Molecular Signatures of Schizophrenia and Insights into Potential Biological Convergence
Abstract
1. Introduction
2. Genetic Risk Architecture of Schizophrenia
2.1. Common Variation (SNPs and PRS)
2.2. Rare Variation
2.2.1. Copy Number Variants (CNVs)
2.2.2. Loss-of-Function Coding Mutation
2.3. Gene Regulation and Functional Genomics
2.4. Systems Biology and Pathway Convergence
2.5. Genetic Subtypes and Network Modeling
3. Epigenetic and Chromatin Regulation in Schizophrenia
3.1. Environmental Influences on the Epigenome
3.2. Epigenetic Inhibitory Alterations in Post-Mortem Brain Tissue
3.3. Histone Modifications and Regulatory Landscapes
3.4. Chromatin Architecture and Long-Range Interactions
3.5. Non-Coding RNAs and Peripheral Epigenetic Signatures
3.6. Therapeutic Potential of Epigenetic Modulation
4. Transcriptomic and RNA-Based Dysregulation
4.1. Alternative Splicing
4.2. Non-Coding RNAs
4.3. Tissue Specificity and Peripheral Transcriptomic Profiles
4.4. Co-Expression Networks and Systems-Level Convergence
5. Proteomic and Functional Phenotypes
5.1. Synaptic and Mitochondrial Proteome Disruption
5.2. Immune Signatures and Peripheral Protein Markers
5.3. Post-Translational and Signaling Modifications
6. Induced Pluripotent Stem Cell Models Elucidate Schizophrenia Pathophysiology
6.1. Early Neurodevelopmental Perturbations and Transcriptional Dysregulation
6.2. Mitochondrial Malfunction and Oxidative Stress
6.3. Synaptic Connectivity and Dendritic Architecture
6.4. Organoid and Interneuron Circuit Deficits
6.5. Oligodendrocyte Precursor Cell Dysfunction
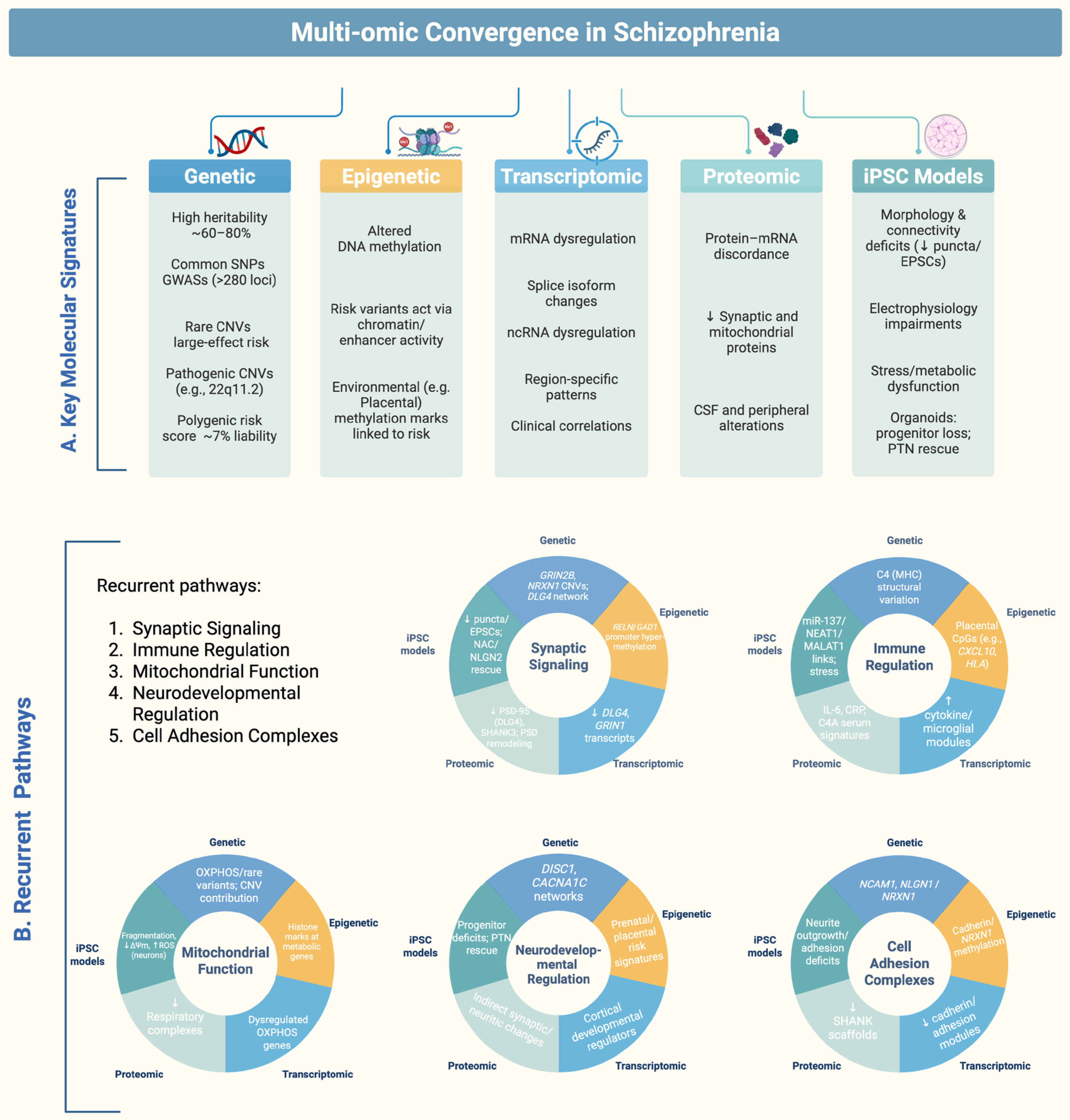
7. Systems Integration: Convergent Molecular Pathways and Cross-Layer Convergence Genes in Schizophrenia
7.1. Synaptic Signaling
7.2. Mitochondrial Bioenergetics
7.3. Cell-Adhesion Complexes
7.4. Immune Regulation
7.5. Neurodevelopmental Regulation
7.6. Cross-Layer Convergence Genes/Proteins
- DLG4 (PSD-95):
- C4A/C4B:
- NRXN1/NLGN1:
- MT-CO1 and ATP5A1 (OXPHOS subunits):
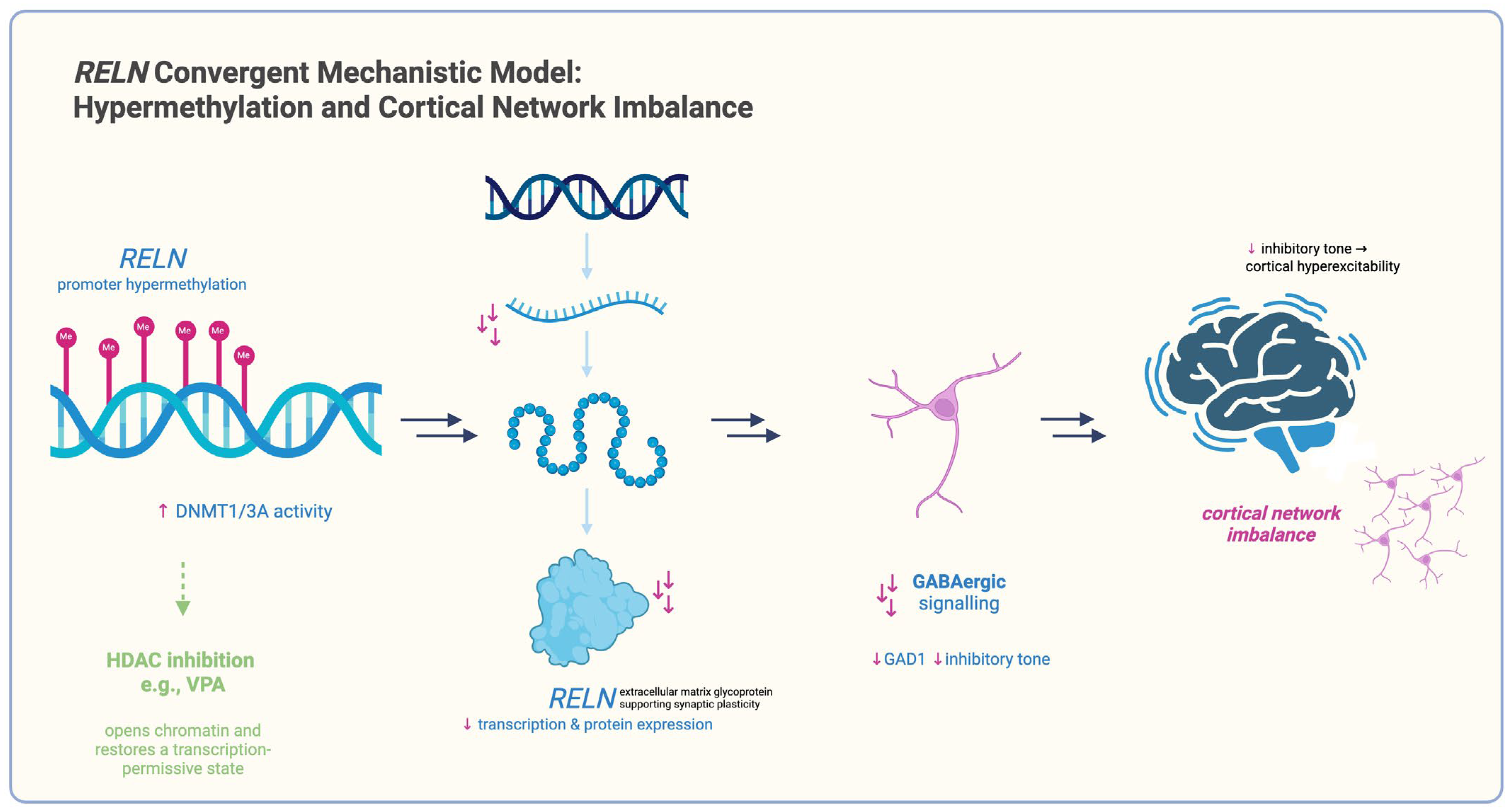
- RELN:
- BRN2 (POU3F2) and PTN (pleiotrophin):
8. Discussion
9. Conclusions and Clinical Outlook
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rubeša, G.; Gudelj, L.; Kubinska, N. Etiology of schizophrenia and therapeutic options. Psychiatr. Danub. 2011, 23, 308–315. [Google Scholar]
- Onaolapo, A.; Onaolapo, O. Schizophrenia Aetiology and Drug Therapy: A Tale of Progressive Demystification and Strides in Management. Adv. Pharmacol. Pharm. 2018, 6, 19–42. [Google Scholar] [CrossRef]
- Hilker, R.; Helenius, D.; Fagerlund, B.; Skytthe, A.; Christensen, K.; Werge, T.M.; Nordentoft, M.; Glenthøj, B. Heritability of Schizophrenia and Schizophrenia Spectrum Based on the Nationwide Danish Twin Register. Biol. Psychiatry 2018, 83, 492–498. [Google Scholar] [CrossRef]
- Hyllested, A. Schizophrenia. Current biological theories on the etiology. Ugeskr. Laeger 1996, 158, 4273–4277. [Google Scholar] [PubMed]
- Walker, E.; Mittal, V.; Tessner, K. Stress and the hypothalamic pituitary adrenal axis in the developmental course of schizophrenia. Annu. Rev. Clin. Psychol. 2008, 4, 189–216. [Google Scholar] [CrossRef] [PubMed]
- Pandarakalam, J.P. The Autoimmune and Infectious Etiological Factors of a Subset of Schizophrenia. Br. J. Med. Pract. 2015, 8, a831. [Google Scholar]
- English, J.A.; Fan, Y.; Föcking, M.; Lopez, L.M.; Hryniewiecka, M.; Wynne, K.; Dicker, P.; Matigian, N.; Cagney, G.; Mackay-Sim, A.; et al. Reduced protein synthesis in schizophrenia patient-derived olfactory cells. Transl. Psychiatry 2015, 5, e663. [Google Scholar] [CrossRef]
- Obiols, J.E.; Vicens-Vilanova, J. Etiología y Signos de Riesgo en la Esquizofrenia. Int. J. Psychol. Psychol. Ther. 2003, 3, 235–250. [Google Scholar]
- Insel, T.R. Rethinking schizophrenia. Nature 2010, 468, 187–193. [Google Scholar] [CrossRef]
- Kahn, R.S.; Sommer, I.E.; Murray, R.M.; Meyer-Lindenberg, A.; Weinberger, D.R.; Cannon, T.D.; O’Donovan, M.; Correll, C.U.; Kane, J.M.; van Os, J.; et al. Schizophrenia. Nat. Rev. Dis. Primers 2015, 1, 15067. [Google Scholar] [CrossRef]
- Ripke, S.; Neale, B.M.; Corvin, A.; Walters, J.T.R.; Farh, K.-H.; Holmans, P.A.; Lee, P.; Bulik-Sullivan, B.; Collier, D.A.; Huang, H.; et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar] [CrossRef]
- Jaffe, A.E.; Gao, Y.; Deep-Soboslay, A.; Tao, R.; Hyde, T.M.; Weinberger, D.R.; Kleinman, J.E. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat. Neurosci. 2016, 19, 40–47. [Google Scholar] [CrossRef]
- Fromer, M.; Roussos, P.; Sieberts, S.K.; Johnson, J.S.; Kavanagh, D.H.; Perumal, T.M.; Ruderfer, D.M.; Oh, E.C.; Topol, A.; Shah, H.R.; et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016, 19, 1442–1453. [Google Scholar] [CrossRef] [PubMed]
- Cariaga-Martinez, A.; Saiz-Ruiz, J.; Alelú-Paz, R. From Linkage Studies to Epigenetics: What We Know and What We Need to Know in the Neurobiology of Schizophrenia. Front. Neurosci. 2016, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Dempster, E.; Viana, J.; Pidsley, R.; Mill, J. Epigenetic Studies of Schizophrenia: Progress, Predicaments, and Promises for the Future. Schizophr. Bull. 2012, 39, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Trubetskoy, V.; Pardiñas, A.; Qi, T.; Panagiotaropoulou, G.; Awasthi, S.; Bigdeli, T.; Bryois, J.; Chen, C.-Y.; Dennison, C.; Hall, L.; et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 2022, 604, 502–508. [Google Scholar] [CrossRef]
