Mechanism of Transcription Factor ChbZIP1 Enhanced Alkaline Stress Tolerance in Chlamydomonas reinhardtii
Abstract
1. Introduction
2. Results
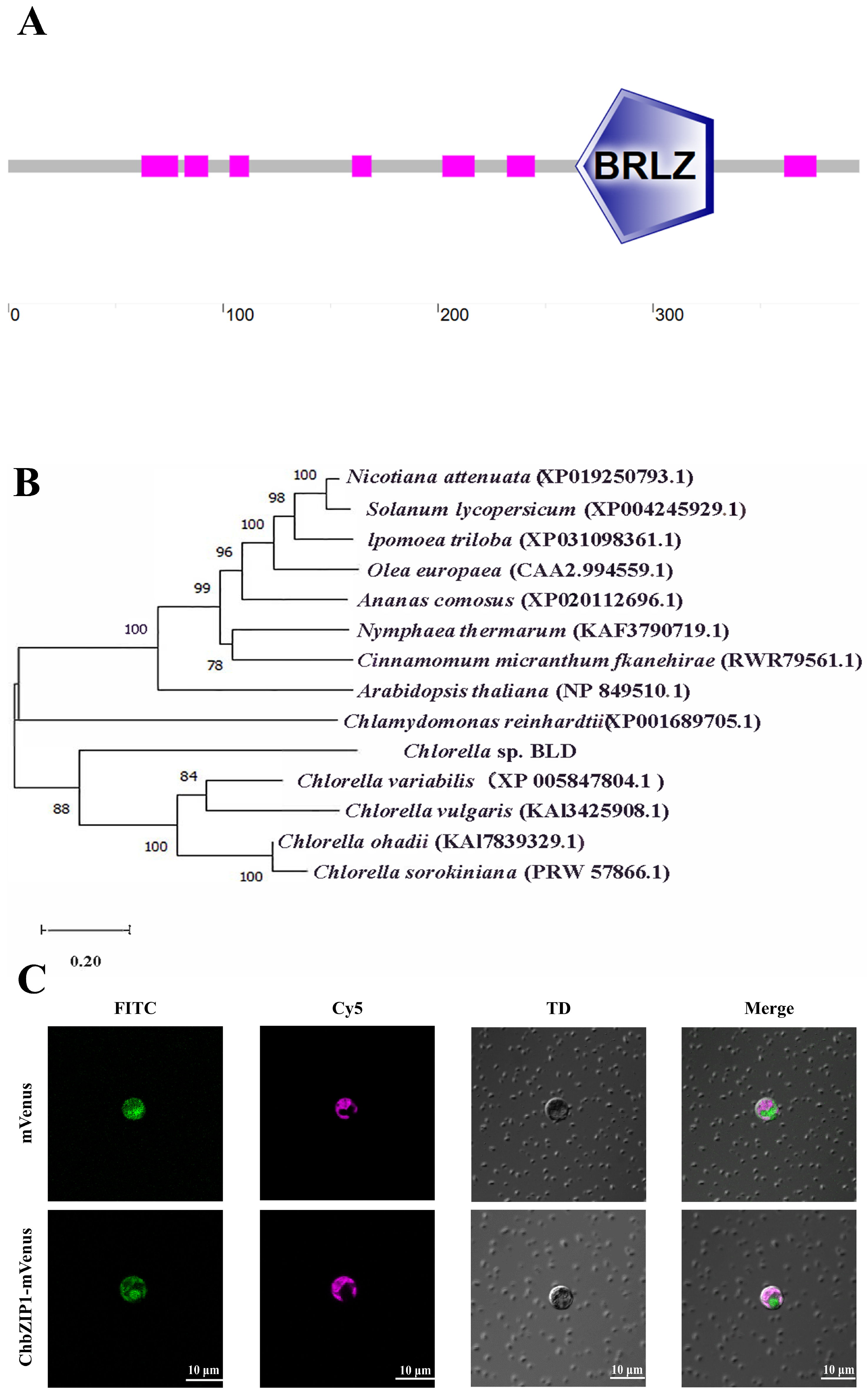
2.1. Identification of the ChbZIP1 from Chlorella sp. BLD
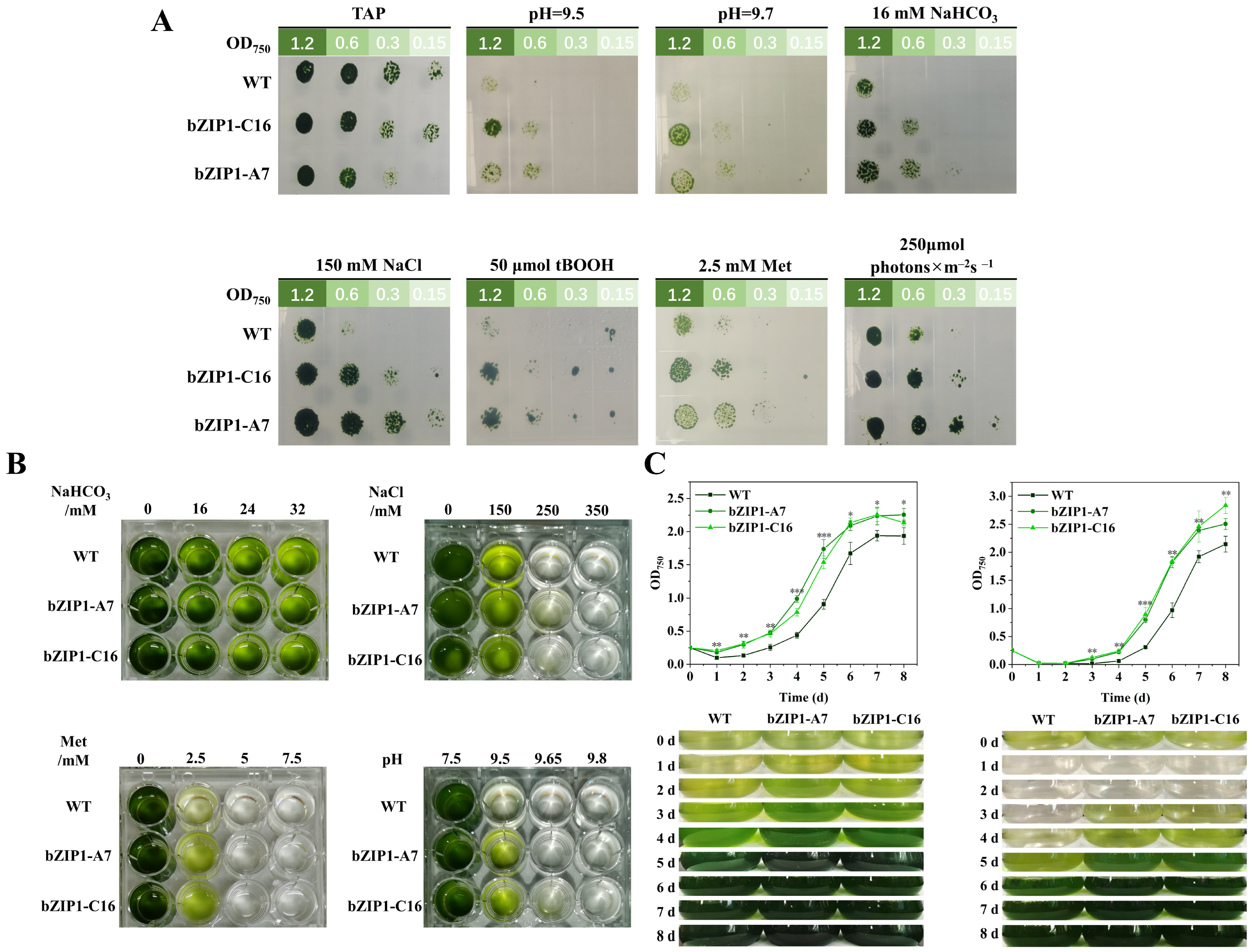
2.2. Heterogeneous Expression of ChbZIP1 Enhances Adaptability to Alkaline Stress in C. reinhardtii
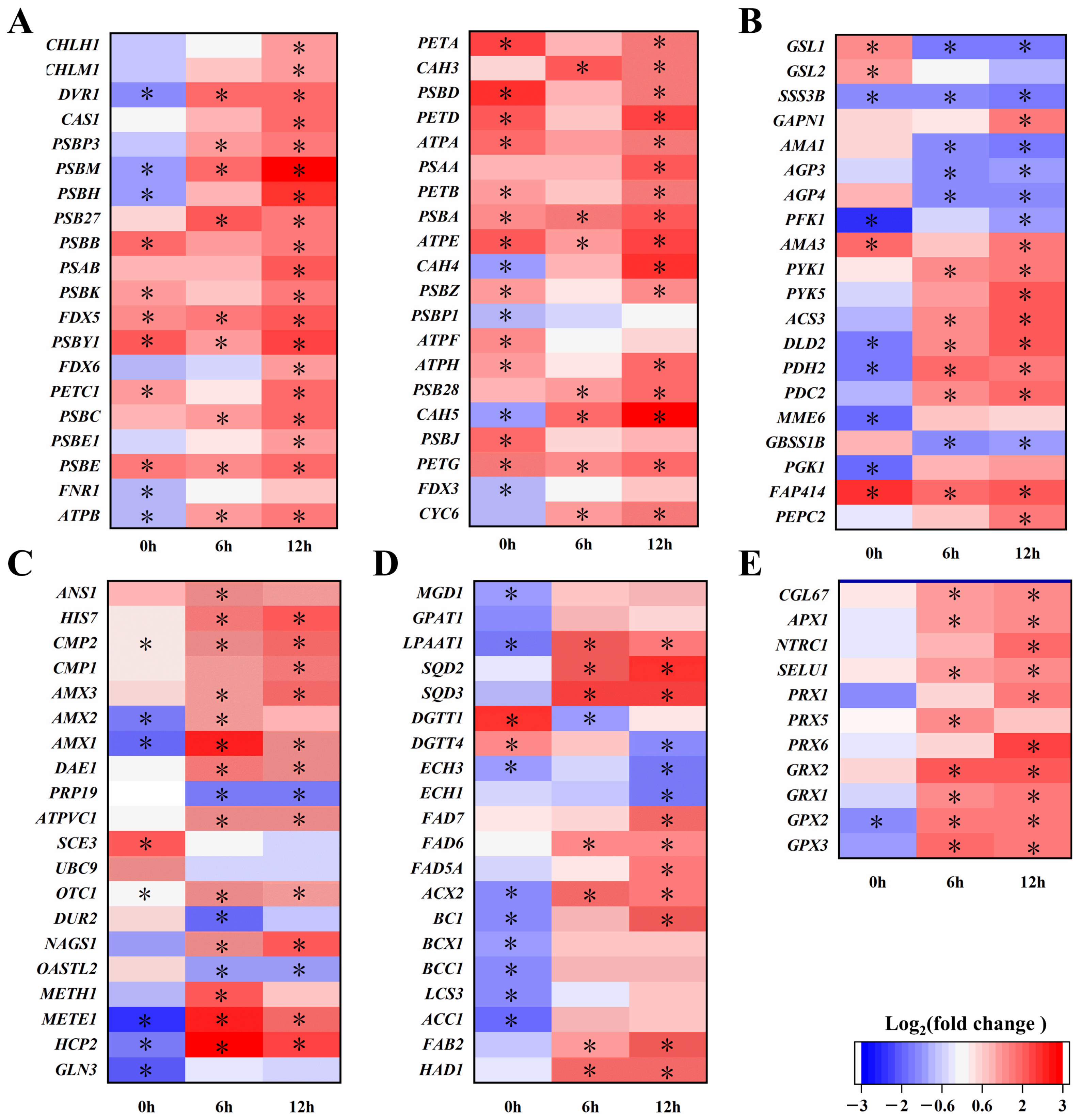
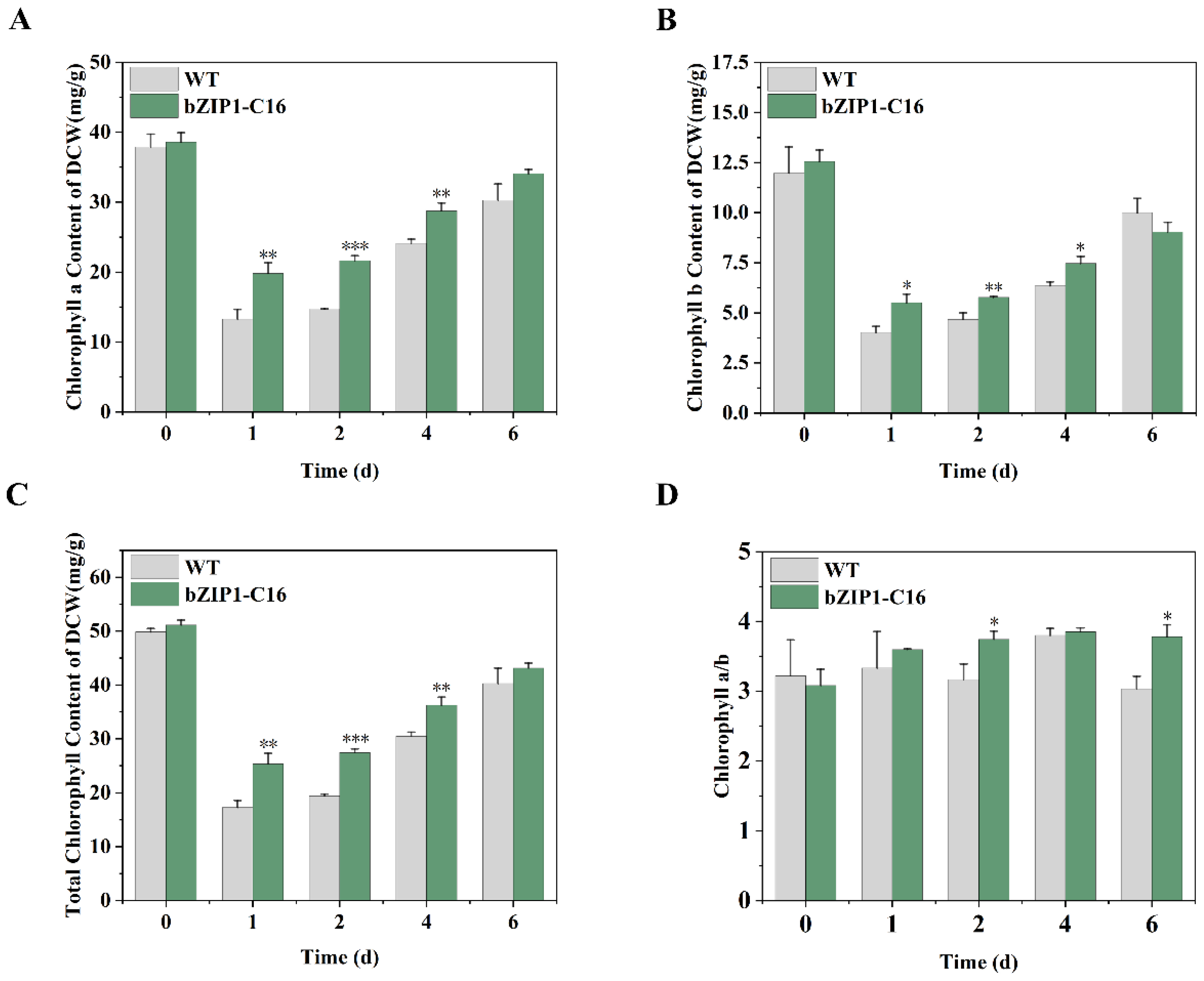
2.3. ChbZIP1 Enhances Photosynthesis of C. Reinhardtii Under Alkaline Stress
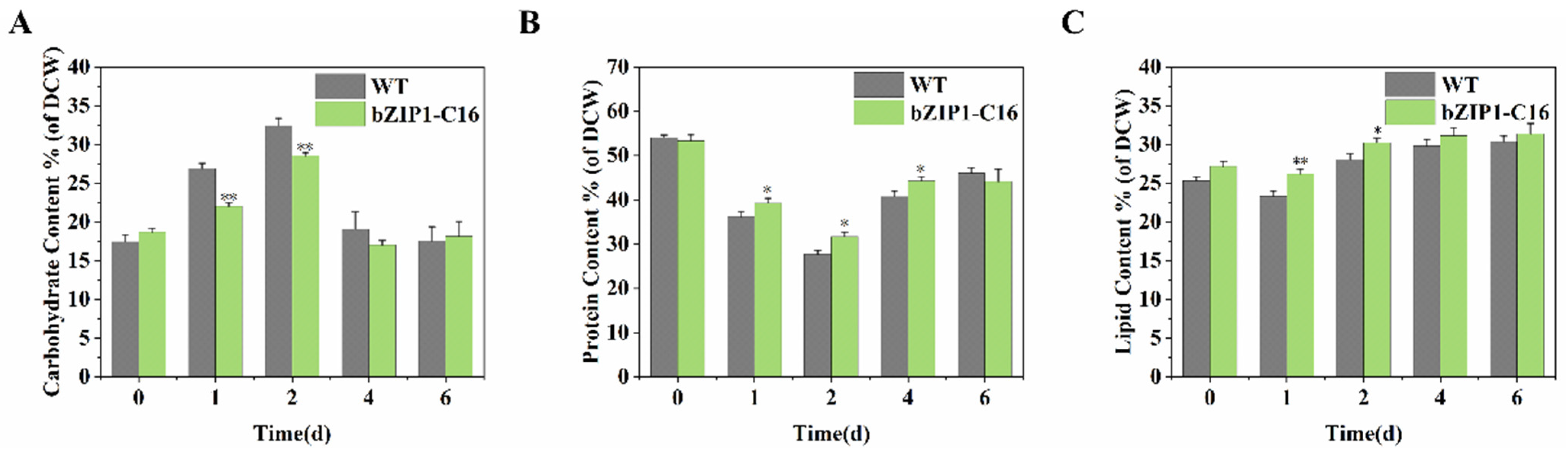
2.4. ChbZIP1 Affects the Cellular Composition of C. reinhardtii Under Alkaline Stress
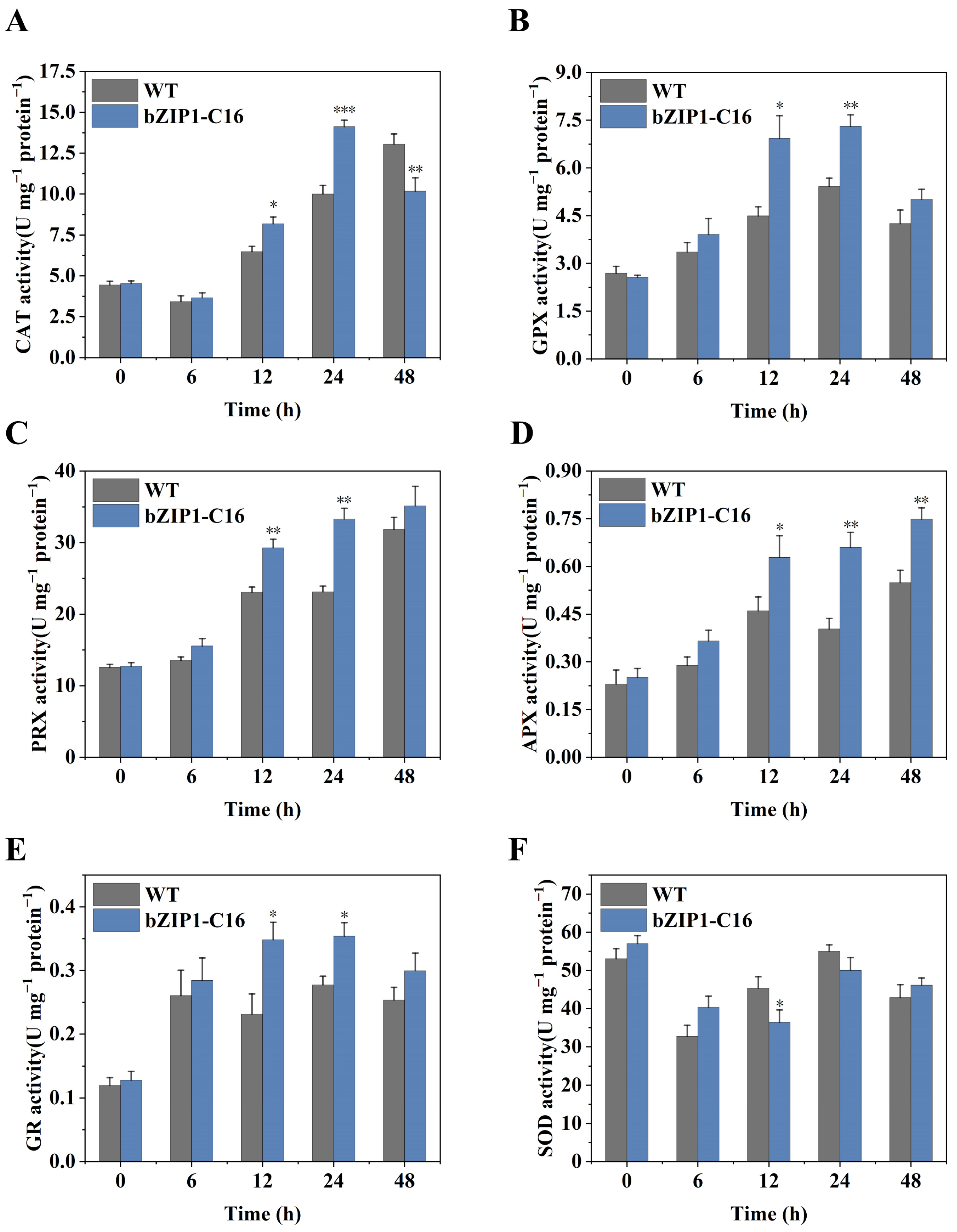
2.5. ChbZIP1 Enhances ROS Detoxification of C. reinhardtii Under Alkaline Stress
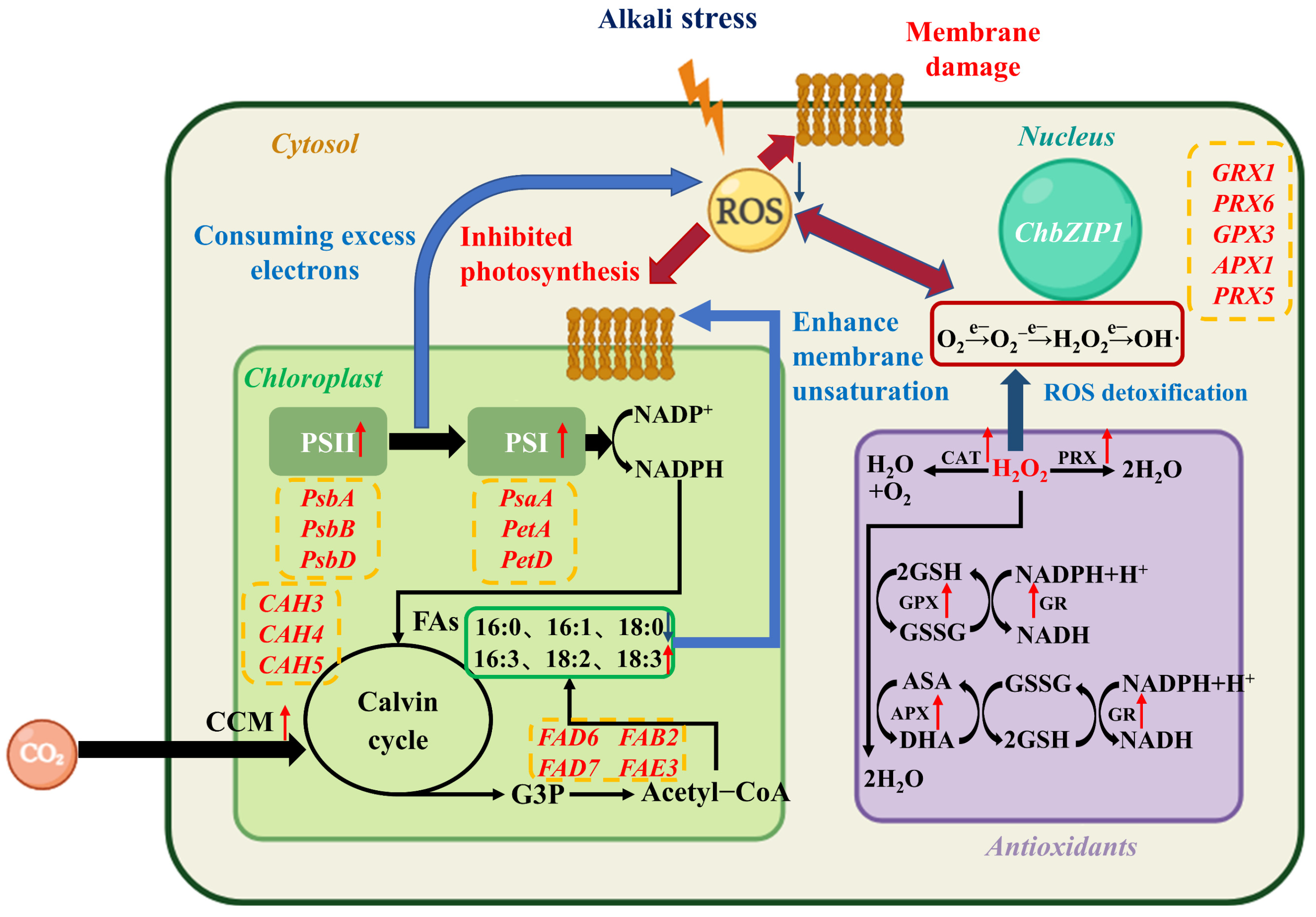
3. Discussion
3.1. ChbZIP1 Improves the Extensive Stress Resistance of C. reinhardtii, Especially the Alkali Resistance
3.2. ChbZIP1 Enhanced Photosynthesis, Enhances Carbon Dioxide Concentration, Promotes Organic Matter Synthesis and Reduces ROS Production
3.3. ChbZIP1 Alters Carbon Flow Distribution to Synthesize Lipids and Increases Fatty Acid Unsaturation to Repair Cell Membrane Structure
3.4. ChbZIP1 Enhances the Detoxification Ability of ROS, Maintains Cell Homeostasis and Reduces Cell Structural Damage
4. Materials and Methods
4.1. Microalgae Strains and Cultivation
4.2. Construction and Verification of C. reinhardtii Transformants
4.3. Subcellular Localization Assay
4.4. Transformants Growth Assay
4.5. RNA Extraction, Transcriptome Sequencing and Analysis
4.6. Verification of Relative Gene Expression Levels by RT-qPCR
4.7. Quantitative Analysis of Main Components
4.8. Determination of Photosynthetic Activity Index
4.9. Determination of Antioxidant Enzyme Activities and Antioxidant Physiological and Biochemical Indicators
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yin, Z.; Zhang, H.; Zhao, Q.; Yoo, M.-J.; Zhu, N.; Yu, J.; Yu, J.; Guo, S.; Miao, Y.; Chen, S.; et al. Physiological and comparative proteomic analyses of saline-alkali NaHCO3-responses in leaves of halophyte Puccinellia tenuiflora. Plant Soil 2019, 437, 137–158. [Google Scholar] [CrossRef]
- Jain, R.M.; Mody, K.H.; Keshri, J.; Jha, B. Biological neutralization and biosorption of dyes of alkaline textile industry wastewater. Mar. Pollut. Bull. 2014, 84, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Guadie, A.; Tizazu, S.; Melese, M.; Guo, W.; Ngo, H.H.; Xia, S. Biodecolorization of textile azo dye using Bacillus sp. strain CH12 isolated from alkaline lake. Biotechnol. Rep. 2017, 15, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.J.; Ma, H.Y.; Wang, Z.J.; Wang, Z.Y.; Bu, Q.Y.; Liu, S.K. A rice LSD1-like-type ZFP gene OsLOL5 enhances saline-alkaline tolerance in transgenic Arabidopsis thaliana, yeast and rice. BMC Genom. 