar] [CrossRef]
- Li, J.; Yang, J.; Xing, R.; Wang, Y. A novel inflammation-related signature for predicting prognosis and characterizing the tumor microenvironment in colorectal cancer. Aging 2023, 15, 2554–2581. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Song, J.; Yu, C.; Zhou, Z.; Liu, X.; Yu, J.; Xu, G.; Yang, J.; He, X.; Bai, X.; et al. Single-cell transcriptomic profiling unravels the adenoma-initiation role of protein tyrosine kinases during colorectal tumorigenesis. Signal Transduct. Target. Ther. 2022, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e3529. [Google Scholar] [CrossRef]
- Luecken, M.D.; Theis, F.J. Current best practices in single-cell RNA-seq analysis: A tutorial. Mol. Syst. Biol. 2019, 15, e8746. [Google Scholar] [CrossRef]
- Huang, A.; Sun, Z.; Hong, H.; Yang, Y.; Chen, J.; Gao, Z.; Gu, J. Novel hypoxia- and lactate metabolism-related molecular subtyping and prognostic signature for colorectal cancer. J. Transl. Med. 2024, 22, 587. [Google Scholar] [CrossRef]
- Liu, J.; Shi, Y.; Zhang, Y. Multi-omics identification of an immunogenic cell death-related signature for clear cell renal cell carcinoma in the context of 3P medicine and based on a 101-combination machine learning computational framework. EPMA J. 2023, 14, 275–305. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Tan, Z.; Chen, X.; Zuo, J.; Fu, S.; Wang, H.; Wang, J. Comprehensive analysis of scRNA-Seq and bulk RNA-Seq reveals dynamic changes in the tumor immune microenvironment of bladder cancer and establishes a prognostic model. J. Transl. Med. 2023, 21, 223. [Google Scholar] [CrossRef] [PubMed]
- Blanche, P.; Dartigues, J.F.; Jacqmin-Gadda, H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat. Med. 2013, 32, 5381–5397. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, L.; Jiang, D.; Yang, M.; Gao, X.; Lv, K.; Xu, W.; Wei, H.; Wan, W.; Xiao, J. A Novel Nomogram for Survival Prediction of Patients with Spinal Metastasis From Prostate Cancer. Spine 2021, 46, E364–E373. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Ma, R.; Qu, X.; Che, X.; Yang, B.; Li, C.; Hou, K.; Guo, T.; Xiao, J.; Liu, Y. Comparative Analysis and in vitro Experiments of Signatures and Prognostic Value of Immune Checkpoint Genes in Colorectal Cancer. Onco Targets Ther. 2021, 14, 3517–3534. [Google Scholar] [CrossRef] [PubMed]
- Maeser, D.; Gruener, R.F.; Huang, R.S. oncoPredict: An R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief. Bioinform. 2021, 22, bbab260. [Google Scholar] [CrossRef] [PubMed]
- Geeleher, P.; Cox, N.; Huang, R.S. pRRophetic: An R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE 2014, 9, e107468. [Google Scholar] [CrossRef] [PubMed]
- Rees, M.G.; Seashore-Ludlow, B.; Cheah, J.H.; Adams, D.J.; Price, E.V.; Gill, S.; Javaid, S.; Coletti, M.E.; Jones, V.L.; Bodycombe, N.E.; et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 2016, 12, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Zhang, L.; Cui, Y.; Mei, J.; Zhang, Z.; Zhang, P. Exploring cellular diversity in lung adenocarcinoma epithelium: Advancing prognostic methods and immunotherapeutic strategies. Cell Prolif. 2024, 57, e13703. [Google Scholar] [CrossRef]
- Li, C.; Wu, H.; Guo, L.; Liu, D.; Yang, S.; Li, S.; Hua, K. Single-cell transcriptomics reveals cellular heterogeneity and molecular stratification of cervical cancer. Commun. Biol. 2022, 5, 1208. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, S.; Hou, S.; Chen, J.; Qi, X. Integrative Single-Cell and Bulk RNA Sequencing Identifies a Macrophage-Related Prognostic Signature for Predicting Prognosis and Therapy Responses in Colorectal Cancer. Int. J. Mol. Sci. 2025, 26, 811. https://doi.org/10.3390/ijms26020811
Xie S, Hou S, Chen J, Qi X. Integrative Single-Cell and Bulk RNA Sequencing Identifies a Macrophage-Related Prognostic Signature for Predicting Prognosis and Therapy Responses in Colorectal Cancer. International Journal of Molecular Sciences. 2025; 26(2):811. https://doi.org/10.3390/ijms26020811
Chicago/Turabian StyleXie, Shaozhuo, Siyu Hou, Jiajia Chen, and Xin Qi. 2025. "Integrative Single-Cell and Bulk RNA Sequencing Identifies a Macrophage-Related Prognostic Signature for Predicting Prognosis and Therapy Responses in Colorectal Cancer" International Journal of Molecular Sciences 26, no. 2: 811. https://doi.org/10.3390/ijms26020811
APA StyleXie, S., Hou, S., Chen, J., & Qi, X. (2025). Integrative Single-Cell and Bulk RNA Sequencing Identifies a Macrophage-Related Prognostic Signature for Predicting Prognosis and Therapy Responses in Colorectal Cancer. International Journal of Molecular Sciences, 26(2), 811. https://doi.org/10.3390/ijms26020811




