Vanadium-Dependent Haloperoxidase Gene Evolution in Brown Algae: Evidence for Horizontal Gene Transfer
Abstract
1. Introduction
2. Result
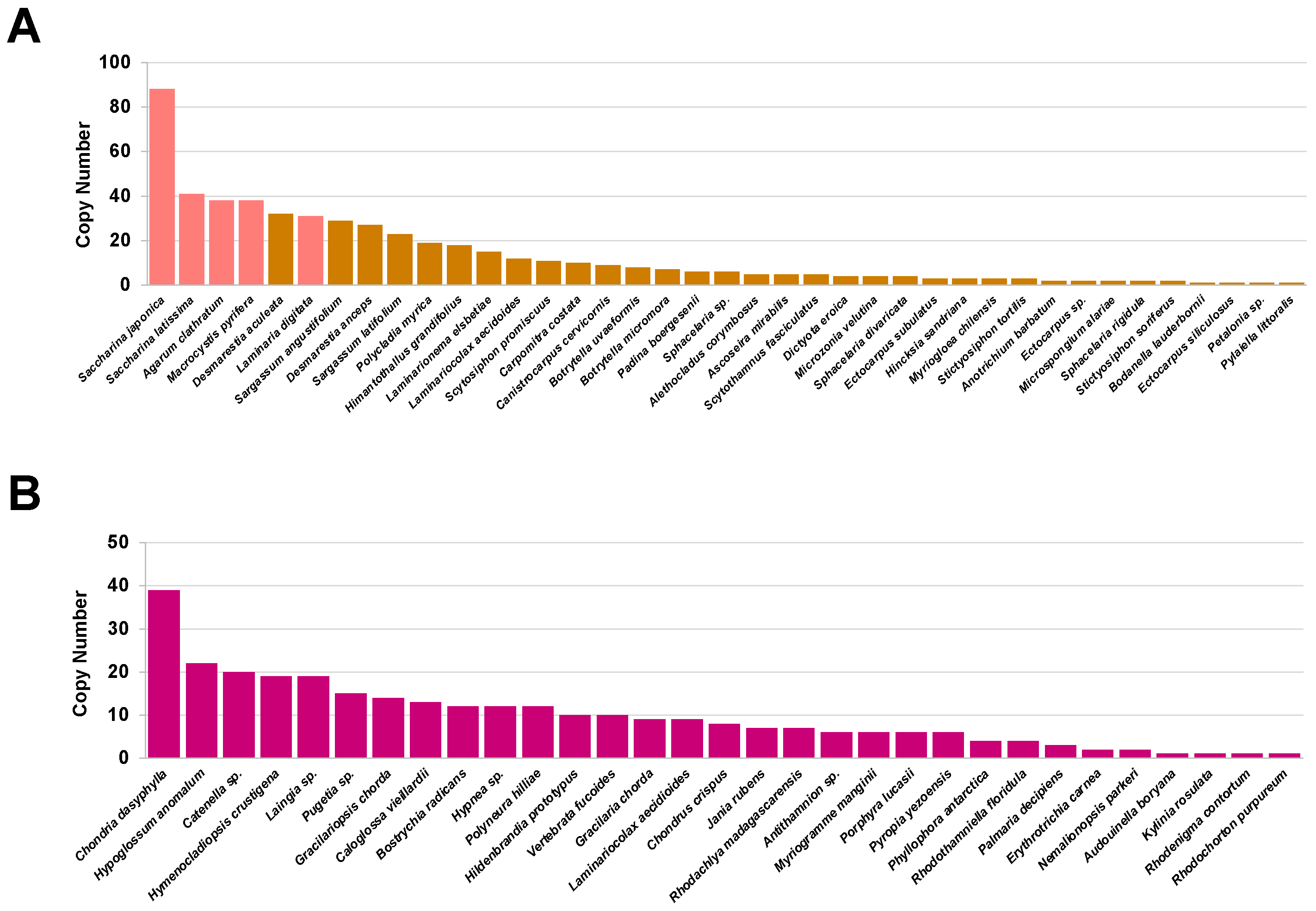
2.1. The V-HPO Gene Family Is Extensively Expanded in the Brown Algae Genome
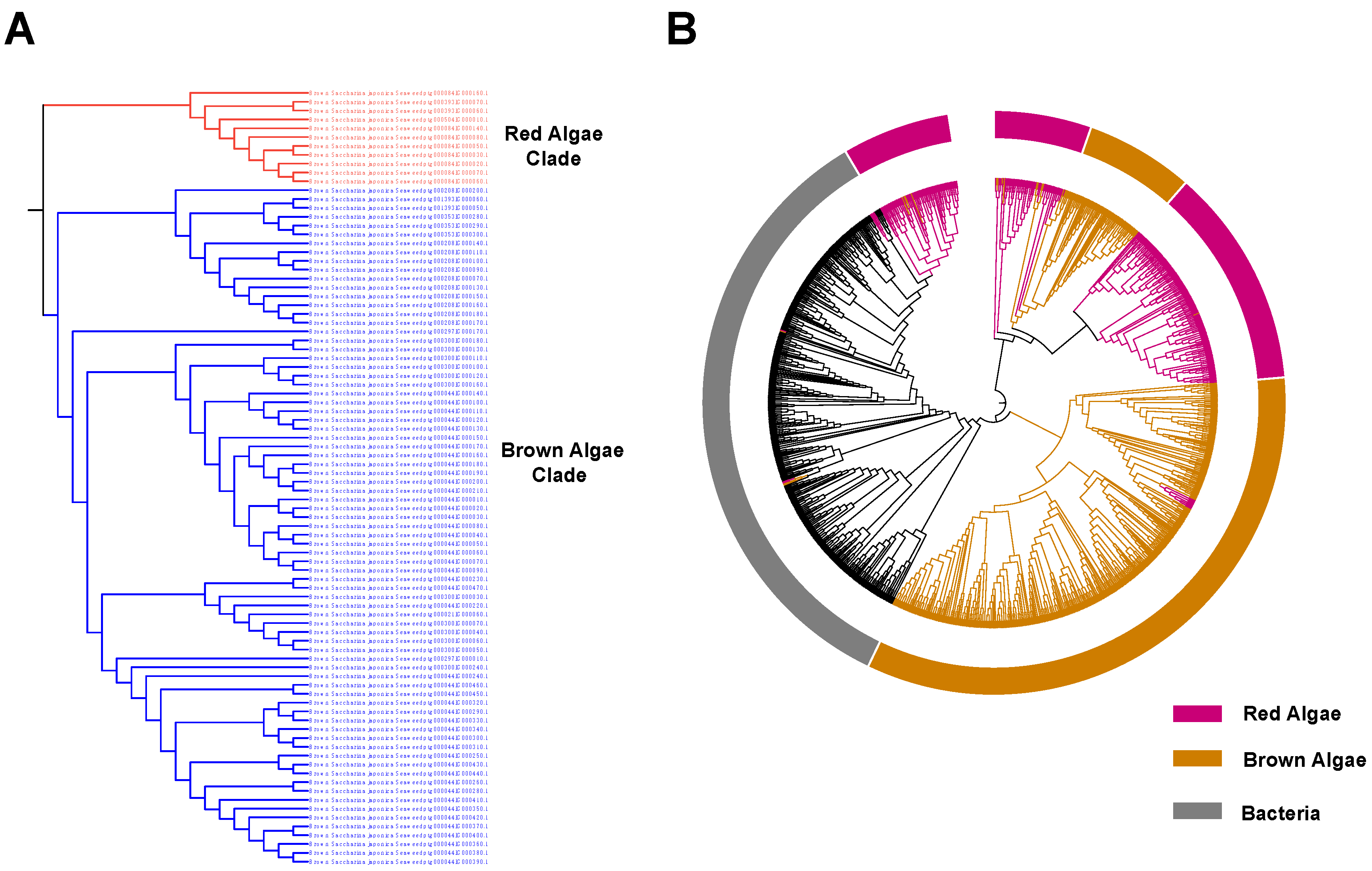
2.2. The V-HPOs in Brown Algae and Red Algae Exhibit Distinct Differences
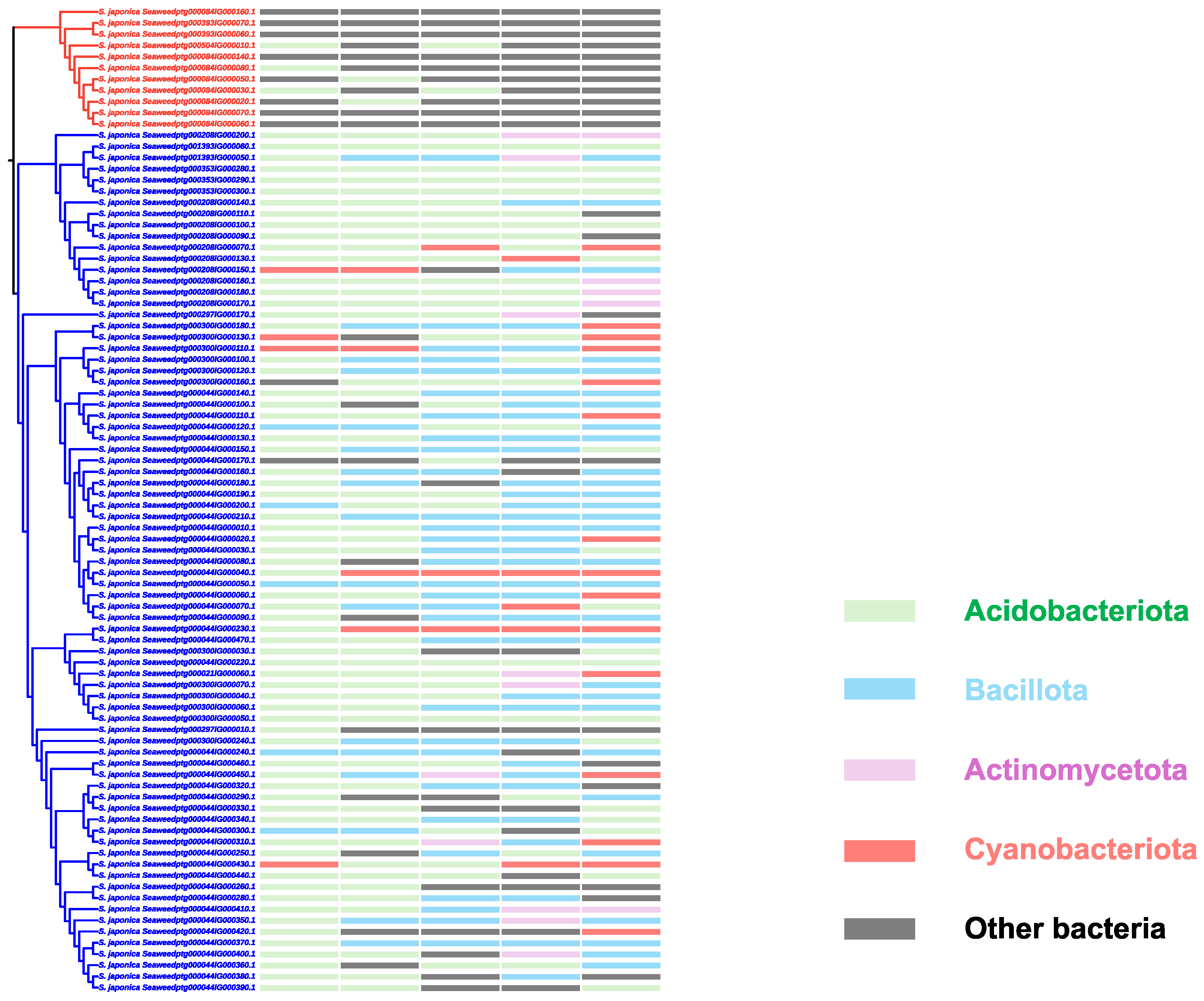
2.3. The Brown Algae Clade V-HPOs in S. japonica Are Probably Horizontally Transferred from Bacteria
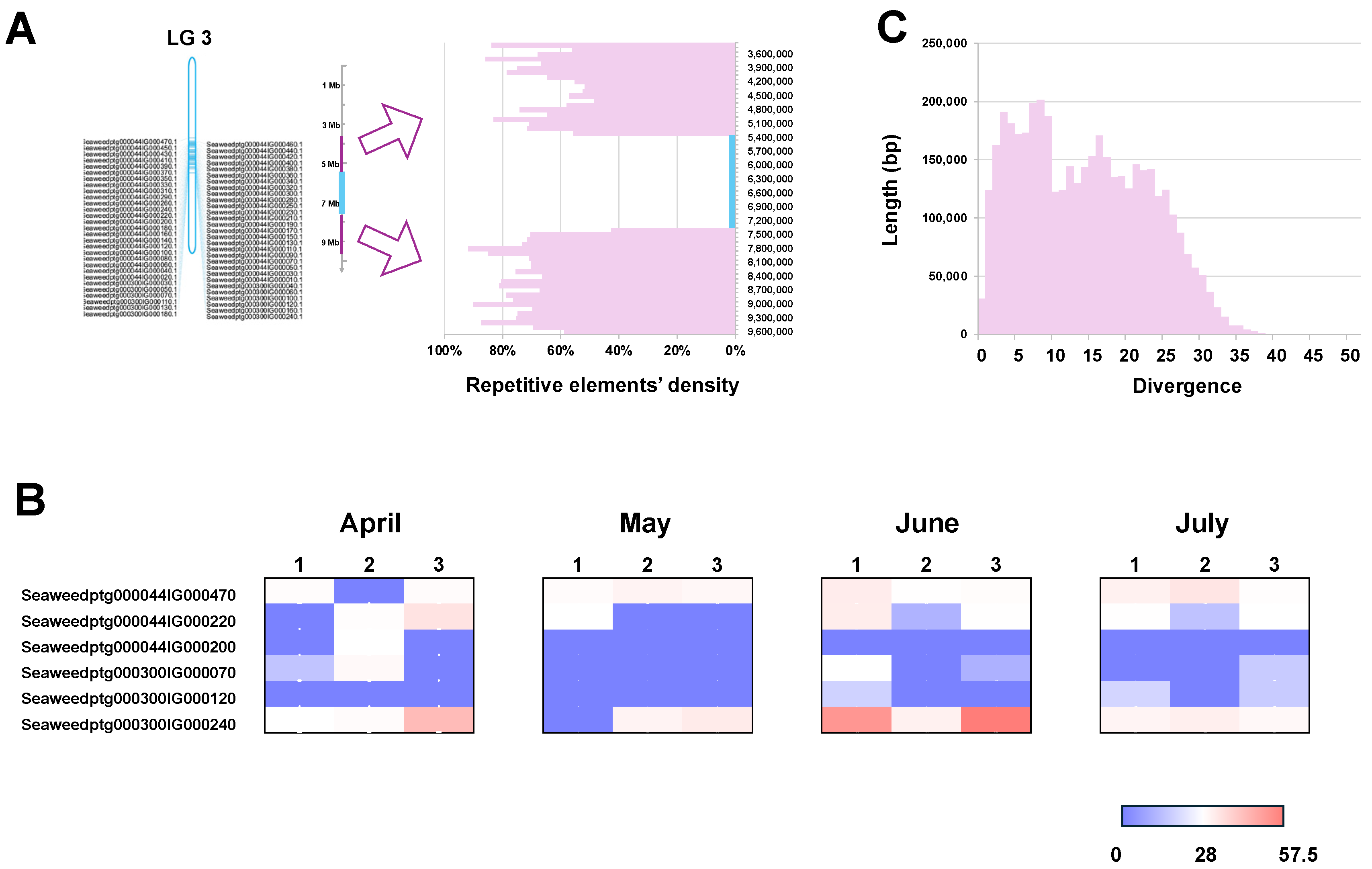
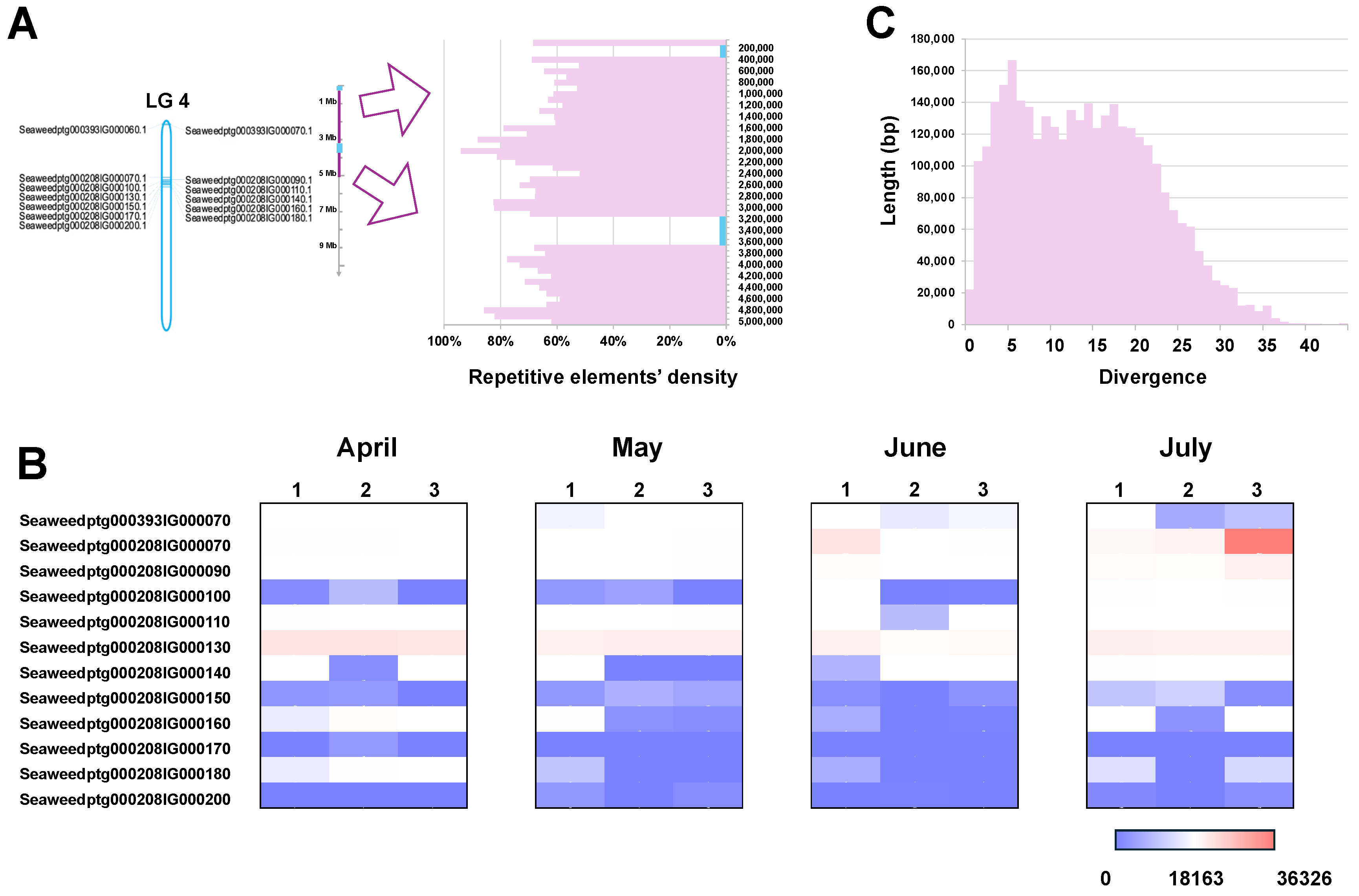
2.4. Tandem Duplication of V-HPOs in the Genome Is Mediated by Active Transposable Elements
3. Discussion
4. Materials and Methods
4.1. Sequence Collection and Phylogenetic Analysis
4.2. Comparative Analyses of Algae and Bacteria V-HPOs
4.3. Repetitive Element Analysis
4.4. Characterization of the Gene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holdt, S.L.; Kraan, S. Bioactive compounds in seaweed: Functional food applications and legislation. J. Appl. Phycol. 2011, 23, 543–597. [Google Scholar] [CrossRef]
- Küpper, F.; Schweigert, N.; Ar Gall, E.; Legendre, J.-M.; Vilter, H.; Kloareg, B. Iodine uptake in Laminariales involves extracellular, haloperoxidase-mediated oxidation of iodide. Planta 1998, 207, 163–171. [Google Scholar] [CrossRef]
- Clark, C.D.; Bassett, B.; Burge, M.R. Effects of kelp supplementation on thyroid function in euthyroid subjects. Endocr. Pract. 2003, 9, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.B.; Boelaert, K. Iodine deficiency and thyroid disorders. Lancet Diabetes Endocrinol. 2015, 3, 286–295. [Google Scholar] [CrossRef]