2016, 17, 142. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Duanmu, H.; Yu, Y.; Chen, C.; Sun, X.; Zhu, P.; Chen, R.; Duan, X.; Li, H.; Cao, L.; et al. GsJ11, identified by genome-wide analysis, facilitates alkaline tolerance in transgenic plants. Plant Cell Tissue Organ Cult. (PCTOC) 2017, 129, 411–430. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, F.; Sun, L.; Yang, L.; Yang, Y.; Wang, Y.; Siddique, K.H.M.; Pang, J. Alkaline Salt Inhibits Seed Germination and Seedling Growth of Canola More Than Neutral Salt. Front. Plant Sci. 2022, 13, 814755. [Google Scholar] [CrossRef]
- Wang, H.; Ahan, J.; Wu, Z.; Shi, D.; Liu, B.; Yang, C. Alteration of nitrogen metabolism in rice variety ’Nipponbare’ induced by alkali stress. Plant Soil. 2012, 355, 131–147. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.-L.; Zhang, R.-X.; Yuan, H.-Y.; Wang, M.-M.; Yang, H.-Y.; Ma, H.-Y.; Liu, D.; Jiang, C.-J.; Liang, Z.-W. Root Damage under Alkaline Stress Is Associated with Reactive Oxygen Species Accumulation in Rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1580. [Google Scholar] [CrossRef]
- Kaiwen, G.; Zisong, X.; Yuze, H.; Qi, S.; Yue, W.; Yanhui, C.; Jiechen, W.; Wei, L.; Huihui, Z. Effects of salt concentration, pH, and their interaction on plant growth, nutrient uptake, and photochemistry of alfalfa (Medicago sativa) leaves. Plant Signal Behav. 2020, 15, 1832373. [Google Scholar] [CrossRef]
- Ye, L.; Zhao, X.; Bao, E.; Cao, K.; Zou, Z. Effects of Arbuscular Mycorrhizal Fungi on Watermelon Growth, Elemental Uptake, Antioxidant, and Photosystem II Activities and Stress-Response Gene Expressions Under Salinity-Alkalinity Stresses. Front. Plant Sci. 2019, 10, 863. [Google Scholar] [CrossRef]
- Hu, L.; Xiang, L.; Zhang, L.; Zhou, X.; Zou, Z.; Hu, X. The Photoprotective Role of Spermidine in Tomato Seedlings under Salinity-Alkalinity Stress. PLoS ONE 2014, 9, e110855. [Google Scholar] [CrossRef]
- Ganapati, R.K.; Naveed, S.A.; Zafar, S.; Wang, W.; Xu, J. Saline-Alkali Tolerance in Rice: Physiological Response, Molecular Mechanism, and QTL Identification and Application to Breeding. Rice Sci. 2022, 29, 412–434. [Google Scholar] [CrossRef]