- Hoffman, G.E.; Hartley, B.J.; Flaherty, E.; Ladran, I.; Gochman, P.; Ruderfer, D.M.; Stahl, E.A.; Rapoport, J.; Sklar, P.; Brennand, K.J. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat. Commun. 2017, 8, 2225. [Google Scholar] [CrossRef]
- Gandal, M.J.; Zhang, P.; Hadjimichael, E.; Walker, R.L.; Chen, C.; Liu, S.; Won, H.; van Bakel, H.; Varghese, M.; Wang, Y.; et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 2018, 362, eaat8127. [Google Scholar] [CrossRef]
- Wang, D.; Liu, S.; Warrell, J.; Won, H.; Shi, X.; Navarro, F.C.P.; Clarke, D.; Gu, M.; Emani, P.; Yang, Y.T.; et al. Comprehensive functional genomic resource and integrative model for the human brain. Science 2018, 362, eaat8464. [Google Scholar] [CrossRef]
- Malhotra, D.; Sebat, J. CNVs: Harbingers of a Rare Variant Revolution in Psychiatric Genetics. Cell 2012, 148, 1223–1241. [Google Scholar] [CrossRef]
- Sullivan, P.F.; Daly, M.J.; O’Donovan, M. Genetic architectures of psychiatric disorders: The emerging picture and its implications. Nat. Rev. Genet. 2012, 13, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Borgmann-Winter, K.E.; Wang, K.; Bandyopadhyay, S.; Torshizi, A.D.; Blair, I.A.; Hahn, C.G. The proteome and its dynamics: A missing piece for integrative multi-omics in schizophrenia. Schizophr. Res. 2020, 217, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, J.M.; Martins-de-Souza, D. The proteome of schizophrenia. npj Schizophr. 2015, 1, 14003. [Google Scholar] [CrossRef]
- Guan, F.; Ni, T.; Zhu, W.; Williams, L.K.; Cui, L.-B.; Li, M.; Tubbs, J.; Sham, P.-C.; Gui, H. Integrative omics of schizophrenia: From genetic determinants to clinical classification and risk prediction. Mol. Psychiatry 2022, 27, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, J.; Perrone-Bizzozero, N.; Calhoun, V.D. A Perspective of the Cross-Tissue Interplay of Genetics, Epigenetics, and Transcriptomics, and Their Relation to Brain Based Phenotypes in Schizophrenia. Front. Genet. 2018, 9, 343. [Google Scholar] [CrossRef]
- Burrack, N.; Yitzhaky, A.; Mizrahi, L.; Wang, M.; Stern, S.; Hertzberg, L. Altered Expression of PDE4 Genes in Schizophrenia: Insights from a Brain and Blood Sample Meta-Analysis and iPSC-Derived Neurons. Genes 2024, 15, 609. [Google Scholar] [CrossRef]
- Stern, S.; Zhang, L.; Wang, M.; Wright, R.; Rosh, I.; Hussein, Y.; Stern, T.; Choudhary, A.; Tripathi, U.; Reed, P.; et al. Monozygotic twins discordant for schizophrenia differ in maturation and synaptic transmission. Mol. Psychiatry 2024, 29, 3208–3222. [Google Scholar] [CrossRef]
- Sarkar, A.; Mei, A.; Paquola, A.C.M.; Stern, S.; Bardy, C.; Klug, J.R.; Kim, S.; Neshat, N.; Kim, H.J.; Ku, M.; et al. Efficient Generation of CA3 Neurons from Human Pluripotent Stem Cells Enables Modeling of Hippocampal Connectivity In Vitro. Cell Stem Cell 2018, 22, 684–697.e9. [Google Scholar] [CrossRef]
- Brennand, K.J.; Simone, A.; Jou, J.; Gelboin-Burkhart, C.; Tran, N.; Sangar, S.; Li, Y.; Mu, Y.; Chen, G.; Yu, D.; et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 2011, 473, 221–225, Erratum in Nature 2011, 479, 556. [Google Scholar] [CrossRef]
- Romanovsky, E.; Choudhary, A.; Peles, D.; Abu-Akel, A.; Stern, S. Uncovering convergence and divergence between autism and schizophrenia using genomic tools and patients’ neurons. Mol. Psychiatry 2025, 30, 1019–1028. [Google Scholar] [CrossRef]
- Birnbaum, R.; Weinberger, D.R. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat. Rev. Neurosci. 2017, 18, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, L.; Choudhary, A.; Ofer, P.; Goldberg, G.; Milanesi, E.; Kelsoe, J.R.; Gurwitz, D.; Alda, M.; Gage, F.H.; Stern, S. Immunoglobulin genes expressed in lymphoblastoid cell lines discern and predict lithium response in bipolar disorder patients. Mol. Psychiatry 2023, 28, 4280–4293. [Google Scholar] [CrossRef] [PubMed]
- Sharma, O.; Nayak, R.; Mizrahi, L.; Rike, W.A.; Choudhary, A.; Hussein, Y.; Rosh, I.; Tripathi, U.; Shemen, A.; Squassina, A.; et al. Predicting Suicide Risk in Bipolar Disorder patients from Lymphoblastoid Cell Lines genetic signatures. bioRxiv 2024. [Google Scholar] [CrossRef]
- Stern, S.; Linker, S.; Vadodaria, K.C.; Marchetto, M.C.; Gage, F.H. Prediction of response to drug therapy in psychiatric disorders. Open Biol. 2018, 8, 180031. [Google Scholar] [CrossRef]
- Marshall, C.R.; Howrigan, D.P.; Merico, D.; Thiruvahindrapuram, B.; Wu, W.; Greer, D.S.; Antaki, D.; Shetty, A.; Holmans, P.A.; Pinto, D.; et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 2017, 49, 27–35, Erratum in Nat Genet. 2017, 49, 1558. [Google Scholar] [CrossRef]
- Purcell, S.M.; Moran, J.L.; Fromer, M.; Ruderfer, D.; Solovieff, N.; Roussos, P.; O’Dushlaine, C.; Chambert, K.; Bergen, S.E.; Kähler, A.; et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014, 506, 185–190. [Google Scholar] [CrossRef]
- Singh, T.; Poterba, T.; Curtis, D.; Akil, H.; Al Eissa, M.; Barchas, J.D.; Bass, N.; Bigdeli, T.B.; Breen, G.; Bromet, E.J.; et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature 2022, 604, 509–516. [Google Scholar] [CrossRef]
- Ursini, G.; Punzi, G.; Chen, Q.; Marenco, S.; Robinson, J.F.; Porcelli, A.; Hamilton, E.G.; Mitjans, M.; Maddalena, G.; Begemann, M.; et al. Convergence of placenta biology and genetic risk for schizophrenia. Nat. Med. 2018, 24, 792–801. [Google Scholar] [CrossRef]