- Bhattacharya, D.; Yoon, H.S.; Hackett, J.D. Photosynthetic eukaryotes unite: Endosymbiosis connects the dots. Bioessays 2004, 26, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.E. Phycology; Cambridge University Press: Cambridge, UK, 2018. [Google Scholar]
- Barsanti, L.; Gualtieri, P. Algae: Anatomy, Biochemistry, and Biotechnology; CRC Press: Boka Raton, FL, USA, 2022. [Google Scholar]
- Küpper, F.C.; Carpenter, L.J.; Mcfiggans, G.B.; Palmer, C.J.; Waite, T.J.; Boneberg, E.-M.; Woitsch, S.; Weiller, M.; Abela, R.; Grolimund, D. Iodide accumulation provides kelp with an inorganic antioxidant impacting atmospheric chemistry. Proc. Natl. Acad. Sci. USA 2008, 105, 6954–6958. [Google Scholar] [CrossRef] [PubMed]
- Küpper, F.C.; Carrano, C.J. Key aspects of the iodine metabolism in brown algae: A brief critical review. Metallomics 2019, 11, 756–764. [Google Scholar] [CrossRef]
- Wever, R.; van der Horst, M.A. The role of vanadium haloperoxidases in the formation of volatile brominated compounds and their impact on the environment. Dalton Trans. 2013, 42, 11778–11786. [Google Scholar] [CrossRef]
- Winter, J.M.; Moore, B.S. Exploring the chemistry and biology of vanadium-dependent haloperoxidases. J. Biol. Chem. 2009, 284, 18577–18581. [Google Scholar] [CrossRef]
- Isupov, M.N.; Dalby, A.R.; Brindley, A.A.; Izumi, Y.; Tanabe, T.; Murshudov, G.N.; Littlechild, J.A. Crystal structure of dodecameric vanadium-dependent bromoperoxidase from the red algae Corallina officinalis. J. Mol. Biol. 2000, 299, 1035–1049. [Google Scholar] [CrossRef] [PubMed]
