Infectious Spleen and Kidney Necrosis Virus ORF093R and ORF102R Regulate Glutamate Metabolic Reprogramming to Support Virus Proliferation by Interacting with c-Myc
Abstract
1. Introduction
2. Results
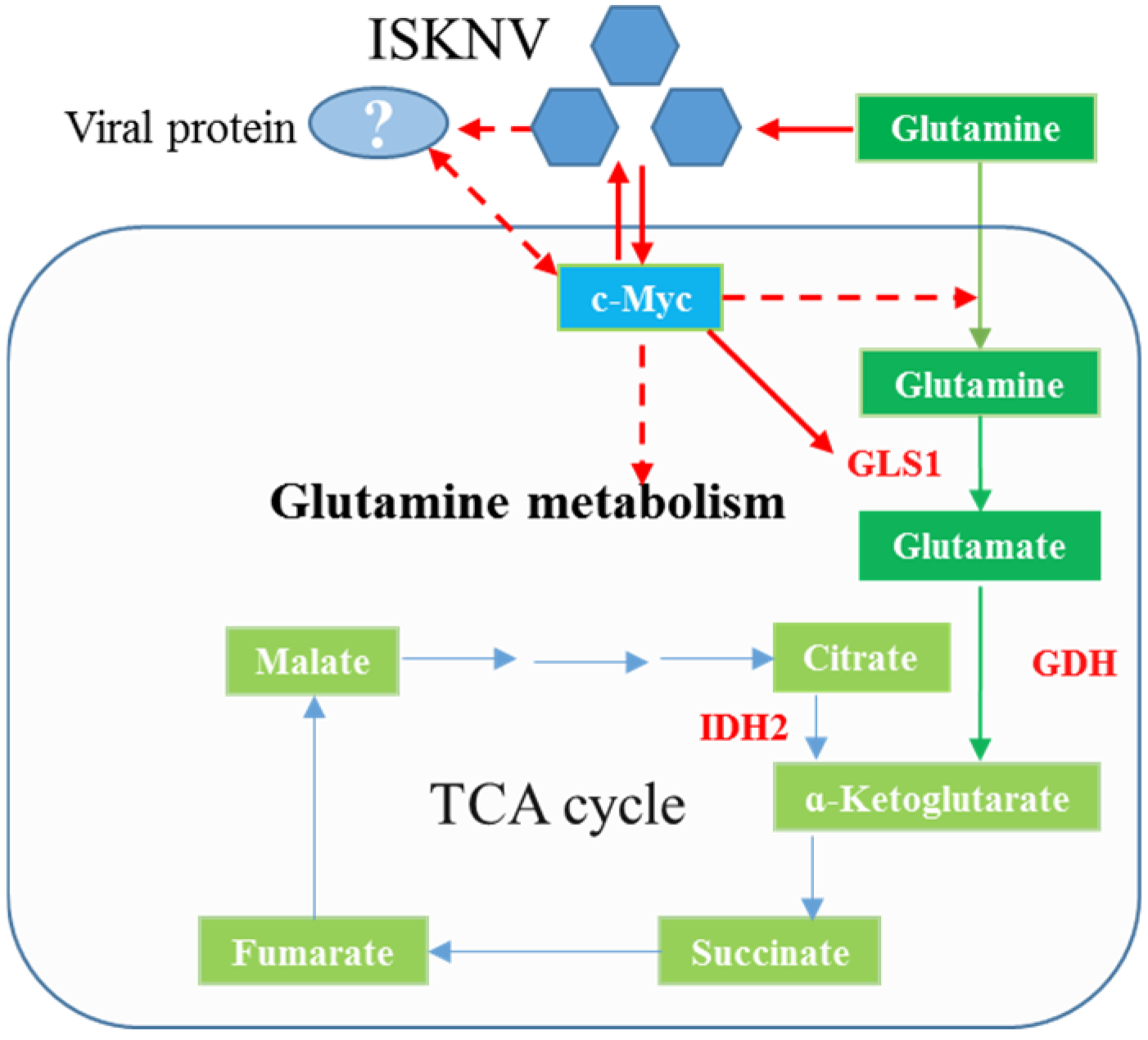
2.1. c-Myc Positively Regulates Glutamine Metabolism
2.2. Screening the ISKNV Proteins’ Interaction with C-Myc
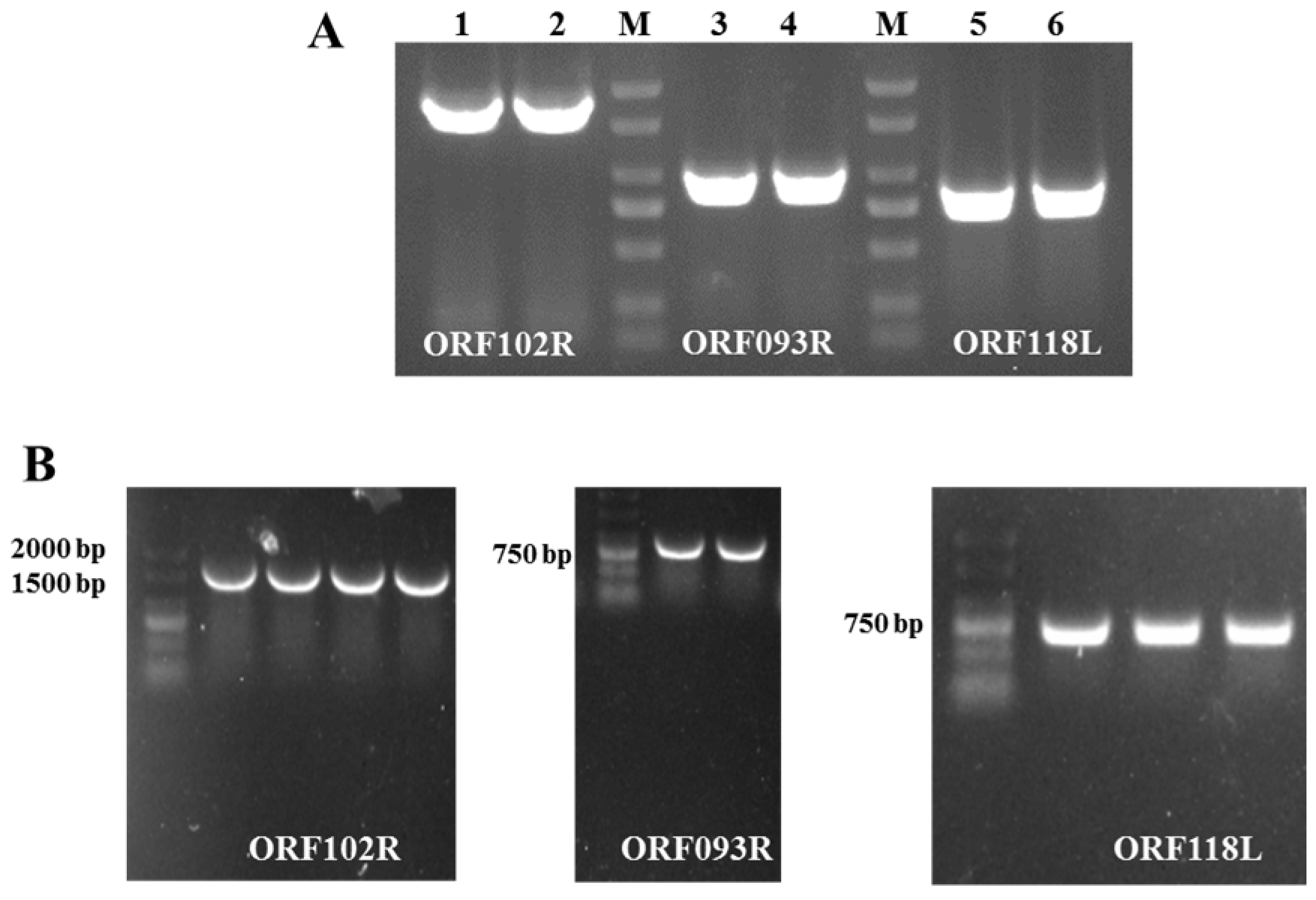
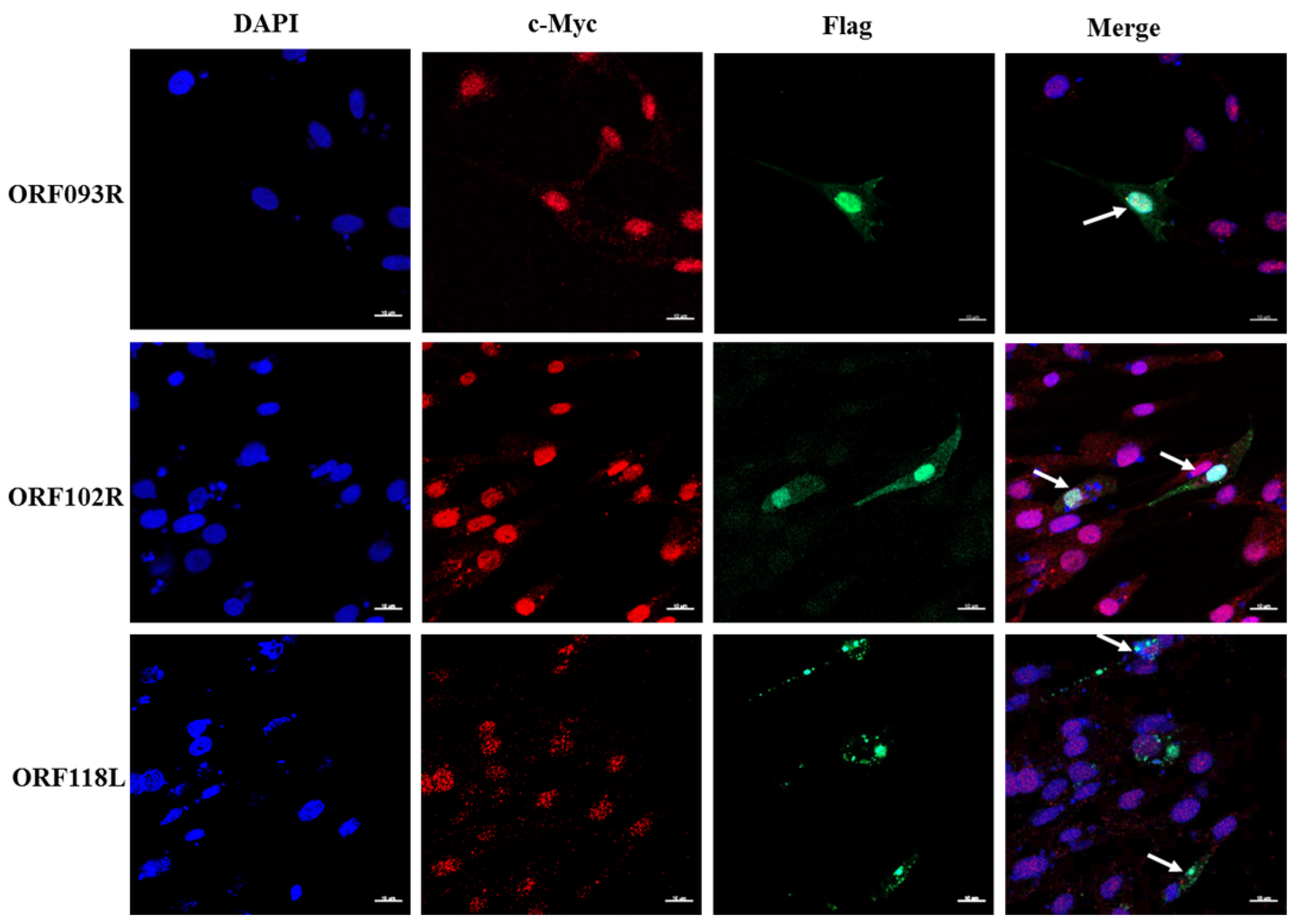
2.3. ORF102R, ORF093R and ORF118L Co-Located with C-Myc
2.4. ISKNV ORF102R and ORF093R Interacted with c-Myc
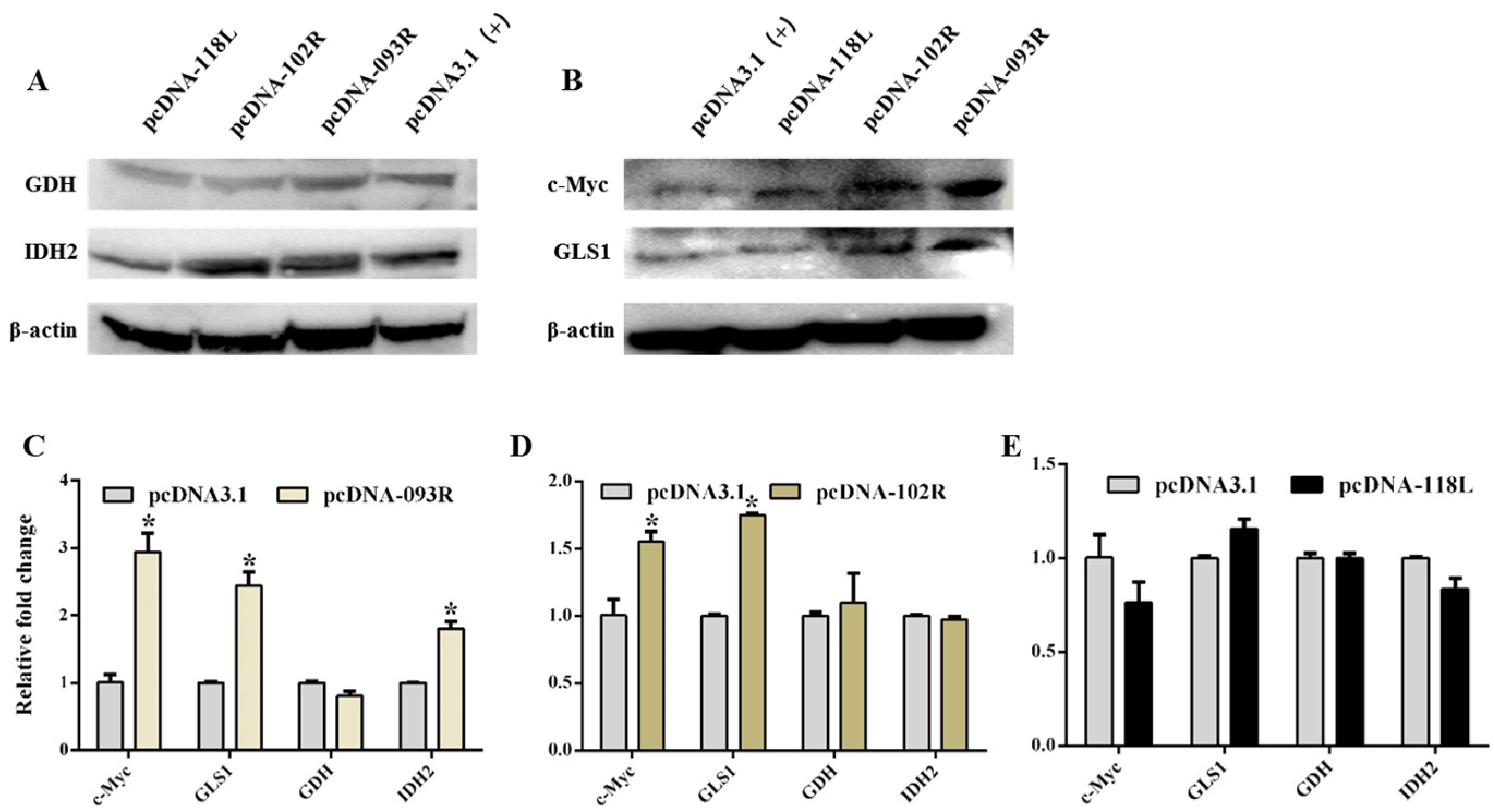
2.5. The Effect of ISKNV ORF093R, ORF102R and ORF118L on Glutamine Metabolism
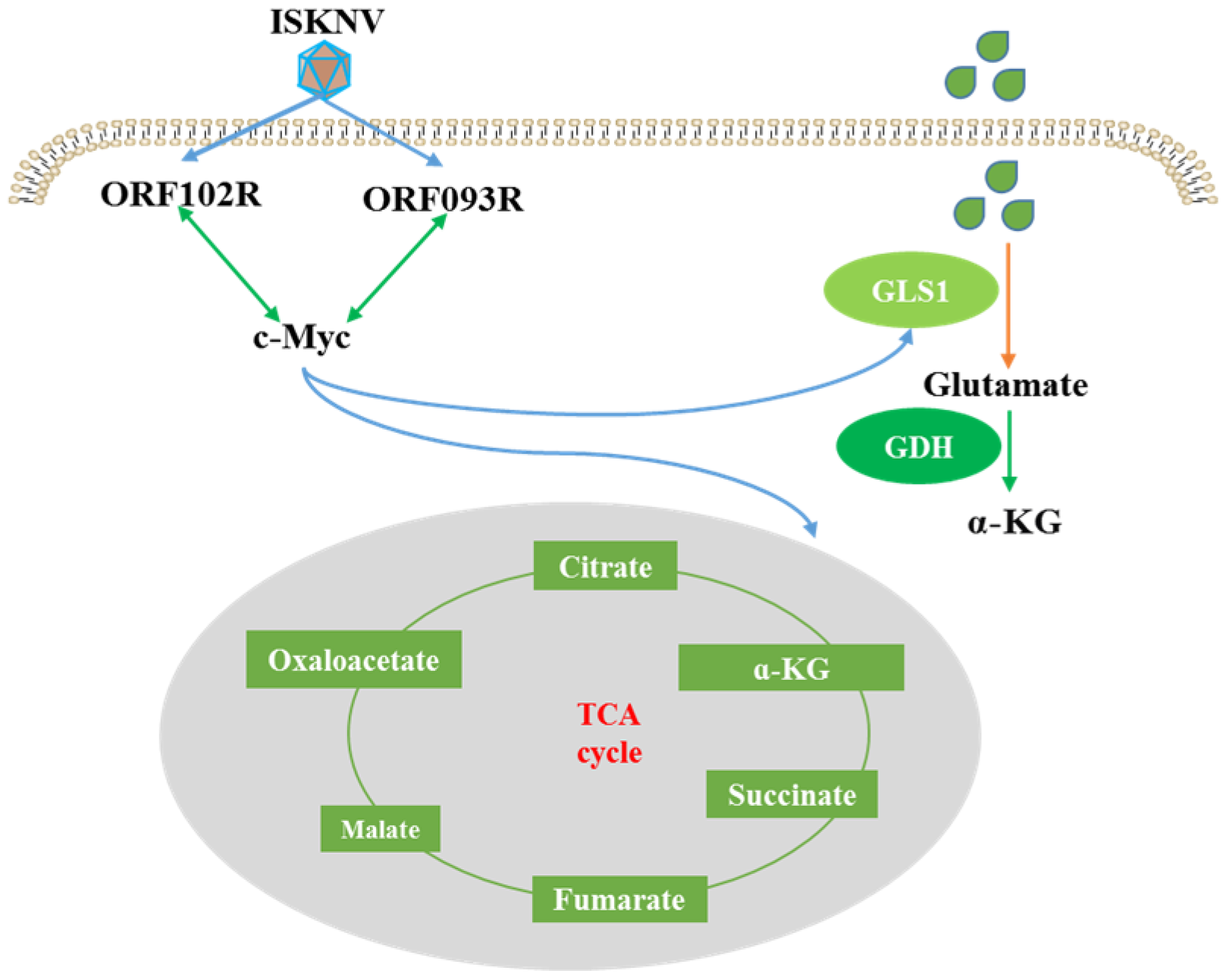
3. Discussion
4. Materials and Methods
4.1. Cells and Viruses
4.2. RNA Extraction and Relative Fluorescence Quantification PCR
4.3. Cell Protein Sample Preparation and Western Blotting
4.4. Immunoprecipitation and Mass Spectrometry
4.5. Eukaryotic Plasmid Construction and Fluorescence Colocalization
4.6. Co-IP
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liang, H.; Zhang, L.; Fu, X.; Lin, Q.; Liu, L.; Niu, Y.; Luo, X.; Huang, Z.; Li, N. Development of a Double-Antibody Sandwich ELISA for Rapid Detection of the MCP Antigen Concentration in Inactivated ISKNV Vaccines. Vaccines 2021, 9, 1264. [Google Scholar] [CrossRef] [PubMed]