- Akbarian, S.; Ruehl, M.G.; Bliven, E.; Luiz, L.A.; Peranelli, A.C.; Baker, S.P.; Roberts, R.C.; Bunney, W.E., Jr.; Conley, R.C.; Jones, E.G.; et al. Chromatin Alterations Associated With Down-regulated Metabolic Gene Expression in the Prefrontal Cortex of Subjects With Schizophrenia. Arch. Gen. Psychiatry 2005, 62, 829–840. [Google Scholar] [CrossRef]
- Gusev, F.E.; Reshetov, D.A.; Mitchell, A.C.; Andreeva, T.V.; Dincer, A.; Grigorenko, A.P.; Fedonin, G.; Halene, T.; Aliseychik, M.; Filippova, E.; et al. Chromatin profiling of cortical neurons identifies individual epigenetic signatures in schizophrenia. Transl. Psychiatry 2019, 9, 256. [Google Scholar] [CrossRef]
- Girdhar, K.; Hoffman, G.E.; Jiang, Y.; Brown, L.; Kundakovic, M.; Hauberg, M.E.; Francoeur, N.J.; Wang, Y.-c.; Shah, H.; Kavanagh, D.H.; et al. Cell-specific histone modification maps in the human frontal lobe link schizophrenia risk to the neuronal epigenome. Nat. Neurosci. 2018, 21, 1126–1136. [Google Scholar] [CrossRef]
- Won, H.; de la Torre-Ubieta, L.; Stein, J.L.; Parikshak, N.N.; Huang, J.; Opland, C.K.; Gandal, M.J.; Sutton, G.J.; Hormozdiari, F.; Lu, D.; et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 2016, 538, 523–527. [Google Scholar] [CrossRef]
- Hoffman, G.E.; Bendl, J.; Girdhar, K.; Schadt, E.E.; Roussos, P. Functional interpretation of genetic variants using deep learning predicts impact on chromatin accessibility and histone modification. Nucleic Acids Res. 2019, 47, 10597–10611. [Google Scholar] [CrossRef] [PubMed]
- Collado-Torres, L.; Burke, E.E.; Peterson, A.; Shin, J.; Straub, R.E.; Rajpurohit, A.; Semick, S.A.; Ulrich, W.S.; Price, A.J.; Valencia, C.; et al. Regional Heterogeneity in Gene Expression, Regulation, and Coherence in the Frontal Cortex and Hippocampus across Development and Schizophrenia. Neuron 2019, 103, 203–216.e8. [Google Scholar] [CrossRef]
- Ruzicka, W.B.; Mohammadi, S.; Fullard, J.F.; Davila-Velderrain, J.; Subburaju, S.; Tso, D.R.; Hourihan, M.; Jiang, S.; Lee, H.-C.; Bendl, J.; et al. Single-cell multi-cohort dissection of the schizophrenia transcriptome. Science 2024, 384, eadg5136. [Google Scholar] [CrossRef] [PubMed]
- Pennington, K.; Beasley, C.L.; Dicker, P.; Fagan, A.; English, J.; Pariante, C.M.; Wait, R.; Dunn, M.J.; Cotter, D.R. Prominent synaptic and metabolic abnormalities revealed by proteomic analysis of the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder. Mol. Psychiatry 2008, 13, 1102–1117. [Google Scholar] [CrossRef] [PubMed]
- Föcking, M.; Lopez, L.M.; English, J.A.; Dicker, P.; Wolff, A.; Brindley, E.; Wynne, K.; Cagney, G.; Cotter, D.R. Proteomic and genomic evidence implicates the postsynaptic density in schizophrenia. Mol. Psychiatry 2014, 20, 424–432. [Google Scholar] [CrossRef]
- MacDonald, M.L.; Garver, M.; Newman, J.; Sun, Z.; Kannarkat, J.; Salisbury, R.; Glausier, J.; Ding, Y.; Lewis, D.A.; Yates, N.; et al. Synaptic Proteome Alterations in the Primary Auditory Cortex of Individuals With Schizophrenia. JAMA Psychiatry 2020, 77, 86. [Google Scholar] [CrossRef]
- Schwarz, E.; Guest, P.C.; Rahmoune, H.; Harris, L.W.; Wang, L.; Leweke, F.M.; Rothermundt, M.; Bogerts, B.; Koethe, D.; Kranaster, L.; et al. Identification of a biological signature for schizophrenia in serum. Mol. Psychiatry 2012, 17, 494–502. [Google Scholar] [CrossRef]
- Domenici, E.; Willé, D.R.; Tozzi, F.; Prokopenko, I.; Miller, S.; McKeown, A.; Brittain, C.; Rujescu, D.; Giegling, I.; Turck, C.W.; et al. Plasma protein biomarkers for depression and schizophrenia by multi analyte profiling of case-control collections. PLoS ONE 2010, 5, e9166. [Google Scholar] [CrossRef]
- Rodrigues, J.E.; Martinho, A.; Santa, C.; Madeira, N.; Coroa, M.; Santos, V.; Martins, M.J.; Pato, C.N.; Macedo, A.; Manadas, A.; et al. Systematic Review and Meta-Analysis of Mass Spectrometry Proteomics Applied to Human Peripheral Fluids to Assess Potential Biomarkers of Schizophrenia. Int. J. Mol. Sci. 2022, 23, 4917. [Google Scholar] [CrossRef]
- Jaros, J.A.J.; Martins-de-Souza, D.; Rahmoune, H.; Rothermundt, M.; Leweke, F.M.; Guest, P.C.; Bahn, S. Protein phosphorylation patterns in serum from schizophrenia patients and healthy controls. J. Proteom. 2012, 76, 43–55. [Google Scholar] [CrossRef]
- Brennand, K.; Savas, J.N.; Kim, Y.; Tran, N.; Simone, A.; Hashimoto-Torii, K.; Beaumont, K.G.; Kim, H.J.; Topol, A.; Ladran, I.; et al. Phenotypic differences in hiPSC NPCs derived from patients with schizo phrenia. Mol. Psychiatry 2014, 20, 361–368. [Google Scholar] [CrossRef]
- de Vrij, F.M.; Bouwkamp, C.G.; Gunhanlar, N.; Shpak, G.; Lendemeijer, B.; Baghdadi, M.; Gopalakrishna, S.; Ghazvini, M.; Li, T.M.; Quadri, M.; et al. Candidate CSPG4 mutations and induced pluripotent stem cell modeling implicate oligodendrocyte progenitor cell dysfunction in familial schizophrenia. Mol. Psychiatry 2019, 24, 757–771. [Google Scholar] [CrossRef] [PubMed]
- Kathuria, A.; Lopez-Lengowski, K.; Watmuff, B.; McPhie, D.; Cohen, B.M.; Karmacharya, R. Synaptic deficits in iPSC-derived cortical interneurons in schizophrenia are mediated by NLGN2 and rescued by N-acetylcysteine. Transl. Psychiatry 2019, 9, 321. [Google Scholar] [CrossRef]