- Colin, C.; Leblanc, C.; Michel, G.; Wagner, E.; Leize-Wagner, E.; Dorsselaer, A.V.; Potin, P. Vanadium-dependent iodoperoxidases in Laminaria digitata, a novel biochemical function diverging from brown algal bromoperoxidases. JBIC J. Biol. Inorg. Chem. 2005, 10, 156–166. [Google Scholar] [CrossRef]
- Rehder, D. Vanadate-dependent peroxidases in macroalgae: Function, applications, and environmental impact. Oceanography 2014, 2, 1000121. [Google Scholar]
- Wever, R.; Krenn, B.; Renirie, R. Chapter Six-Marine vanadium-dependent haloperoxidases, their isolation, characterization, and application. In Methods Enzymol; Moore, B.S., Ed.; Academic Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Balakirev, E.S.; Krupnova, T.N.; Ayala, F.J. Symbiotic associations in the phenotypically-diverse brown alga Saccharina japonica. PLoS ONE 2012, 7, e39587. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Zhang, X.; Chang, L.; Xiao, L.; Ahmad, R.; Saha, M.; Wu, H.; Wang, G. Dynamic shift of the epibacterial communities on commercially cultivated Saccharina japonica from mature sporophytes to sporelings and juvenile sporophytes. J. Appl. Phycol. 2021, 33, 1171–1179. [Google Scholar] [CrossRef]
- Hu, Z.M.; Shan, T.F.; Zhang, Q.S.; Liu, F.L.; Jueterbock, A.; Wang, G.; Sun, Z.M.; Wang, X.Y.; Chen, W.Z.; Critchley, A.T. Kelp breeding in China: Challenges and opportunities for solutions. Rev. Aquac. 2024, 16, 855–871. [Google Scholar] [CrossRef]
- Matveeva, T.V.; Otten, L. Widespread occurrence of natural genetic transformation of plants by Agrobacterium. Plant Mol. Biol. 2019, 101, 415–437. [Google Scholar] [CrossRef]