- Alathari, S.; Joseph, A.; Bolaños, L.M.; Studholme, D.J.; Jeffries, A.R.; Appenteng, P.; Duodu, K.A.; Sawyerr, E.B.; Paley, R.; Tyler, C.R.; et al. In field use of water samples for genomic surveillance of infectious spleen and kidney necrosis virus (ISKNV) infecting tilapia fish in Lake Volta, Ghana. PeerJ 2024, 12, e17605. [Google Scholar] [CrossRef] [PubMed]
- Bhowmick, N.; Posadas, E.; Ellis, L.; Freedland, S.J.; Di Vizio, D.; Freeman, M.R.; Theodorescu, D.; Figlin, R.; Gong, J. Targeting Glutamine Metabolism in Prostate Cancer. Front. Biosci.-Elite 2023, 15, 2. [Google Scholar] [CrossRef]
- Fu, X.; Hu, X.; Li, N.; Zheng, F.; Dong, X.; Duan, J.; Lin, Q.; Tu, J.; Zhao, L.; Huang, Z.; et al. Glutamine and glutaminolysis are required for efficient replication of infectious spleen and kidney necrosis virus in Chinese perch brain cells. Oncotarget 2017, 8, 2400–2412. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Li, N.; Niu, Y.; Lin, Q.; Luo, X.; Liang, H.; Liu, L.; Fu, X. Characterization and function of mandarin fish c-Myc during viral infection process. Fish Shellfish Immunol. 2022, 120, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Liu, S.; Li, N.; Zuo, S.; Niu, Y.; Lin, Q.; Liang, H.; Luo, X.; Fu, X. Mandarin Fish (Siniperca chuatsi) p53 Regulates Glutaminolysis Induced by Virus via the p53/miR145-5p/c-Myc Pathway in Chinese Perch Brain Cells. Microbiol. Spectr. 2022, 10, e02727-21. [Google Scholar] [CrossRef]