- Kathuria, A.; Lopez-Lengowski, K.; Jagtap, S.S.; McPhie, D.; Perlis, R.H.; Cohen, B.M.; Karmacharya, R. Transcriptomic Landscape and Functional Characterization of Induced Pluripotent Stem Cell–Derived Cerebral Organoids in Schizophrenia. JAMA Psychiatry 2020, 77, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Notaras, M.; Lodhi, A.; Dündar, F.; Collier, P.; Sayles, N.M.; Tilgner, H.; Greening, D.; Colak, D. Schizophrenia is defined by cell-specific neuropathology and multiple neurodevelopmental mechanisms in patient-derived cerebral organoids. Mol. Psychiatry 2022, 27, 1416–1434. [Google Scholar] [CrossRef]
- Robicsek, O.; Karry, R.; Petit, I.; Salman-Kesner, N.; Müller, F.J.; Klein, E.; Aberdam, D.; Ben-Shachar, D. Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Mol. Psychiatry 2013, 18, 1067–1076. [Google Scholar] [CrossRef]
- Pedrosa, E.; Sandler, V.; Shah, A.; Carroll, R.; Chang, C.; Rockowitz, S.; Guo, X.; Zheng, D.; Lachman, H.M. Development of Patient-Specific Neurons in Schizophrenia Using Induced Pluripotent Stem Cells. J. Neurogenet. 2011, 25, 88–103. [Google Scholar] [CrossRef]
- Sekar, A.; Bialas, A.R.; de Rivera, H.; Davis, A.; Hammond, T.R.; Kamitaki, N.; Tooley, K.; Presumey, J.; Baum, M.; Van Doren, V.; et al. Schizophrenia risk from complex variation of complement component 4. Nature 2016, 530, 177–183, Erratum in Nature 2022, 601, E4–E5. [Google Scholar] [CrossRef]
- Sanders, A.R.; Drigalenko, E.I.; Duan, J.; Moy, W.; Freda, J.; Göring, H.H.H.; Gejman, P.V. Transcriptome sequencing study implicates immune-related genes differentially expressed in schizophrenia: New data and a meta-analysis. Transl. Psychiatry 2017, 7, e1093. [Google Scholar] [CrossRef]
- Föcking, M.; Pollak, T.; Dicker, P.; Cagney, G.; Winter, I.; Kahn, R.; McGuire, P.; Cotter, D. O1.7. PROTEOMIC ANALYSIS OF BLOOD BASED SAMPLES FROM THE OPTiMiSE (OPTIMIZATION OF TREATMENT AND MANAGEMENT OF SCHIZOPHRENIA IN EUROPE) STUDY POINT TOWARDS COMPLEMENT PATHWAY PROTEIN CHANGES. Schizophr. Bull. 2018, 44, S74–S75. [Google Scholar] [CrossRef]
- Upthegrove, R.; Manzanares-Teson, N.; Barnes, N.M. Cytokine function in medication-naive first episode psychosis: A systematic review and meta-analysis. Schizophr. Res. 2014, 155, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Khandaker, G.M.; Pearson, R.M.; Zammit, S.; Lewis, G.; Jones, P.B. Association of Serum Interleukin 6 and C-Reactive Protein in Childhood With Depression and Psychosis in Young Adult Life: A Population-Based Longitudinal Study. JAMA Psychiatry 2014, 71, 1121–1128. [Google Scholar] [CrossRef] [PubMed]
- Prabakaran, S.; Swatton, J.E.; Ryan, M.M.; Huffaker, S.J.; Huang, J.-J.; Griffin, J.L.; Wayland, M.; Freeman, T.; Dudbridge, F.; Lilley, K.S.; et al. Mitochondrial dysfunction in schizophrenia: Evidence for compromised brain metabolism and oxidative stress. Mol. Psychiatry 2004, 9, 684–697. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Sportelli, V.; Ziller, M.; Spengler, D.; Hoffmann, A. Tracing Early Neurodevelopment in Schizophrenia with Induced Pluripote nt Stem Cells. Cells 2018, 7, 140. [Google Scholar] [CrossRef]
- Kato, H.; Kimura, H.; Kushima, I.; Takahashi, N.; Aleksic, B.; Ozaki, N. The genetic architecture of schizophrenia: Review of large-scale genetic studies. J. Hum. Genet. 2023, 68, 175–182. [Google Scholar] [CrossRef]
- Yang, H.; Sun, W.; Li, J.; Zhang, X. Epigenetics factors in schizophrenia: Future directions for etiologic and therapeutic study approaches. Ann. Gen. Psychiatry 2025, 24, 21. [Google Scholar] [CrossRef]
- Priyanka; Kumar, R.; Kumar, V.; Kumar, A.; Rana, S.S. Deciphering transcriptomic signatures in schizophrenia, bipolar disorder, and major depressive disorder. Front. Psychiatry 2025, 16, 1574458. [Google Scholar] [CrossRef]
- Hong, Y.; Yang, Q.; Song, H.; Ming, G.L. Opportunities and limitations for studying neuropsychiatric disorders using patient-derived induced pluripotent stem cells. Mol. Psychiatry 2023, 28, 1430–1439. [Google Scholar] [CrossRef]
- Owen, M.J.; Sawa, A.; Mortensen, P.B. Schizophrenia. Lancet 2016, 388, 86–97. [Google Scholar] [CrossRef]
- Shifman, S.; Johannesson, M.; Bronstein, M.; Chen, S.X.; Collier, D.A.; Craddock, N.J.; Kendler, K.S.; Li, T.; O’Donovan, M.; O’Neill, F.A.; et al. Genome-Wide Association Identifies a Common Variant in the Reelin Gene That Increases the Risk of Schizophrenia Only in Women. PLoS Genet. 2008, 4, e28. [Google Scholar] [CrossRef]
- Purcell, S.M.; Wray, N.R.; Stone, J.L.; Visscher, P.M.; O’Donovan, M.C.; Sullivan, P.F.; Sklar, P.; Purcell, S.M.; Stone, J.L.; Sullivan, P.F.; et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009, 460, 748–752. [Google Scholar] [CrossRef] [PubMed]
- Legge, S.E.; Santoro, M.L.; Periyasamy, S.; Okewole, A.; Arsalan, A.; Kowalec, K. Genetic architecture of schizophrenia: A review of major advancements. Psychol. Med. 2021, 51, 2168–2177. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.; Chen, C.Y.; Li, Z.; Martin, A.R.; Bryois, J.; Ma, X.; Gaspar, H.; Ikeda, M.; Benyamin, B.; Brown, B.C.; et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 2019, 51, 1670–1678. [Google Scholar] [CrossRef]
- Ruan, Y.; Lin, Y.-F.; Feng, Y.-C.A.; Chen, C.-Y.; Lam, M.; Guo, Z.; Stanley Global Asia Initiatives; He, L.; Sawa, A.; Martin, A.R.; et al. Improving polygenic prediction in ancestrally diverse populations. Nat. Genet. 2022, 54, 573–580, Erratum in Nat Genet. 2022, 54, 1259. [Google Scholar] [CrossRef] [PubMed]
- Pardiñas, A.F.; Holmans, P.; Pocklington, A.J.; Escott-Price, V.; Ripke, S.; Carrera, N.; Legge, S.E.; Bishop, S.; Cameron, D.; Hamshere, M.L.; et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 2018, 50, 381–389, Erratum in Nat Genet. 2019, 51, 1193. [Google Scholar] [CrossRef]