- Nelson, D.R.; Mystikou, A.; Jaiswal, A.; Rad-Menendez, C.; Preston, M.J.; de Boever, F.; El Assal, D.C.; Daakour, S.; Lomas, M.W.; Twizere, J.-C. Macroalgal deep genomics illuminate multiple paths to aquatic, photosynthetic multicellularity. Mol. Plant 2024, 17, 747–771. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Zhang, X.; Miao, M.; Fan, X.; Zheng, Y.; Xu, D.; Wang, J.; Zhou, L.; Wang, D.; Gao, Y. Saccharina genomes provide novel insight into kelp biology. Nat. Commun. 2015, 6, 6986. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.R.; Chance, R.; Bell, T.; Jones, O.; Loades, D.C.; May, R.; Tinel, L.; Weddell, K.; Widdicombe, C.; Carpenter, L.J. Iodide, iodate & dissolved organic iodine in the temperate coastal ocean. Front. Mar. Sci. 2024, 11, 1277595. [Google Scholar]
- Carpenter, L.J.; Chance, R.J.; Sherwen, T.; Adams, T.J.; Ball, S.M.; Evans, M.J.; Hepach, H.; Hollis, L.D.; Hughes, C.; Jickells, T.D. Marine iodine emissions in a changing world. Proc. R. Soc. A 2021, 477, 20200824. [Google Scholar] [CrossRef] [PubMed]
- Bergamini, C.M.; Gambetti, S.; Dondi, A.; Cervellati, C. Oxygen, reactive oxygen species and tissue damage. Curr. Pharm. Des. 2004, 10, 1611–1626. [Google Scholar] [CrossRef] [PubMed]
- Amachi, S. Microbial contribution to global iodine cycling: Volatilization, accumulation, reduction, oxidation, and sorption of iodine. Microbes Environ. 2008, 23, 269–276. [Google Scholar] [CrossRef]