- Liu, S.; Li, N.; Lin, Q.; Liu, L.; Niu, Y.; Liang, H.; Huang, Z.; Fu, X. Glutaminase 1 in mandarin fish Siniperca chuatsi: Molecular characterization, expression pattern and function involving in virus replication. Aquaculture 2020, 519, 734924. [Google Scholar] [CrossRef]
- Xu, K.; Ding, J.; Zhou, L.; Li, D.; Luo, J.; Wang, W.; Shang, M.; Lin, B.; Zhou, L.; Zheng, S. SMYD2 Promotes Hepatocellular Carcinoma Progression by Reprogramming Glutamine Metabolism via c-Myc/GLS1 Axis. Cells 2022, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, X.; Wu, L.; Pei, M.; Li, H.; Jiang, Y. miR-145 inhibits glutamine metabolism through c-myc/GLS1 pathways in ovarian cancer cells. Cell Biol. Int. 2019, 43, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Purhonen, J.; Klefström, J.; Kallijärvi, J. MYC—An emerging player in mitochondrial diseases. Front. Cell Dev. Biol. 2023, 11, 1257651. [Google Scholar] [CrossRef]
- Dittmer, D.P.; Sanchez, E.L.; Carroll, P.A.; Thalhofer, A.B.; Lagunoff, M. Latent KSHV Infected Endothelial Cells Are Glutamine Addicted and Require Glutaminolysis for Survival. PLOS Pathog. 2015, 11, e1005052. [Google Scholar]
- Smallwood, H.S.; Duan, S.; Morfouace, M.; Rezinciuc, S.; Shulkin, B.L.; Shelat, A.; Zink, E.E.; Milasta, S.; Bajracharya, R.; Oluwaseum, A.J.; et al. Targeting Metabolic Reprogramming by Influenza Infection for Therapeutic Intervention. Cell Rep. 2017, 19, 1640–1653. [Google Scholar] [CrossRef]
