Preexisting Genetic Background Primes the Responses of Human Neurons to Amyloid β
Abstract
1. Introduction
2. Results
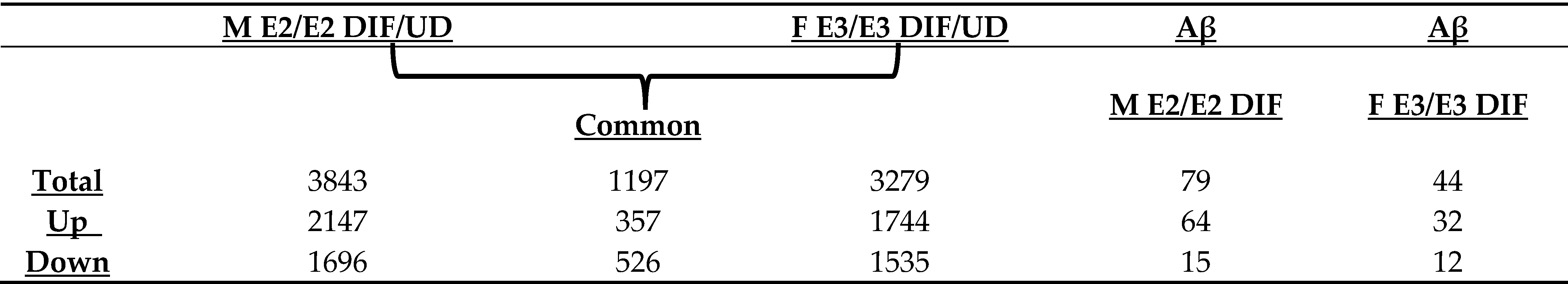
2.1. Genome-Wide RNA-Seq Analysis Identified a Substantial Number of Genes That Were Up- or Down-Regulated in the Same Manner During Neuronal Differentiation of Two Genetically Distinct iPSC Cell Lines
2.2. Aβ-Induced Genes Are Predominantly Suppressed During Neuronal Differentiation
2.3. Pathway Analysis Shows That Aβ Induces a Network of Genes Promoting Inflammatory Responses and Cell Migration Selectively in M E2/E2 Neurons
3. Discussion
4. Materials and Methods
4.1. iPSC Cell Culture and Differentiation
 |
4.2. Oligomeric Aβ Preparation
4.3. Immunocytochemistry
4.4. RNA Sequencing (RNA-Seq) Analysis
4.5. Gene Ontology (GO) and Network Analysis of Transcriptomic Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s Disease |
| NFT | Neurofibrillary Tangles |
| EOAD | Early Onset Alzheimer’s Disease |
| LOAD | Late Onset Alzheimer’s Disease |
| APP | Amyloid Precursor Protein |
| PSEN1 | Presenilin 1 |
| PSEN2 | Presenilin 2 |
| APOE | Apolipoprotein E gene/alleles |
| Aβ | Amyloid Beta |
| BACE1 | Β-Secretase |
| iPSC | Induced pluripotent stem cells |
| RNA | RNA Sequencing |
| CTF-β | C-terminal domain fragment beta |
| ESC | Embryonic stem cells |
References
- Alzheimer’s Association. 2024 Alzheimer’s Disease Facts and Figures. Alzheimers Dement 2024, 20, 3708–3821. [Google Scholar] [CrossRef]
- WHO. Dementia: Key Facts; World Health Organization: Geneva, Switzerland, 2025. [Google Scholar]
- Alzheimer, A. Neuere Arbeiten Über Die Dementia Senilis Und Die Auf Atheromatöser Gefässerkrankung Basierenden Gehirnkrankheiten. Monatsschrift Für Psychiatr. Und Neurol. 2009, 3, 101–115. [Google Scholar] [CrossRef]
- Alzheimer, A. Uber Eigenartige Erkrankung Der Hirnrinde. All Z Psychiatr 1907, 64, 146–148. [Google Scholar]
- Leslie, C.; Masliah, E. Molecular Mechanisms of Neurodegeneration in Alzheimer’s Disease. Human. Mol. Genet. 2010, 19, R12–R20. [Google Scholar]
- Selkoe, D.J. Alzheimer’s Disease. Cold Spring Harb. Perspect. Biol. 2011, 3, a004457. [Google Scholar] [CrossRef] [PubMed]
- Kaj, B.; de Leon, M.J.; Zetterberg, H. Alzheimer’s Disease. Lancet 2006, 368, 387–403. [Google Scholar]
- Valdez-Gaxiola, C.A.; Rosales-Leycegui, F.; Gaxiola-Rubio, A.; Moreno-Ortiz, J.M.; Figuera, L.E. Early- and Late-Onset Alzheimer’s Disease: Two Sides of the Same Coin? Diseases 2024, 12, 110. [Google Scholar] [CrossRef]