- Carpenter, L.J.; Nightingale, P.D. Chemistry and release of gases from the surface ocean. Chem. Rev. 2015, 115, 4015–4034. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, L.J.; Macdonald, S.M.; Shaw, M.D.; Kumar, R.; Saunders, R.W.; Parthipan, R.; Wilson, J.; Plane, J.M. Atmospheric iodine levels influenced by sea surface emissions of inorganic iodine. Nat. Geosci. 2013, 6, 108–111. [Google Scholar] [CrossRef]
- Javier, L.H.; Benzekri, H.; Gut, M.; Claros, M.G.; van Bergeijk, S.; Canavate, J.P.; Manchado, M. Characterization of Iodine-Related molecular processes in the marine microalga Tisochrysis lutea (Haptophyta). Front. Mar. Sci. 2018, 5, 134. [Google Scholar] [CrossRef]
- Gribble, G.W. Biological activity of recently discovered halogenated marine natural products. Mar. Drugs 2015, 13, 4044–4136. [Google Scholar] [CrossRef]
- Grzanka, M.; Smoleń, S.; Kováčik, P. Effect of vanadium on the uptake and distribution of organic and inorganic forms of iodine in sweetcorn plants during early-stage development. Agronomy 2020, 10, 1666. [Google Scholar] [CrossRef]
- Denoeud, F.; Godfroy, O.; Cruaud, C.; Heesch, S.; Nehr, Z.; Tadrent, N.; Couloux, A.; Brillet-Guéguen, L.; Delage, L.; Mckeown, D. Evolutionary genomics of the emergence of brown algae as key components of coastal ecosystems. Cell 2024, 187, 6943.e39–6965.e39. [Google Scholar] [CrossRef] [PubMed]
- Cock, J.M.; Sterck, L.; Rouzé, P.; Scornet, D.; Allen, A.E.; Amoutzias, G.; Anthouard, V.; Artiguenave, F.; Aury, J.-M.; Badger, J.H. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 2010, 465, 617–621. [Google Scholar] [CrossRef]
- Lerminiaux, N.A.; Cameron, A.D. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef]