- Yu, Y.; Clippinger, A.J.; Alwine, J.C. Viral effects on metabolism: Changes in glucose and glutamine utilization during human cytomegalovirus infection. Trends Microbiol. 2011, 19, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Yuneva, M.; Zamboni, N.; Oefner, P.; Sachidanandam, R.; Lazebnik, Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J. Cell Biol. 2007, 178, 93–105. [Google Scholar] [CrossRef]
- Blaheta, R.A.; Weich, E.; Marian, D.; Bereiter-Hahn, J.; Jones, J.; Jonas, D.; Michaelis, M.; Doerr, H.W.; Cinatl, J. Human Cytomegalovirus Infection Alters PC3 Prostate Carcinoma Cell Adhesion to Endothelial Cells, Extracellular Matrix. Neoplasia 2006, 8, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Thai, M.; Thaker, S.K.; Feng, J.; Du, Y.; Hu, H.; Wu, T.T.; Graeber, T.G.; Braas, D.; Christofk, H.R. MYC-induced reprogramming of glutamine catabolism supports optimal virus replication. Nat. Commun. 2015, 6, 8873. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Petrashen, A.P.; Sanders, J.A.; Peterson, A.L.; Sedivy, J.M. SLC1A5 glutamine transporter is a target of MYC and mediates reduced mTORC1 signaling and increased fatty acid oxidation in long-lived Myc hypomorphic mice. Aging Cell. 2019, 18, e12947. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Sayed, N.; Ditsworth, D.; Thompson, C.B. Brick by brick: Metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 2008, 18, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.-A.; Hong, J.; Asaka, R.; Asaka, S.; Hsu, F.-C.; Rahmanto, Y.S.; Jung, J.-G.; Chen, Y.-W.; Yen, T.-T.; Tomaszewski, A.; et al. Inhibition of the MYC-Regulated Glutaminase Metabolic Axis Is an Effective Synthetic Lethal Approach for Treating Chemoresistant Ovarian Cancers. Cancer Res. 2020, 80, 4514–4526. [Google Scholar] [CrossRef] [PubMed]