- Lindemose, S.; O’Shea, C.; Jensen, M.K.; Skriver, K. Structure, Function and Networks of Transcription Factors Involved in Abiotic Stress Responses. Int. J. Mol. Sci. 2013, 14, 5842–5878. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Zhou, J.; Deng, R.-Y.; Zhao, H.-X.; Li, C.-L.; Chen, H.; Suzuki, T.; Park, S.-U.; Wu, Q. Overexpression of a tartary buckwheat R2R3-MYB transcription factor gene, FtMYB9, enhances tolerance to drought and salt stresses in transgenic Arabidopsis. J. Plant Physiol. 2017, 214, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Yu, Y.; Ding, X.; Zhu, D.; Yang, F.; Liu, B.; Sun, X.; Duan, X.; Yin, K.; Zhu, Y. The Glycine soja NAC transcription factor GsNAC019 mediates the regulation of plant alkaline tolerance and ABA sensitivity. Plant Mol. Biol. 2017, 95, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liu, A.; Duan, X.; Wang, S.; Sun, X.; Duanmu, H.; Zhu, D.; Chen, C.; Cao, L.; Xiao, J.; et al. GsERF6, an ethylene-responsive factor from Glycine soja, mediates the regulation of plant bicarbonate tolerance in Arabidopsis. Planta 2016, 244, 681–698. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Duan, X.; Ding, X.; Chen, C.; Zhu, D.; Yin, K.; Cao, L.; Song, X.; Zhu, P.; Li, Q.; et al. A novel AP2/ERF family transcription factor from Glycine soja, GsERF71, is a DNA binding protein that positively regulates alkaline stress tolerance in Arabidopsis. Plant Mol. Biol. 2017, 94, 509–530. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Li, X.; Xu, C.; Ajab, Z.; Liu, Q.; Majeed, Z.; Guan, Q. Analysis of drought and salt-alkali tolerance in tobacco by overexpressing WRKY39 gene from Populus trichocarpa. Plant Signal Behav. 2021, 16, 1918885. [Google Scholar] [CrossRef]
- Zhang, C.; Li, C.; Liu, J.; Lv, Y.; Yu, C.; Li, H.; Zhao, T.; Liu, B. The OsABF1 transcription factor improves drought tolerance by activating the transcription of COR413-TM1 in rice. J. Exp. Bot. 2017, 68, 4695–4707. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.D.; Singh, N.; Ahuja, P.S.; Reddy, T.V. Abscisic acid response element binding factor 1 is required for establishment of Arabidopsis seedlings during winter. Mol. Biol. Rep. 2011, 38, 5147–5159. [Google Scholar] [CrossRef]
- Kong, Y.; Chen, S.; Yang, Y.; An, C. ABA-insensitive (ABI) 4 and ABI5 synergistically regulate DGAT1 expression in Arabidopsis seedlings under stress. FEBS Lett. 2013, 587, 3076–3082. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhu, J.; Yuan, W.; Wang, Y.; Hu, P.; Jiao, C.; Xia, H.; Wang, D.; Cai, Q.; Li, J.; et al. Genome-wide characterization of bZIP transcription factors and their expression patterns in response to drought and salinity stress in Jatropha curcas. Int. J. Biol. Macromol. 2021, 181, 1207–1223. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, C.; Li, Z.; Sun, J.; Wang, D.; Xu, L.; Li, X.; Guo, Y. Identification and Analysis of bZIP Family Genes in Potato and Their Potential Roles in Stress Responses. Front. Plant Sci. 2021, 12, 637343. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Jung, J.-M.; Jung, S.; Park, Y.-K.; Tsang, Y.F.; Lin, K.-Y.A.; Choi, Y.-E.; Kwon, E.E. Biodiesel from microalgae: Recent progress and key challenges. Prog. Energy Combust. Sci. 2022, 93, 101020. [Google Scholar] [CrossRef]
- Díaz, V.; Leyva-Díaz, J.C.; Almécija, M.C.; Poyatos, J.M.; del Mar Muñío, M.; Martín-Pascual, J. Microalgae bioreactor for nutrient removal and resource recovery from wastewater in the paradigm of circular economy. Bioresour. Technol. 2022, 363, 127968. [Google Scholar] [CrossRef]
- Scaife, M.A.; Nguyen, G.; Rico, J.; Lambert, D.; Helliwell, K.E.; Smith, A.G. Establishing Chlamydomonas reinhardtii as an industrial biotechnology host. Plant J. 2015, 82, 532–546. [Google Scholar] [CrossRef]
- Zhang, M.-P.; Wang, M.; Wang, C. Nuclear transformation of Chlamydomonas reinhardtii: A review. Biochimie 2021, 181, 1–11. [Google Scholar] [CrossRef]
- Kong, Q.-x.; Li, L.; Martinez, B.; Chen, P.; Ruan, R. Culture of Microalgae Chlamydomonas reinhardtii in Wastewater for Biomass Feedstock Production. Appl. Biochem. Biotechnol. 2010, 160, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Hasan, R.; Zhang, B.; Wang, L.; Shahbazi, A. Bioremediation of Swine Wastewater and Biofuel Potential by usingChlorella vulgaris, Chlamydomonas reinhardtii, and Chlamydomonasdebaryana. J. Pet. Environ. Biotechnol. 2014, 5, 1–5. [Google Scholar] [CrossRef]
- Qi, F.; Xu, Y.; Yu, Y.; Liang, X.; Zhang, L.; Zhao, H.; Wang, H. Enhancing growth of Chlamydomonas reinhardtii and nutrient removal in diluted primary piggery wastewater by elevated CO2 supply. Water Sci. Technol. 2017, 75, 2281–2290. [Google Scholar] [CrossRef]
- Ochoa-Alfaro, A.E.; Gaytán-Luna, D.E.; González-Ortega, O.; Zavala-Arias, K.G.; Paz-Maldonado, L.M.T.; Rocha-Uribe, A.; Soria-Guerra, R.E. pH effects on the lipid and fatty acids accumulation in Chlamydomonas reinhardtii. Biotechnol. Prog. 2019, 35, e2891. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Miao, X. Carbon flow conversion induces alkali resistance and lipid accumulation under alkaline conditions based on transcriptome analysis in Chlorella sp. BLD. Chemosphere 2021, 265, 129046. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Show, P.-L.; Miao, X. Transcription Factor ChbZIP1 from Alkaliphilic Microalgae Chlorella sp. BLD Enhancing Alkaline Tolerance in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 2387. [Google Scholar] [CrossRef]