- Schönknecht, G.; Weber, A.P.; Lercher, M.J. Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution. Bioessays 2014, 36, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Burki, F.; Okamoto, N.; Pombert, J.-F.; Keeling, P.J. The evolutionary history of haptophytes and cryptophytes: Phylogenomic evidence for separate origins. Proc. R. Soc. B Biol. Sci. 2012, 279, 2246–2254. [Google Scholar] [CrossRef]
- Domozych, D.S.; Ciancia, M.; Fangel, J.U.; Mikkelsen, M.D.; Ulvskov, P.; Willats, W.G. The cell walls of green algae: A journey through evolution and diversity. Front. Plant Sci. 2012, 3, 82. [Google Scholar] [CrossRef]
- Song, W.; Wemheuer, B.; Steinberg, P.D.; Marzinelli, E.M.; Thomas, T. Contribution of horizontal gene transfer to the functionality of microbial biofilm on a macroalgae. ISME J. 2021, 15, 807–817. [Google Scholar] [CrossRef]
- Li, F.-W.; Villarreal, J.C.; Kelly, S.; Rothfels, C.J.; Melkonian, M.; Frangedakis, E.; Ruhsam, M.; Sigel, E.M.; Der, J.P.; Pittermann, J. Horizontal transfer of an adaptive chimeric photoreceptor from bryophytes to ferns. Proc. Natl. Acad. Sci. USA 2014, 111, 6672–6677. [Google Scholar] [CrossRef]
- Kielak, A.M.; Cipriano, M.A.; Kuramae, E.E. Acidobacteria strains from subdivision 1 act as plant growth-promoting bacteria. Arch. Microbiol. 2016, 198, 987–993. [Google Scholar] [CrossRef]
- Quaiser, A.; Ochsenreiter, T.; Lanz, C.; Schuster, S.C.; Treusch, A.H.; Eck, J.; Schleper, C. Acidobacteria form a coherent but highly diverse group within the bacterial domain: Evidence from environmental genomics. Mol. Microbiol. 2003, 50, 563–575. [Google Scholar] [CrossRef]