- Townsley, K.G.; Li, A.; Deans, P.J.M.; Fullard, J.F.; Yu, A.; Cartwright, S.; Zhang, W.; Wang, M.; Voloudakis, G.; Girdhar, K.; et al. Convergent impact of schizophrenia risk genes. bioRxiv 2022. [Google Scholar] [CrossRef]
- Roussos, P.; Katsel, P.; Davis, K.L.; Siever, L.J.; Haroutunian, V. A System-Level Transcriptomic Analysis of Schizophrenia Using Postmortem Brain Tissue Samples. Arch. Gen. Psychiatry 2012, 69, 1205–1213. [Google Scholar] [CrossRef]
- Arnedo, J.; Svrakic, D.M.; Val, C.d.; Romero-Zaliz, R.; Hernández-Cuervo, H.; Molecular Genetics of Schizophrenia Consortium; Fanous, A.H.; Pato, M.T.; Pato, C.N.; Erausquin, G.A.d.; et al. Uncovering the Hidden Risk Architecture of the Schizophrenias: Confirmation in Three Independent Genome-Wide Association Studies. Am. J. Psychiatry 2015, 172, 139–153. [Google Scholar] [CrossRef]
- Migdalska-Richards, A.; Mill, J. Epigenetic studies of schizophrenia: Current status and future directions. Curr. Opin. Behav. Sci. 2019, 25, 102–110. [Google Scholar] [CrossRef]
- Roth, T.L.; Lubin, F.D.; Sodhi, M.; Kleinman, J.E. Epigenetic mechanisms in schizophrenia. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2009, 1790, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Svrakic, D.M.; Zorumski, C.F.; Svrakic, N.M.; Zwir, I.; Cloninger, C.R. Risk architecture of schizophrenia: The role of epigenetics. Curr. Opin. Psychiatry 2013, 26, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Meyer, U.; Feldon, J. Epidemiology-driven neurodevelopmental animal models of schizophrenia. Prog. Neurobiol. 2010, 90, 285–326. [Google Scholar] [CrossRef]
- Brown, A.S.; Derkits, E.J. Prenatal Infection and Schizophrenia: A Review of Epidemiologic and Translational Studies. Am. J. Psychiatry 2010, 167, 261–280. [Google Scholar] [CrossRef]
- Bale, T.L.; Baram, T.Z.; Brown, A.S.; Goldstein, J.M.; Insel, T.R.; McCarthy, M.M.; Nemeroff, C.B.; Reyes, T.M.; Simerly, R.B.; Susser, E.S.; et al. Early Life Programming and Neurodevelopmental Disorders. Biol. Psychiatry 2010, 68, 314–319. [Google Scholar] [CrossRef]
- Punzi, G.; Bharadwaj, R.; Ursini, G. Neuroepigenetics of Schizophrenia. Prog. Mol. Biol. Transl. Sci. 2018, 158, 195–226. [Google Scholar] [CrossRef]
- Costa, E.; Dong, E.; Grayson, D.R.; Guidotti, A.; Ruzicka, W.; Veldic, M. Reviewing the role of DNA (cytosine-5) methyltransferase overexpression in the cortical GABAergic dysfunction associated with psychosis vulnerability. Epigenetics 2007, 2, 29–36. [Google Scholar] [CrossRef]
- Gavin, D.P.; Sharma, R.P. Histone modifications, DNA methylation, and Schizophrenia. Neurosci. Biobehav. Rev. 2010, 34, 882–888. [Google Scholar] [CrossRef]
- Nishioka, M.; Bundo, M.; Kasai, K.; Iwamoto, K. DNA methylation in schizophrenia: Progress and challenges of epigenetic studies. Genome Med. 2012, 4, 96. [Google Scholar] [CrossRef]
- Chen, Q.; Li, D.; Jin, W.; Shi, Y.; Li, Z.; Ma, P.; Sun, J.; Chen, S.; Li, P.; Lin, P. Research Progress on the Correlation Between Epigenetics and Schizophrenia. Front. Neurosci. 2021, 15, 688727. [Google Scholar] [CrossRef] [PubMed]
- Gavin, D.P.; Kartan, S.; Chase, K.; Grayson, D.R.; Sharma, R.P. Reduced baseline acetylated histone 3 levels, and a blunted response to HDAC inhibition in lymphocyte cultures from schizophrenia subjects. Schizophr. Res. 2008, 103, 330–332. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, A.; Dong, E.; Kundakovic, M.; Satta, R.; Grayson, D.R.; Costa, E. Characterization of the action of antipsychotic subtypes on valproate-induced chromatin remodeling. Trends Pharmacol. Sci. 2009, 30, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Covington, H.E.; Maze, I.; LaPlant, Q.C.; Vialou, V.F.; Ohnishi, Y.N.; Berton, O.; Fass, D.M.; Renthal, W.; Rush, A.J.; Wu, E.Y.; et al. Antidepressant Actions of Histone Deacetylase Inhibitors. J. Neurosci. 2009, 29, 11451–11460. [Google Scholar] [CrossRef]
- Simonini, M.V.; Camargo, L.M.; Dong, E.; Maloku, E.; Veldic, M.; Costa, E.; Guidotti, A. The benzamide MS-275 is a potent, long-lasting brain region-selective inhibitor of histone deacetylases. Proc. Natl. Acad. Sci. USA 2006, 103, 1587–1592. [Google Scholar] [CrossRef]
- Day, J.J.; Sweatt, J.D. Epigenetic Mechanisms in Cognition. Neuron 2011, 70, 813–829. [Google Scholar] [CrossRef]
- Nestler, E.J.; Peña, C.J.; Kundakovic, M.; Mitchell, A.; Akbarian, S. Epigenetic Basis of Mental Illness. Neurosci. 2016, 22, 447–463. [Google Scholar] [CrossRef]
- Liu, Y.; Chang, X.; Hahn, C.G.; Gur, R.E.; Sleiman, P.A.M.; Hakonarson, H. Non-coding RNA dysregulation in the amygdala region of schizophrenia patients contributes to the pathogenesis of the disease. Transl. Psychiatry 2018, 8, 44. [Google Scholar] [CrossRef]
- Wu, J.Q.; Wang, X.; Beveridge, N.J.; Tooney, P.A.; Scott, R.J.; Carr, V.J.; Cairns, M.J. Transcriptome Sequencing Revealed Significant Alteration of Cortical Promoter Usage and Splicing in Schizophrenia. PLoS ONE 2012, 7, e36351. [Google Scholar] [CrossRef]
- Cohen, O.S.; McCoy, S.Y.; Middleton, F.A.; Bialosuknia, S.; Zhang-James, Y.; Liu, L.; Tsuang, M.T.; Faraone, S.V.; Glatt, S.J. Transcriptomic analysis of postmortem brain identifies dysregulated splicing events in novel candidate genes for schizophrenia. Schizophr. Res. 2012, 142, 188–199. [Google Scholar] [CrossRef]