- Crozet, P.; Navarro, F.J.; Willmund, F.; Mehrshahi, P.; Bakowski, K.; Lauersen, K.J.; Pérez-Pérez, M.-E.; Auroy, P.; Gorchs Rovira, A.; Sauret-Gueto, S.; et al. Birth of a Photosynthetic Chassis: A MoClo Toolkit Enabling Synthetic Biology in the Microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2018, 7, 2074–2086. [Google Scholar] [CrossRef] [PubMed]
- Baier, T.; Wichmann, J.; Kruse, O.; Lauersen, K.J. Intron-containing algal transgenes mediate efficient recombinant gene expression in the green microalga Chlamydomonas reinhardtii. Nucleic Acids Res. 2018, 46, 6909–6919. [Google Scholar] [CrossRef]
- Fu, W.; Nelson, D.R.; Mystikou, A.; Daakour, S.; Salehi-Ashtiani, K. Advances in microalgal research and engineering development. Curr. Opin. Biotechnol. 2019, 59, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Hou, X.; Liang, X. Response Mechanisms of Plants Under Saline-Alkali Stress. Front. Plant Sci. 2021, 12, 667458. [Google Scholar] [CrossRef] [PubMed]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Seo, D.H.; Shin, H.; Kim, H.J.; Kim, C.M.; Jang, G. The Role of Stress-Responsive Transcription Factors in Modulating Abiotic Stress Tolerance in Plants. Agronomy 2020, 10, 788. [Google Scholar] [CrossRef]
- Wu, S.; Zhu, P.; Jia, B.; Yang, J.; Shen, Y.; Cai, X.; Sun, X.; Zhu, Y.; Sun, M. A Glycine soja group S2 bZIP transcription factor GsbZIP67 conferred bicarbonate alkaline tolerance in Medicago sativa. BMC Plant Biol. 2018, 18, 234. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, F.; Ma, Y.; Dang, H.; Hu, X. Transcription Factor SlAREB1 Is Involved in the Antioxidant Regulation under Saline-Alkaline Stress in Tomato. Antioxidants 2022, 11, 1673. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, P.; Wang, X.; Zhang, Z.; Wang, Y. Functional Identification of Malus halliana MhbZIP23 Gene Demonstrates That It Enhances Saline–Alkali Stress Tolerance in Arabidopsis thaliana. Plants 2024, 13, 1803. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, C.; Zhu, Q.; Guo, F.; Chai, R.; Wang, M.; Deng, X.; Dong, T.; Meng, X.; Zhu, M. Genome- and transcriptome-wide systematic characterization of bZIP transcription factor family identifies promising members involved in abiotic stress response in sweetpotato. Sci. Hortic. 2022, 303, 111185. [Google Scholar] [CrossRef]
- Gai, Y.; Li, L.; Liu, B.; Ma, H.; Chen, Y.; Zheng, F.; Sun, X.; Wang, M.; Jiao, C.; Li, H. Distinct and essential roles of bZIP transcription factors in the stress response and pathogenesis in Alternaria alternata. Microbiol. Res. 2022, 256, 126915. [Google Scholar] [CrossRef]