- Tapparel, C.; Hafner, A.; Meurs, N.; Garner, A.; Azar, E.; Kannan, A.; Passalacqua, K.D.; Nagrath, D.; Wobus, C.E. Norovirus NS1/2 protein increases glutaminolysis for efficient viral replication. PLoS Pathog. 2024, 20, e1011909. [Google Scholar]
- Fontaine, K.A.; Camarda, R.; Lagunoff, M. Vaccinia Virus Requires Glutamine but Not Glucose for Efficient Replication. J. Virol. 2014, 88, 4366–4374. [Google Scholar] [CrossRef] [PubMed]
- Chambers, J.W.; Maguire, T.G.; Alwine, J.C. Glutamine Metabolism Is Essential for Human Cytomegalovirus Infection. J. Virol. 2010, 84, 1867–1873. [Google Scholar] [CrossRef] [PubMed]
- Ritter, J.B.; Wahl, A.S.; Freund, S.; Genzel, Y.; Reichl, U. Metabolic effects of influenza virus infection in cultured animal cells: Intra- and extracellular metabolite profiling. BMC Syst. Biol. 2010, 4, 61. [Google Scholar] [CrossRef] [PubMed]
- Tambay, V.; Raymond, V.-A.; Bilodeau, M. MYC Rules: Leading Glutamine Metabolism toward a Distinct Cancer Cell Phenotype. Cancers 2021, 13, 4484. [Google Scholar] [CrossRef] [PubMed]
- Quek, L.-E.; van Geldermalsen, M.; Guan, Y.F.; Wahi, K.; Mayoh, C.; Balaban, S.; Pang, A.; Wang, Q.; Cowley, M.J.; Brown, K.K.; et al. Glutamine addiction promotes glucose oxidation in triple-negative breast cancer. Oncogene 2022, 41, 4066–4078. [Google Scholar] [CrossRef]