- Sanders, A.R.; Göring, H.H.H.; Duan, J.; Drigalenko, E.I.; Moy, W.; Freda, J.; He, D.; Shi, J.; MGS; Gejman, P.V. Transcriptome study of differential expression in schizophrenia. Hum. Mol. Genet. 2013, 22, 5001–5014. [Google Scholar] [CrossRef] [PubMed]
- Olde Loohuis, N.F.; Nadif Kasri, N.; Glennon, J.C.; van Bokhoven, H.; Hébert, S.S.; Kaplan, B.B.; Martens, G.J.; Aschrafi, A. The schizophrenia risk gene MIR137 acts as a hippocampal gene network node orchestrating the expression of genes relevant to nervous system development and function. Prog. Neuropsychopharmacol. Biol. Psychiatry 2017, 73, 109–118. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Collins, A.L.; Kim, Y.; Bloom, R.J.; Kelada, S.N.; Sethupathy, P.; Sullivan, P.F. Transcriptional targets of the schizophrenia risk gene MIR137. Transl. Psychiatry 2014, 4, e404. [Google Scholar] [CrossRef]
- Geaghan, M.P.; Atkins, J.R.; Brichta, A.M.; Tooney, P.A.; Scott, R.J.; Carr, V.J.; Cairns, M.J. Alteration of miRNA-mRNA interactions in lymphocytes of individuals with schizophrenia. J. Psychiatr. Res. 2019, 112, 89–98. [Google Scholar] [CrossRef]
- Perkins, D.O.; Olde Loohuis, L.; Barbee, J.; Ford, J.; Jeffries, C.D.; Addington, J.; Bearden, C.E.; Cadenhead, K.S.; Cannon, T.D.; Cornblatt, B.A.; et al. Polygenic Risk Score Contribution to Psychosis Prediction in a Target Population of Persons at Clinical High Risk. Am. J. Psychiatry 2020, 177, 155–163. [Google Scholar] [CrossRef]
- Moreau, M.P.; Bruse, S.E.; David-Rus, R.; Buyske, S.; Brzustowicz, L.M. Altered MicroRNA Expression Profiles in Postmortem Brain Samples from Individuals with Schizophrenia and Bipolar Disorder. Biol. Psychiatry 2011, 69, 188–193. [Google Scholar] [CrossRef]
- Rodrigues-Amorim, D.; Rivera-Baltanás, T.; Vallejo-Curto, M.d.C.; Rodriguez-Jamardo, C.; de las Heras, E.; Barreiro-Villar, C.; Blanco-Formoso, M.; Fernández-Palleiro, P.; Álvarez-Ariza, M.; López, M.; et al. Proteomics in Schizophrenia: A Gateway to Discover Potential Biomarkers of Psychoneuroimmune Pathways. Front. Psychiatry 2019, 10, 885. [Google Scholar] [CrossRef]
- Ramsey, J.M.; Schwarz, E.; Guest, P.C.; van Beveren, N.J.M.; Leweke, F.M.; Rothermundt, M.; Bogerts, B.; Steiner, J.; Bahn, S. Distinct Molecular Phenotypes in Male and Female Schizophrenia Patients. PLoS ONE 2013, 8, e78729. [Google Scholar] [CrossRef]
- Tomasik, J.; Smits, S.L.; Leweke, F.M.; Eljasz, P.; Pas, S.; Kahn, R.S.; Osterhaus, A.D.M.E.; Bahn, S.; de Witte, L.D. Virus discovery analyses on post-mortem brain tissue and cerebrospinal fluid of schizophrenia patients. Schizophr. Res. 2018, 197, 605–606. [Google Scholar] [CrossRef]
- Davalieva, K.; Maleva Kostovska, I.; Dwork, A.J. Proteomics Research in Schizophrenia. Front. Cell. Neurosci. 2016, 16, 18. [Google Scholar] [CrossRef]
- Goldman, S.A.; Kuypers, N.J. How to make an oligodendrocyte. Development 2015, 142, 3983–3995. [Google Scholar] [CrossRef]
- Cerneckis, J.; Cai, H.; Shi, Y. Induced pluripotent stem cells (iPSCs): Molecular mechanisms of induction and applications. Signal Transduct. Target. Ther. 2024, 9, 112. [Google Scholar] [CrossRef]
| Variant Class | Tissue/ Sample | Example Loci/ Genes | Frequency | Effect Size | Main Pathways | References |
|---|---|---|---|---|---|---|
| SNPs | Peripheral blood (germline DNA; 36,989 cases/113,075 controls) | CACNA1C; MIR137; GRIN2A | High minor allele frequency (MAF) 20–50% | Small per-allele effect: 1.06–1.12× | Synaptic signaling; calcium channel regulation | [11,16] |
| CNVs | Peripheral blood (germline DNA; >21,000 cases/>20,000 controls) | 22q11.2 deletion; 16p11.2 duplication | Rare ~0.3% | Moderate–large 9–30× | Neurodevelopment immune modulation | [20] |
| Rare LoF coding mutations | Peripheral blood WES (germline DNA; 6000 cases/6000 controls) | SETD1A; RBM12; GRIN2A; TRIO; CACNA1G | Very rare <0.1% cases | Large (high penetrance) OR: 3–50× | Chromatin remodeling transcriptional regulation synaptogenesis; glutamatergic signaling; ion channel regulation | [13,37,77] |
| Regulatory eQTLs | DLPFC post-mortem (RNA-seq/eQTL; 467 donors) | Non-coding SNPs modulating DRD2 | Common minor allele frequency (MAF) ~10–30% | Small ~1.1 | Gene expression modulation | [12,13] |
| PRS | Peripheral blood genotyping (arrays; PRS derived from >320,000 | Aggregate of 104–106 SNPs | Present in all ancestries | Cumulative across many loci | Pleiotropic effects on neurodevelopment and immunity | [74] |
| Epigenetic Mechanism | Sample/Tissue | Key Examples | Impact on Gene Regulation | Main Pathways | References |
|---|---|---|---|---|---|
| DNA Methylation | – Post-mortem PFCs (BA9, n = 14 cases vs. 14 controls) – Post-mortem DLPFCs (n = 15 vs. 15) – Placentae (n = 157) – PBMCs (n = 20 vs. 20) | – RELN and GAD1 promoter hypermethylation in BA9 and DLPFC – DNMT1/3A upregulation in PFC – Placental DMRs at immune (CXCL10, HLA) and oxidative stress loci mediating PRS × obstetric complications – DRD2 methylation in blood | Transcriptional repression | GABAergic signaling; immune regulation | [38,88,89,90] |
| Histone Modifications | – PFC neuronal nuclei (BA9; 10 cases vs. 10 controls) – DLPFC ChIP-seq (n = 236 donors) | – ↑ H3R17me at metabolic gene promoters with concomitant mRNA downregulation – 3000+ DLPFC enhancers with altered H3K27ac – Enrichment of risk variants at combined H3K4me3 + H3K27ac peaks in excitatory neurons | Altered chromatin accessibility | Synaptic plasticity; immune response | [12,39,40] |