- Tellechea, P.; Pujol, N.; Esteve-Belloch, P.; Echeveste, B.; García-Eulate, M.; Arbizu, J.; Riverol, M. Early- and Late-Onset Alzheimer Disease: Are They the Same Entity? Neurologia 2018, 33, 244–253. [Google Scholar] [CrossRef]
- Mendez, M.F. Early-Onset Alzheimer Disease and Its Variants. Continuum (Minneap. Minn.) 2019, 25, 34–51. [Google Scholar] [CrossRef]
- Bird, T.D. Genetic Aspects of Alzheimer Disease. Genet. Med. 2008, 10, 231–239. [Google Scholar] [CrossRef]
- Cacace, R.; Sleegers, K.; Van Broeckhoven, C. Molecular Genetics of Early-Onset Alzheimer’s Disease Revisited. Alzheimer’s Dement. 2016, 12, 733–748. [Google Scholar] [CrossRef]
- Ayodele, T.; Rogaeva, E.; Kurup, J.T.; Beecham, G.; Reitz, C. Early-Onset Alzheimer’s Disease: What Is Missing in Research? Curr. Neurol. Neurosci. Rep. 2021, 21, 4. [Google Scholar] [CrossRef] [PubMed]
- Hoogmartens, J.; Cacace, R.; Van Broeckhoven, C. Insight into the Genetic Etiology of Alzheimer’s Disease: A Comprehensive Review of the Role of Rare Variants. Alzheimers Dement. 2021, 13, e12155. [Google Scholar] [CrossRef] [PubMed]
- Mielke, M.M. Sex and Gender Differences in Alzheimer’s Disease Dementia. Psychiatr. Times 2018, 35, 14–17. [Google Scholar] [PubMed]
- van der Lee, S.J.; Wolters, F.J.; Ikram, M.K.; Hofman, A.; Ikram, M.A.; Amin, N.; van Duijn, C.M. The Effect of Apoe and Other Common Genetic Variants on the Onset of Alzheimer’s Disease and Dementia: A Community-Based Cohort Study. Lancet Neurol. 2018, 17, 434–444, Erratum in Lancet Neurol. 2018, 17, 495. [Google Scholar] [CrossRef]
- Deming, Y.; Vasiljevic, E.; Morrow, A.; Miao, J.; Van Hulle, C.; Jonaitis, E.; Ma, Y.; Whitenack, V.; Kollmorgen, G.; Wild, N.; et al. Neuropathology-Based Apoe Genetic Risk Score Better Quantifies Alzheimer’s Risk. Alzheimers Dement. 2023, 19, 3406–3416. [Google Scholar] [CrossRef]
- Huang, Y.; Mahley, R.W. Apolipoprotein E: Structure and Function in Lipid Metabolism, Neurobiology, and Alzheimer’s Diseases. Neurobiol. Dis. 2014, 72 Pt. A, 3–12. [Google Scholar] [CrossRef]
- Troutwine, B.R.; Hamid, L.; Lysaker, C.R.; Strope, T.A.; Wilkins, H.M. Apolipoprotein E and Alzheimer’s Disease. Acta Pharm. Sin. B 2022, 12, 496–510. [Google Scholar] [CrossRef]
- Jackson, R.J.; Hyman, B.T.; Serrano-Pozo, A. Multifaceted Roles of Apoe in Alzheimer Disease. Nat. Rev. Neurol. 2024, 20, 457–474. [Google Scholar] [CrossRef]
- Cho, H.; Seo, S.W.; Kim, J.-H.; Suh, M.K.; Lee, J.-H.; Choe, Y.S.; Lee, K.-H.; Kim, J.S.; Kim, G.H.; Noh, Y.; et al. Amyloid Deposition in Early Onset Versus Late Onset Alzheimer’s Disease. J. Alzheimer’s Dis. 2013, 35, 813–821. [Google Scholar] [CrossRef]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. Nia-Aa Research Framework: Toward a Biological Definition of Alzheimer’s Disease. Alzheimer Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef] [PubMed]