- Wicker, C.A.; Hunt, B.G.; Krishnan, S.; Aziz, K.; Parajuli, S.; Palackdharry, S.; Elaban, W.R.; Wise-Draper, T.M.; Mills, G.B.; Waltz, S.E.; et al. Glutaminase inhibition with telaglenastat (CB-839) improves treatment response in combination with ionizing radiation in head and neck squamous cell carcinoma models. Cancer Lett. 2021, 502, 180–188. [Google Scholar] [CrossRef]
- Hensley, C.T.; Wasti, A.T.; DeBerardinis, R.J. Glutamine and cancer: Cell biology, physiology, and clinical opportunities. J. Clin. Investig. 2013, 123, 3678–3684. [Google Scholar] [CrossRef]
- Marhl, M. What do stimulated beta cells have in common with cancer cells? BioSystems 2024, 242, 105257. [Google Scholar] [CrossRef]
- Shi, F.; He, Y.; Li, J.; Tang, M.; Li, Y.; Xie, L.; Zhao, L.; Hu, J.; Luo, X.; Zhou, M.; et al. Wild-type IDH2 contributes to Epstein–Barr virus-dependent metabolic alterations and tumorigenesis. Mol. Metab. 2020, 36, 100966. [Google Scholar] [CrossRef]
- Mosavi, L.K.; Cammett, T.J.; Desrosiers, D.C.; Peng, Z. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2009, 13, 1435–1448. [Google Scholar] [CrossRef]
- Fu, X.; Li, N.; Lai, Y.; Luo, X.; Wang, Y.; Shi, C.; Huang, Z.; Wu, S.; Su, J. A novel fish cell line derived from the brain of Chinese perch Siniperca chuatsi: Development and characterization. J. Fish Biol. 2015, 86, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Lin, Q.; Liang, H.; Liu, L.; Huang, Z.; Li, N.; Su, J. The biological features and genetic diversity of novel fish rhabdovirus isolates in China. Arch. Virol. 2017, 162, 2829–2834. [Google Scholar] [CrossRef] [PubMed]