| Chromatin Looping (3D Contacts) | – Promoter-capture Hi-C in adult DLPFCs (3 cases, 3 controls) – In situ Hi-C in adult PFC (5 donors) – Hi-C in iPSC-derived neurons | – Schizophrenia loci contacting GRIN2B, MEF2C, and C4A genes – Enrichment of risk SNPs at loop anchors genome-wide – One-third of non-coding SNPs link to synaptic and chromatin-remodeling gene promoters | Long-range regulation of risk genes | Synaptic pruning; neurodevelopment | [19,40,87] |
| Non-Coding RNAs (miRNA and lncRNA) | – DLPFC tissue and paired plasma (30 cases vs. 30 controls; 60 × 60) | – ↓ miR-137 in both DLPFC and plasma, derepressing SYN2 and IL6R – ↑ NEAT1/MALAT1 lncRNAs correlated with CRP and oxidative markers – Organoid studies: miR-137 correction rescues arborization deficits | Post-transcriptional modulation; network rewiring | Neuronal differentiation; immune signaling | [87,91] |
| Peripheral Epigenetic Signatures | – PBMCs (20 antipsychotic-naïve cases vs. 20 controls) – Saliva (25 vs. 25) – Olfactory epithelium (small pilot) | – Global hypomethylation in PBMCs – DRD2 promoter hypermethylation in blood – ST6GALNAC1 promoter hypermethylation in saliva correlated with IL-6 (r = 0.56) | Potential peripheral biomarkers | Neuroimmune regulation; diagnostic utility | [90,91] |
| Transcriptomic Feature | Sample/Tissue | Key Examples | Functional Impact | Main Pathways | References |
|---|---|---|---|---|---|
| Differential Gene Expression | DLPFC (245 schizophrenia patients vs. 279 controls); hippocampus (48 vs. 48) | 245 DEGs in DLPFC; 48 DEGs in hippocampus | Altered mRNA abundance | Synaptic signaling; mitochondrial function; immune response | [13,44] |
| Alternative Splicing and Isoform Shifts | Frontal and temporal cortices (258 schizophrenia patients vs. 301 controls); DLPFC BA46 (100 vs. 100); BA10 (40 vs. 40) | 3803 dysregulated isoforms; 515 splicing events (e.g., DCLK1 and PLP1); disrupted exon usage in ENAH and CPNE3; many events map to eQTLs | Isoform-specific expression changes | Neurodevelopment; neurotransmission; myelination | [12,18,99,100] |
| Non-Coding RNA Dysregulation | Amygdalae (13 schizophrenia patients vs. 14 controls); LCLs (20 vs. 20); DLPFCs (258 vs. 301); PBMCs (36 vs. 15) | ↓ miRNAs (miR-1307, miR-34 family, and miR-137); ↓ DICER1 expression; lncRNA co-expression modules | Post-transcriptional regulation; network rewiring | Neuronal maturation; immune modulation | [18,98,101,102,104] |
| Tissue-Specific and Peripheral Signatures | PBMCs (36 schizophrenia patients vs. 15 controls); LCLs (20 vs. 20) | Upregulation of immune-related genes in PBMCs and LCLs | Systemic transcriptional alterations | Neuroimmune signaling; biomarker potential | [61,104] |
| Co-Expression Network Perturbations | DLPFCs (258 schizophrenia patients vs. 279 controls); frontal and temporal cortices (258 vs. 301) | Synaptic, glial, and immune modules identified by WGCNA; modules enriched for GWAS risk variants | Coordinated dysregulation of gene clusters | Synaptic transmission; glial function; immunity | [13,18,77] |
| Section | Sample/Tissue | Key Examples | Functional Impact | Main Pathways | Ref. |
|---|---|---|---|---|---|
| Synaptic proteome | Anterior cingulate cortex (ACC; 20 SZ vs. 20 CTRLs) | Differential PSD proteins (e.g., DNM1, AP2B1) | Vesicle-cycling/plasticity changes | Synaptic signaling | [7] |
| Primary auditory cortex (A1; 48 SZ vs. 48 CTRLs) | ↓ PSD markers (PSD-95/SHANK3) confined to synaptic fraction | Synaptic dysfunction; altered vesicle/plasticity machinery | Synaptic signaling | [48] | |
| Mitochondrial/energetic proteome | DLPFC/ACC cortex (post-mortem) | (post-mortem) Altered respiratory-chain (Complex I–V) and energy-metabolism proteins | Synaptic–energetic coupling deficits; cellular energetics changes | Mitochondrial OXPHOS; metabolic pathways | [46] |
| Immune and Inflammatory Markers | Serum (multi-cohort case–control) | 34-analyte serum signature distinguishing SZ from controls/other disorders; cross-cohort classification (∼60–75%) | Systemic immune activation; diagnostic signal | Cytokine, growth-factor and endocrine networks | [49] |
| Serum (first-episode, antipsychotic-naïve; sex-stratified) | Sex-specific molecular profiles (16 molecules differing by sex across four cohorts) | Hormonal/inflammatory dysregulation; sex effects | Immune & endocrine biomarkers | [108] | |
| Plasma (large discovery/replication) | Multi-analyte plasma panel distinguishing SZ and depression in large collections | Diagnostic stratification (discovery + validation) | Immune/inflammatory & growth-factor networks | [50] | |
| Trial cohort (OPTiMiSE) | Complement-pathway changes in blood proteomics; explored as predictors of antipsychotic response | Treatment-response prediction signal | Complement activation | [62] | |
| Post-Translational Modifications | Serum phosphoproteome (50 SZ vs. 50 CTRL) | Altered phosphorylation of signaling and acute-phase proteins (e.g., Akt1/STAT3 pathways; coagulation/synaptic scaffolding sites) | Dysregulated signaling; immune–coagulation crosstalk | Protein phosphorylation; signal transduction | [52] |
| Main Pathways | Model/System and Format | Cohort (Schizophrenia Patients vs. CTRLs) | Key Molecular Findings | Functional Consequence | References |
|---|---|---|---|---|---|