- Hammers, D.B.; Eloyan, A.; Thangarajah, M.; Taurone, A.; Beckett, L.; Gao, S.; Polsinelli, A.J.; Kirby, K.; Dage, J.L.; Nudelman, K.; et al. Differences in Baseline Cognitive Performance between Participants with Early-Onset and Late-Onset Alzheimer’s Disease: Comparison of Leads and Adni. Alzheimer’s Dement. 2025, 21, e14218. [Google Scholar] [CrossRef] [PubMed]
- Coronel, R.; Bernabeu-Zornoza, A.; Palmer, C.; Muñiz-Moreno, M.; Zambrano, A.; Cano, E.; Liste, I. Role of Amyloid Precursor Protein (App) and Its Derivatives in the Biology and Cell Fate Specification of Neural Stem Cells. Mol. Neurobiol. 2018, 55, 7107–7117. [Google Scholar] [CrossRef] [PubMed]
- Takahisa, K.; Xu, H.; Bu, G. Apoe and Aβ in Alzheimer’s Disease: Accidental Encounters or Partners? Neuron 2014, 81, 740–754. [Google Scholar]
- Gonneaud, J.; Arenaza-Urquijo, E.M.; Fouquet, M.; Perrotin, A.; Fradin, S.; de La Sayette, V.; Eustache, F.; Chételat, G. Relative Effect of Apoe Ε4 on Neuroimaging Biomarker Changes across the Lifespan. Neurology 2016, 87, 1696–1703. [Google Scholar] [CrossRef]
- Fleisher, A.S.; Chen, K.; Liu, X.; Ayutyanont, N.; Roontiva, A.; Thiyyagura, P.; Protas, H.; Joshi, A.D.; Sabbagh, M.; Sadowsky, C.H.; et al. Apolipoprotein E Ε4 and Age Effects on Florbetapir Positron Emission Tomography in Healthy Aging and Alzheimer Disease. Neurobiol. Aging 2013, 34, 1–12. [Google Scholar] [CrossRef]
- Liu, C.-C.; Zhao, N.; Fu, Y.; Wang, N.; Linares, C.; Tsai, C.-W.; Bu, G. Apoe4 Accelerates Early Seeding of Amyloid Pathology. Neuron 2017, 96, 1024–1032.e3. [Google Scholar]
- Liu, C.-C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer Disease: Risk, Mechanisms and Therapy. Nat. Rev. Neurol. 2013, 9, 106–118, Correction in Nat. Rev. Neurol. 2013, 9, 184. [Google Scholar] [CrossRef]
- Bernabeu-Zornoza, A.; Coronel, R.; Palmer, C.; Monteagudo, M.; Zambrano, A.; Liste, I. Physiological and Pathological Effects of Amyloid-Β Species in Neural Stem Cell Biology. Neural Regen. Res. 2019, 14, 2035–2042. [Google Scholar] [CrossRef]
- Fontana, I.C.; Zimmer, A.R.; Rocha, A.S.; Gosmann, G.; Souza, D.O.; Lourenco, M.V.; Ferreira, S.T.; Zimmer, E.R. Amyloid-Β Oligomers in Cellular Models of Alzheimer’s Disease. J. Neurochem. 2020, 155, 348–369. [Google Scholar] [CrossRef]
- Tolar, M.; Hey, J.; Power, A.; Abushakra, S. Neurotoxic Soluble Amyloid Oligomers Drive Alzheimer’s Pathogenesis and Represent a Clinically Validated Target for Slowing Disease Progression. Int. J. Mol. Sci. 2021, 22, 6355. [Google Scholar] [CrossRef]