| Protein IDs | Intensity | MW (kDa) | Predicted | Description |
|---|---|---|---|---|
| E2CU38 | 68,329 | 53.68 | 102R | Ankyrin repeat-containing |
| Q4KS43 | 624,010 | 25.3 | 118L | Novel viral envelope |
| Q8QUM5 | 7561.5 | 22.65 | 085R | Unknown |
| M1SWU3 | 14,918 | 28.95 | 087R | Ribonuclease III family |
| Q5YF04 | 221,650 | 32.7 | 093R | Transmembrane protein |
| E2CU56 | 26,090 | 32.7 | 111R | C3HC4 type |
| Primer Name | Sequence(5′–3′) |
|---|---|
| OFR 102R-F | (KpnI) CGGGGTACCGCCACCATGGCAGCAAACCAGACCATTG |
| OFR 102R-R | (BamHI) CGCGGATCCTTA CTTATCGTCGTCATCCTTGTAATCCGTTCCGCGCACACAAG (56 °C) |
| OFR 093R-F | (KpnI) CGGGGTACCGCCACCATGACATCGAGGCATACATCAGACGC |
| OFR 093R-R | (BamHI) CGCGGATCCTTA CTTATCGTCGTCATCCTTGTAATCGAGCCGTATGGACTTGTGC (58 °C) |
| OFR 118L-F | (KpnI) CGGGGTACCGCCACCATGGCATTCGCTACTACAATGAGAGT |
| OFR 118L-R | (EcoRI) CGGAATTCTTA CTTATCGTCGTCATCCTTGTAATCCACTTGACCAAAGAGGTCCT (56 °C) |
| 18S-F | CATTCGTATTGTGCCGCTAGA |
| 18S-R | CAAATGCTTTCGCTTTGGTC |
| c-Myc | ACCGCCACCTTCATCCTCTTCC (MN759310.1) |
| c-Myc | CGCCGCTCCTCCTCATCCTC |
| GLS1-F | TCCTGCGGCATGTACGACTTCT (MN267067) |
| GLS1-R | CCAGCTTGTCCAGTGGAGGTGA |
| GDH-F | AGGTCCGTCACTATGCCGATGC |
| GDH-R | AGATCCTCCACCAGCTTGTCCTC |