- Calzadilla, P.I.; Carvalho, F.E.L.; Gomez, R.; Lima Neto, M.C.; Signorelli, S. Assessing photosynthesis in plant systems: A cornerstone to aid in the selection of resistant and productive crops. Environ. Exp. Bot. 2022, 201, 104950. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef]
- Liu, J.; Hou, H.; Zhao, L.; Sun, Z.; Li, H. Protective Effect of foliar application of sulfur on photosynthesis and antioxidative defense system of rice under the stress of Cd. Sci. Total Environ. 2020, 710, 136230. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.Y.; Kim, H.; Shim, D.; Jang, S.; Yamaoka, Y.; Shin, S.; Yamano, T.; Kajikawa, M.; Jin, E.; Fukuzawa, H.; et al. The Chlamydomonas bZIP transcription factor BLZ8 confers oxidative stress tolerance by inducing the carbon-concentrating mechanism. Plant Cell 2022, 34, 910–926. [Google Scholar] [CrossRef]
- Mansour, M.M.F.; Salama, K.H.A. Cellular Mechanisms of Plant Salt Tolerance. In Microorganisms in Saline Environments: Strategies and Functions; Giri, B., Varma, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 169–210. [Google Scholar]
- Jeandet, P.; Formela-Luboińska, M.; Labudda, M.; Morkunas, I. The Role of Sugars in Plant Responses to Stress and Their Regulatory Function during Development. Int. J. Mol. Sci. 2022, 23, 5161. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Ufer, G.; Bartels, D. Lipid signalling in plant responses to abiotic stress. Plant Cell Env. 2016, 39, 1029–1048. [Google Scholar] [CrossRef] [PubMed]
- Miyasaka, H.; Ikeda, K. Osmoregulating mechanism of the halotolerant green alga Chlamydomonas, strain HS-5. Plant Sci. 1997, 127, 91–96. [Google Scholar] [CrossRef]
- Li, Q.; Shen, W.; Zheng, Q.; Fowler, D.B.; Zou, J. Adjustments of lipid pathways in plant adaptation to temperature stress. Plant Signal Behav. 2016, 11, e1058461. [Google Scholar] [CrossRef]
- He, M.; Ding, N.Z. Plant Unsaturated Fatty Acids: Multiple Roles in Stress Response. Front. Plant Sci. 2020, 11, 562785. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y.; Shin, S.; Choi, B.Y.; Kim, H.; Jang, S.; Kajikawa, M.; Yamano, T.; Kong, F.; Légeret, B.; Fukuzawa, H.; et al. The bZIP1 Transcription Factor Regulates Lipid Remodeling and Contributes to ER Stress Management in Chlamydomonas reinhardtii. Plant Cell 2019, 31, 1127–1140. [Google Scholar] [CrossRef]