- Canevari, L.; Abramov, A.Y.; Duchen, M.R. Toxicity of Amyloid Β Peptide: Tales of Calcium, Mitochondria, and Oxidative Stress. Neurochem. Res. 2004, 29, 637–650. [Google Scholar] [CrossRef]
- Bode, D.C.; Baker, M.D.; Viles, J.H. Ion Channel Formation by Amyloid-β42 Oligomers but Not Amyloid-β40 in Cellular Membranes. J. Biol. Chem. 2017, 292, 1404–1413. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jin, M.; Koeglsperger, T.; Shepardson, N.E.; Shankar, G.M.; Selkoe, D.J. Soluble Aβ Oligomers Inhibit Long-Term Potentiation through a Mechanism Involving Excessive Activation of Extrasynaptic Nr2b-Containing Nmda Receptors. J. Neurosci. 2011, 31, 6627–6638. [Google Scholar] [CrossRef] [PubMed]
- Zott, B.; Simon, M.M.; Hong, W.; Unger, F.; Chen-Engerer, H.J.; Frosch, M.P.; Sakmann, B.; Walsh, D.M.; Konnerth, A. A Vicious Cycle of Β Amyloid-Dependent Neuronal Hyperactivation. Science 2019, 365, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Christopher, H.D.; Swanson, C.J.; Aisen, P.; Bateman, R.J.; Chen, C.; Gee, M.; Kanekiyo, M.; Li, D.; Reyderman, L.; Cohen, S.; et al. Lecanemab in Early Alzheimer’s Disease. N. Engl. J. Med. 2023, 388, 9–21. [Google Scholar]
- Hossain, F.; Husna, A.U.; Kharel, M. Use of Lecanemab for the Treatment of Alzheimer’s Disease: A Systematic Review. Brain Behav. 2024, 14, e3592. [Google Scholar] [CrossRef]
- Gueorguieva, I.; Chow, K.; Chua, L.; Shcherbinin, S.; Zimmer, J.A.; Evans, C.D.; Wang, H.; Nery, E.S.M.; Brooks, D.A.; Sims, J.R. Donanemab Exposure-Response in Early Symptomatic Alzheimer’s Disease. Alzheimer’s Dement. 2025, 21, e70491. [Google Scholar] [CrossRef]
- Weglinski, C.; Jeans, A. Amyloid-Β in Alzheimer’s Disease—Front and Centre after All? Neuronal Signal 2023, 7, Ns20220086. [Google Scholar] [CrossRef]
- Lapasset, L.; Milhavet, O.; Prieur, A.; Besnard, E.; Babled, A.; Aït-Hamou, N.; Leschik, J.; Pellestor, F.; Ramirez, J.-M.; De Vos, J.; et al. Rejuvenating Senescent and Centenarian Human Cells by Reprogramming through the Pluripotent State. Genes. Dev. 2011, 25, 2248–2253. [Google Scholar] [CrossRef]
- Lee, J.; Bignone, P.A.; Coles, L.; Liu, Y.; Snyder, E.; Larocca, D. Induced Pluripotency and Spontaneous Reversal of Cellular Aging in Supercentenarian Donor Cells. Biochem. Biophys. Res. Commun. 2020, 525, 563–569. [Google Scholar] [CrossRef]
- Aversano, S.; Caiazza, C.; Caiazzo, M. Induced Pluripotent Stem Cell-Derived and Directly Reprogrammed Neurons to Study Neurodegenerative Diseases: The Impact of Aging Signatures. Front. Aging Neurosci. 2022, 14, 1069482. [Google Scholar] [CrossRef] [PubMed]