| Neurodevelopment and Transcriptional Dysregulation | Forebrain NPCs (2D culture) | 4 schizophrenia patients vs. 4 CTRLs | ↓ NCAM1/NRXN1/NLGN1 (1.5–1.7×), ↑ antioxidant enzymes (2.2×); miR-137 ↑ 1.8×, miR-9 ↓ 1.4×; SOX2 ↓ 40%, PAX6 ↓ 30% | Migration −35%, ROS +28%, MAP2 onset delayed ~7 d | [53,66] |
| Mitochondrial Dysfunction and Oxidative Stress | 2D neurons (dopaminergic and glutamatergic) and 3D organoids | 3 schizophrenia patients vs. 2 CTRLs (neurons); 8 schizophrenia patients vs. 8 CTRLs (organoids) | Mito fragmentation +30%; ΔΨm −25%; ROS +35%Basal OCR −22%; ATP-linked OCR −28% | Neurite length −20%; spike rate −40% | [58] |
| Synaptic Connectivity and Dendritic Architecture | 2D cortical neurons | 4 schizophrenia patients (incl. 22q11.2del) vs. 3 CTRLs | PSD-95 puncta −40%; dendritic intersections −35%; OCT4/NANOG persistence; Syn1 ↓ 32%, NRXN1↓28% | sEPSC frequency −50%; loxapine rescue PSD-95 +25%; EPSC +30% | [29,59] |
| Circuit-Level Vulnerabilities | Interneuron co-cultures (2D) and cerebral organoids (3D) | 9 schizophrenia patients vs. 9 CTRLs (interneurons); 9 schizophrenia patients vs. 5 CTRLs (organoids; n = 25) | VGAT+ puncta −30%; GAD67 −42%; gephyrin −38%; NLGN2 −45%BRN2 −50%; PTN −60% | Firing rate rescue +50% (NLGN2/NAC); progenitor survival +40%, NeuN+ neurons ×2 (PTN) | [55,57] |
| Oligodendrocyte Precursor Dysfunction | NG2+ OPCs (2D culture) | 3 CSPG4-mut schizophrenia patients vs. 3 siblings | NG2 high-mannose ×3; MBP −45%; PLP1 −50%; SOX10/OLIG2 −30% | In vivo FA −15% (DTI) | [54] |
| Pathway | Genetics | Epigenetics | Transcriptomics | Proteomics | iPSC Models |
|---|---|---|---|---|---|
| Synaptic Signaling | CACNA1C, GRIN2A, and DLG2 GWAS loci [11,16]; SETD1A LoF [37] | ↓ H3K27ac/H3K4me3 at synaptic enhancers [12,40] | Disrupted synaptic co-expression modules; isoform shifts [13,18] | ↓ PSD-95/SHANK3; phospho-Akt1 alterations [47,48,52] | ↓ PSD-95 puncta and sEPSCs; loxapine rescue [29,55] |
| Mitochondrial Bioenergetics | Mito-ETC gene variants; 16p11.2 CNV [20,21] | H3R17me at metabolic promoters [39] | Downregulated OXPHOS transcripts [44] | ↓ Complex I–V subunits; altered mitochondrial proteins [47] | Mito fragmentation, ↓ ΔΨm, ↑ ROS [58]; ↓ OCR in organoids [56] |
| Cell-Adhesion Complexes | NRXN1/NLGN1 CNVs; NCAM1/BRN2 risk loci [11,20] | RELN promoter hypermethylation [88,89] | Dysregulated protocadherins and adhesion isoforms [18] | Altered AP2B1/DNM1 phosphorylation [48,109] | ↓ NCAM1/NRXN1/NLGN1 in NPCs [53]; OPC NG2 misprocessing [54] |
| Immune Regulation | C4A/C4B MHC variation [60] | Placental/blood immune-gene DMRs [38,90] | Upregulated cytokine/microglial modules [18,104] | IL-6, CFI, and C4A serum signatures [49,62] | Rescue of complement/cytokine defects by PTN or NAC [57] |
| Neurodevelopmental Regulation | 22q11.2del; POU3F2/BRN2, PTN risk loci [57] | Placental DMRs at PAX6/SOX2; developmental histone marks [38] | Disrupted NPC and neuronal differentiation modules [53,66] | ↓ BRN2, PTN in organoid proteomes [57] | NPC migration delays; miR-137/PAX6 imbalance; organoid progenitor loss [53,66] |
| Gene/Protein | Genetics | Epigenetics/Chromatin | Transcriptomics | Proteomics | iPSC Models |
|---|---|---|---|---|---|
| DLG4 (PSD-95) | (Not a GWAS hit) | ↓ H3K27ac at DLG4 enhancer in DLPFC [12] | ↓ DLG4 mRNA in DLPFC [13] | ↓ PSD-95 in ACC and A1 cortices [47,48] | ↓ PSD-95 puncta and sEPSC frequency in cortical neurons; rescued by loxapine [29] |
| C4A/C4B | Complex structural variation in MHC confers risk [60] | Enriched H3K4me3/H3K27ac at C4 loci in neurons [40] | ↑ C4A within immune co-expression modules [18] | ↑ Serum C4A/C4B in treatment responders [62] | — |
| NRXN1/NLGN1 | NRXN1 deletions and NLGN1 GWAS signals [20] | — | ↓ NRXN1 and NLGN1 transcripts in NPCs [53] | — | ↓ Presynaptic puncta in cortical [29] and glutamatergic neurons [55,59] |
| MT-CO1/ATP5A1 | Rare variants in ETC genes; 16p11.2 CNV [20,21] | — | ↓ OXPHOS transcripts in cortex [44] | ↓ Complex I–V subunits in ACC [47] | ↑ Mitochondrial fragmentation, ↓ ΔΨm, ↑ ROS in neurons [58] |
| RELN | — | Promoter hypermethylation in BA9 [88,89] | ↓ RELN mRNA in DLPFC [88,89] | — | VPA restores H3K9ac at RELN promoter and increases mRNA [93] |
| POU3F2 (BRN2)/PTN | POU3F2/PTN risk loci in schizophrenia organoids [57] | Placental DMRs at PAX6/SOX2, altered developmental histone marks [38] | Disrupted NPC and neuronal differentiation modules [53,66] | ↓ BRN2 and PTN proteins in organoids [57] | Exogenous PTN or BRN2 rescues progenitor survival and neuronal output [57] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saada, M.; Stern, S. Molecular Signatures of Schizophrenia and Insights into Potential Biological Convergence. Int. J. Mol. Sci. 2025, 26, 9830. https://doi.org/10.3390/ijms26199830
Saada M, Stern S. Molecular Signatures of Schizophrenia and Insights into Potential Biological Convergence. International Journal of Molecular Sciences. 2025; 26(19):9830. https://doi.org/10.3390/ijms26199830
Chicago/Turabian StyleSaada, Malak, and Shani Stern. 2025. "Molecular Signatures of Schizophrenia and Insights into Potential Biological Convergence" International Journal of Molecular Sciences 26, no. 19: 9830. https://doi.org/10.3390/ijms26199830
APA StyleSaada, M., & Stern, S. (2025). Molecular Signatures of Schizophrenia and Insights into Potential Biological Convergence. International Journal of Molecular Sciences, 26(19), 9830. https://doi.org/10.3390/ijms26199830