- Kwon, S.; Kang, N.K.; Koh, H.G.; Shin, S.E.; Lee, B.; Jeong, B.R.; Chang, Y.K. Enhancement of biomass and lipid productivity by overexpression of a bZIP transcription factor in Nannochloropsis salina. Biotechnol. Bioeng. 2018, 115, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Sui, N.; Wang, Y.; Liu, S.; Yang, Z.; Wang, F.; Wan, S. Transcriptomic and Physiological Evidence for the Relationship between Unsaturated Fatty Acid and Salt Stress in Peanut. Front. Plant Sci. 2018, 9, 7. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Liu, L.; Barkla, B.J. Membrane Lipid Remodeling in Response to Salinity. Int. J. Mol. Sci. 2019, 20, 4264. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.L.; Li, F.; Su, N.; Sun, X.L.; Zhao, S.J.; Meng, Q.W. The increase in unsaturation of fatty acids of phosphatidylglycerol in thylakoid membrane enhanced salt tolerance in tomato. Photosynthetica 2010, 48, 400–408. [Google Scholar] [CrossRef]
- Zhang, L.; Liao, C.; Yang, Y.; Wang, Y.-Z.; Ding, K.; Huo, D.; Hou, C. Response of lipid biosynthesis in Chlorella pyrenoidosa to intracellular reactive oxygen species level under stress conditions. Bioresour. Technol. 2019, 287, 121414. [Google Scholar] [CrossRef]
- Neelam, S.; Subramanyam, R. Alteration of photochemistry and protein degradation of photosystem II from Chlamydomonas reinhardtii under high salt grown cells. J. Photochem. Photobiol. B 2013, 124, 63–70. [Google Scholar] [CrossRef]
- Orthesin, N.; Hidayat, I.T.; Wahyuni, W.T.; Syafitri, U.D.; Herbani, Y.; Sari, Y.W. Optimization of Chlorophyll Extraction from Dried Spirulina platensis Using Low Power Microwave Assisted Extraction Method. IOP Conf. Ser. Earth Environ. Sci. 2024, 1359, 012021. [Google Scholar] [CrossRef]


| Fatty Acid (%) | 0 h | 6 h | 12 h | 24 h | 48 h | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | bZIP1-C16 | WT | bZIP1-C16 | WT | bZIP1-C16 | WT | bZIP1-C16 | WT | bZIP1-C16 | |
| C14:0 | 0.37 ± 0.03 | 0.26 ± 0.02 ** | 0.3 ± 0.06 | 0.27 ± 0.02 | 0.41 ± 0.03 | 0.40 ± 0.04 | 0.58 ± 0.02 | 0.45 ± 0.04 * | 0.52 ± 0.02 | 0.47 ± 0.02 |
| C16:0 | 34.61 ± 1.28 | 38.94 ± 1.81 | 40.49 ± 1 | 35.17 ± 0.82 ** | 39.8 ± 0.69 | 33.54 ± 0.49 *** | 33.5 ± 0.1 | 28.31 ± 0.62 *** | 34.3 ± 0.69 | 29.85 ± 0.6 ** |
| C16:1 | 2.66 ± 0.25 | 3.17 ± 0.07 * | 4.3 ± 0.23 | 3.86 ± 0.06 | 4.05 ± 0.18 | 3.08 ± 0.17 ** | 3.68 ± 0.19 | 2.95 ± 0.14 * | 3.88 ± 0.08 | 2.53 ± 0.05 *** |
| C16:2 | 1.12 ± 0.03 | 0.79 ± 0.03 *** | 1.45 ± 0.08 | 1.62 ± 0.11 | 0.74 ± 0.04 | 0.7 ± 0.06 | 1.03 ± 0.05 | 0.93 ± 0.03 | 1.03 ± 0.05 | 1.35 ± 0.11 * |
| C16:3n6 | 0.60 ± 0.06 | 0.45 ± 0.01 ** | 1.03 ± 0.04 | 1.2 ± 0.15 | 0.56 ± 0.02 | 0.54 ± 0.02 | 0.6 ± 0.02 | 0.46 ± 0.04 ** | 0.55 ± 0.01 | 0.61 ± 0.05 |
| C16:3n3 | 1.06 ± 0.03 | 1.09 ± 0.02 | 1.77 ± 0.22 | 2.54 ± 0.08 ** | 2.45 ± 0.14 | 3.32 ± 0.17 ** | 3.54 ± 0.38 | 4.52 ± 0.07 * | 2.46 ± 0.11 | 2.25 ± 0.08 |