| IDH2-F | GTCATCAGTGTGGTCACGGTACG |
| IDH2-R | TGGAGATGGACGGAGACGAGATG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, Y.; Ye, C.; Lin, Q.; Liang, H.; Luo, X.; Ma, B.; Li, N.; Fu, X. Infectious Spleen and Kidney Necrosis Virus ORF093R and ORF102R Regulate Glutamate Metabolic Reprogramming to Support Virus Proliferation by Interacting with c-Myc. Int. J. Mol. Sci. 2025, 26, 718. https://doi.org/10.3390/ijms26020718
Niu Y, Ye C, Lin Q, Liang H, Luo X, Ma B, Li N, Fu X. Infectious Spleen and Kidney Necrosis Virus ORF093R and ORF102R Regulate Glutamate Metabolic Reprogramming to Support Virus Proliferation by Interacting with c-Myc. International Journal of Molecular Sciences. 2025; 26(2):718. https://doi.org/10.3390/ijms26020718
Chicago/Turabian StyleNiu, Yinjie, Caimei Ye, Qiang Lin, Hongru Liang, Xia Luo, Baofu Ma, Ningqiu Li, and Xiaozhe Fu. 2025. "Infectious Spleen and Kidney Necrosis Virus ORF093R and ORF102R Regulate Glutamate Metabolic Reprogramming to Support Virus Proliferation by Interacting with c-Myc" International Journal of Molecular Sciences 26, no. 2: 718. https://doi.org/10.3390/ijms26020718
APA StyleNiu, Y., Ye, C., Lin, Q., Liang, H., Luo, X., Ma, B., Li, N., & Fu, X. (2025). Infectious Spleen and Kidney Necrosis Virus ORF093R and ORF102R Regulate Glutamate Metabolic Reprogramming to Support Virus Proliferation by Interacting with c-Myc. International Journal of Molecular Sciences, 26(2), 718. https://doi.org/10.3390/ijms26020718