- Ward, N.L.; Challacombe, J.F.; Janssen, P.H.; Henrissat, B.; Coutinho, P.M.; Wu, M.; Xie, G.; Haft, D.H.; Sait, M.; Badger, J. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 2009, 75, 2046–2056. [Google Scholar] [CrossRef]
- Sjöstrand, J.; Arvestad, L.; Lagergren, J.; Sennblad, B. GenPhyloData: Realistic simulation of gene family evolution. BMC Bioinform. 2013, 14, 1–5. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, L.D.; Young, E.; Delaney, J.; Ruhnau, F.; Ritchie, K.B.; Paul, J.H. High frequency of horizontal gene transfer in the oceans. Science 2010, 330, 50. [Google Scholar] [CrossRef]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef]
- Keeling, P.J.; Palmer, J.D. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 2008, 9, 605–618. [Google Scholar] [CrossRef]
- Boto, L. Horizontal gene transfer in evolution: Facts and challenges. Proc. R. Soc. B Biol. Sci. 2010, 277, 819–827. [Google Scholar] [CrossRef]
- Brown, J.R. Ancient horizontal gene transfer. Nat. Rev. Genet. 2003, 4, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Feschotte, C.; Pritham, E.J. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007, 41, 331–368. [Google Scholar] [CrossRef] [PubMed]
- Krasileva, K.V. The role of transposable elements and DNA damage repair mechanisms in gene duplications and gene fusions in plant genomes. Curr. Opin. Plant Biol. 2019, 48, 18–25. [Google Scholar] [CrossRef]