| C16:4 | 11.04 ± 0.51 | 10.21 ± 0.25 | 6.39 ± 0.52 | 6.19 ± 0.37 | 4.82 ± 0.26 | 5.8 ± 0.85 | 9.5 ± 0.42 | 9.56 ± 0.53 | 9.72 ± 0.31 | 10.03 ± 0.63 |
| C18:0 | 8.21 ± 0.35 | 8.42 ± 0.34 | 9.93 ± 1.16 | 7.72 ± 0.48 | 10.77 ± 0.21 | 8.82 ± 0.17 *** | 9.55 ± 0.31 | 8.04 ± 0.19 ** | 8.75 ± 0.19 | 6.79 ± 0.14 *** |
| C18:1 | 5.59 ± 0.09 | 4.49 ± 0.17 ** | 6.37 ± 1.05 | 7.02 ± 0.53 | 6.24 ± 0.56 | 7.19 ± 0.43 | 5.31 ± 0.06 | 7.5 ± 0.26 *** | 5.85 ± 0.16 | 7.26 ± 0.41 * |
| C18:2 | 7.00 ± 0.32 | 5.74 ± 0.43 * | 3.79 ± 0.15 | 5.44 ± 0.09 *** | 3.47 ± 0.3 | 4.67 ± 0.45 * | 3.75 ± 0.17 | 5 ± 0.45 ** | 4.67 ± 0.14 | 6.88 ± 0.32 *** |
| C18:3n6 | 3.83 ± 0.21 | 3.20 ± 0.07 * | 3.1 ± 0.17 | 4.7 ± 0.33 ** | 4.09 ± 0.13 | 4.36 ± 0.3 | 4.53 ± 0.38 | 5.17 ± 0.11 | 4.43 ± 0.3 | 5.97 ± 0.54 * |
| C18:3n3 | 22.35 ± 0.53 | 21.70 ± 0.46 | 19.36 ± 1.08 | 22.4 ± 0.31 * | 19.75 ± 0.54 | 24.55 ± 0.59 ** | 21.81 ± 0.47 | 23.63 ± 0.35 * | 20.04 ± 0.25 | 22.93 ± 0.65 ** |
| C18:4 | 1.56 ± 0.1 | 1.55 ± 0.08 | 1.71 ± 0.41 | 1.88 ± 0.09 | 2.86 ± 0.06 | 3.02 ± 0.11 | 2.61 ± 0.03 | 3.47 ± 0.05 *** | 3.81 ± 0.19 | 3.08 ± 0.07 ** |
| SFA | 43.18 ± 1.51 | 47.62 ± 0.75 * | 50.72 ± 2.02 | 43.16 ± 0.6 ** | 50.97 ± 0.83 | 42.77 ± 0.58 *** | 43.64 ± 0.42 | 36.8 ± 0.57 | 43.56 ± 0.84 | 37.11 ± 0.6 |
| MUFA | 8.25 ± 0.09 | 7.66 ± 0.07 | 10.67 ± 1.26 | 10.87 ± 0.57 | 10.29 ± 0.4 | 10.27 ± 0.58 | 8.99 ± 0.22 | 10.46 ± 0.34 | 9.73 ± 0.23 | 9.78 ± 0.4 |
| PUFA | 48.56 ± 1.41 | 44.72 ± 0.34 * | 38.61 ± 2.07 | 45.97 ± 0.49 ** | 38.74 ± 0.74 | 46.96 ± 0.32 *** | 47.37 ± 0.54 | 52.74 ± 0.52 * | 46.71 ± 0.66 | 53.11 ± 1 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, A.; Wang, R.; Miao, X. Mechanism of Transcription Factor ChbZIP1 Enhanced Alkaline Stress Tolerance in Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2025, 26, 769. https://doi.org/10.3390/ijms26020769
Wang A, Wang R, Miao X. Mechanism of Transcription Factor ChbZIP1 Enhanced Alkaline Stress Tolerance in Chlamydomonas reinhardtii. International Journal of Molecular Sciences. 2025; 26(2):769. https://doi.org/10.3390/ijms26020769
Chicago/Turabian StyleWang, Ao, Rui Wang, and Xiaoling Miao. 2025. "Mechanism of Transcription Factor ChbZIP1 Enhanced Alkaline Stress Tolerance in Chlamydomonas reinhardtii" International Journal of Molecular Sciences 26, no. 2: 769. https://doi.org/10.3390/ijms26020769
APA StyleWang, A., Wang, R., & Miao, X. (2025). Mechanism of Transcription Factor ChbZIP1 Enhanced Alkaline Stress Tolerance in Chlamydomonas reinhardtii. International Journal of Molecular Sciences, 26(2), 769. https://doi.org/10.3390/ijms26020769