- Ahmed, M.; Liang, P. Transposable elements are a significant contributor to tandem repeats in the human genome. Int. J. Genom. 2012, 2012, 947089. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.; Potter, S.C.; Finn, R.D. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Bao, Z.; Eddy, S.R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002, 12, 1269–1276. [Google Scholar] [CrossRef]
- Price, A.L.; Jones, N.C.; Pevzner, P.A. De novo identification of repeat families in large genomes. Bioinformatics 2005, 21, i351–i358. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 1–6. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A user-friendly online tool for drawing genetic maps. Mol. Hortic. 2021, 1, 1–4. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Z.; Zhang, J.; Duan, D. Vanadium-Dependent Haloperoxidase Gene Evolution in Brown Algae: Evidence for Horizontal Gene Transfer. Int. J. Mol. Sci. 2025, 26, 716. https://doi.org/10.3390/ijms26020716
Yuan Z, Zhang J, Duan D. Vanadium-Dependent Haloperoxidase Gene Evolution in Brown Algae: Evidence for Horizontal Gene Transfer. International Journal of Molecular Sciences. 2025; 26(2):716. https://doi.org/10.3390/ijms26020716
Chicago/Turabian StyleYuan, Zihao, Jie Zhang, and Delin Duan. 2025. "Vanadium-Dependent Haloperoxidase Gene Evolution in Brown Algae: Evidence for Horizontal Gene Transfer" International Journal of Molecular Sciences 26, no. 2: 716. https://doi.org/10.3390/ijms26020716
APA StyleYuan, Z., Zhang, J., & Duan, D. (2025). Vanadium-Dependent Haloperoxidase Gene Evolution in Brown Algae: Evidence for Horizontal Gene Transfer. International Journal of Molecular Sciences, 26(2), 716. https://doi.org/10.3390/ijms26020716