- Kinney, J.W.; Bemiller, S.M.; Murtishaw, A.S.; Leisgang, A.M.; Salazar, A.M.; Lamb, B.T. Inflammation as a Central Mechanism in Alzheimer’s Disease. Alzheimers Dement. 2018, 4, 575–590. [Google Scholar] [CrossRef] [PubMed]
- Nasb, M.; Tao, W.; Chen, N. Alzheimer’s Disease Puzzle: Delving into Pathogenesis Hypotheses. Aging Dis. 2024, 15, 43–73. [Google Scholar] [PubMed]
- Wang, C.; Zong, S.; Cui, X.; Wang, X.; Wu, S.; Wang, L.; Liu, Y.; Lu, Z. The Effects of Microglia-Associated Neuroinflammation on Alzheimer’s Disease. Front. Immunol. 2023, 14, 1117172. [Google Scholar] [CrossRef]
- Young-Pearse, T.L.; Bai, J.; Chang, R.; Zheng, J.B.; LoTurco, J.J.; Selkoe, D.J. A Critical Function for Beta-Amyloid Precursor Protein in Neuronal Migration Revealed by in Utero Rna Interference. J. Neurosci. 2007, 27, 14459–14469. [Google Scholar] [CrossRef]
- Pfundstein, G.; Nikonenko, A.G.; Sytnyk, V. Amyloid Precursor Protein (App) and Amyloid Β (Aβ) Interact with Cell Adhesion Molecules: Implications in Alzheimer’s Disease and Normal Physiology. Front. Cell Dev. Biol. 2022, 10, 969547. [Google Scholar] [CrossRef]
- Hohsfield, L.A.; Humpel, C. Migration of Blood Cells to Β-Amyloid Plaques in Alzheimer’s Disease. Exp. Gerontol. Exp. Gerontol. 2015, 65, 8–15. [Google Scholar] [CrossRef]
- Duara, R.; Barker, W. Heterogeneity in Alzheimer’s Disease Diagnosis and Progression Rates: Implications for Therapeutic Trials. Neurotherapeutics 2022, 19, 8–25. [Google Scholar] [CrossRef]
- Ferreira, D.; Wahlund, L.O.; Westman, E. The Heterogeneity within Alzheimer’s Disease. Aging 2018, 10, 3058–3060. [Google Scholar] [CrossRef]
- Gordon, A.; Benzinger, T.L.S.; Sotiras, A.; Gordon, B.A. Unraveling Alzheimer’s Disease Heterogeneity: A Comparative Analysis Using Hydra and Chimera. Alzheimer’s Dement. 2024, 20, e094135. [Google Scholar]
- Arenaza-Urquijo, E.M.; Boyle, R.; Casaletto, K.; Anstey, K.J.; Vila-Castelar, C.; Colverson, A.; Palpatzis, E.; Eissman, J.M.; Ng, T.K.S.; Raghavan, S.; et al. Sex and Gender Differences in Cognitive Resilience to Aging and Alzheimer’s Disease. Alzheimers Dement. 2024, 20, 5695–5719. [Google Scholar] [CrossRef]
- Wang, Q.; Tian, J.; Chen, H.; Du, H.; Guo, L. Amyloid Beta-Mediated Kif5a Deficiency Disrupts Anterograde Axonal Mitochondrial Movement. Neurobiol. Dis. 2019, 127, 410–418. [Google Scholar] [CrossRef]
- Marchenko, S.; Flanagan, L. Immunocytochemistry: Human Neural Stem Cells. J. Vis. Exp. 2007, 267. [Google Scholar] [CrossRef]

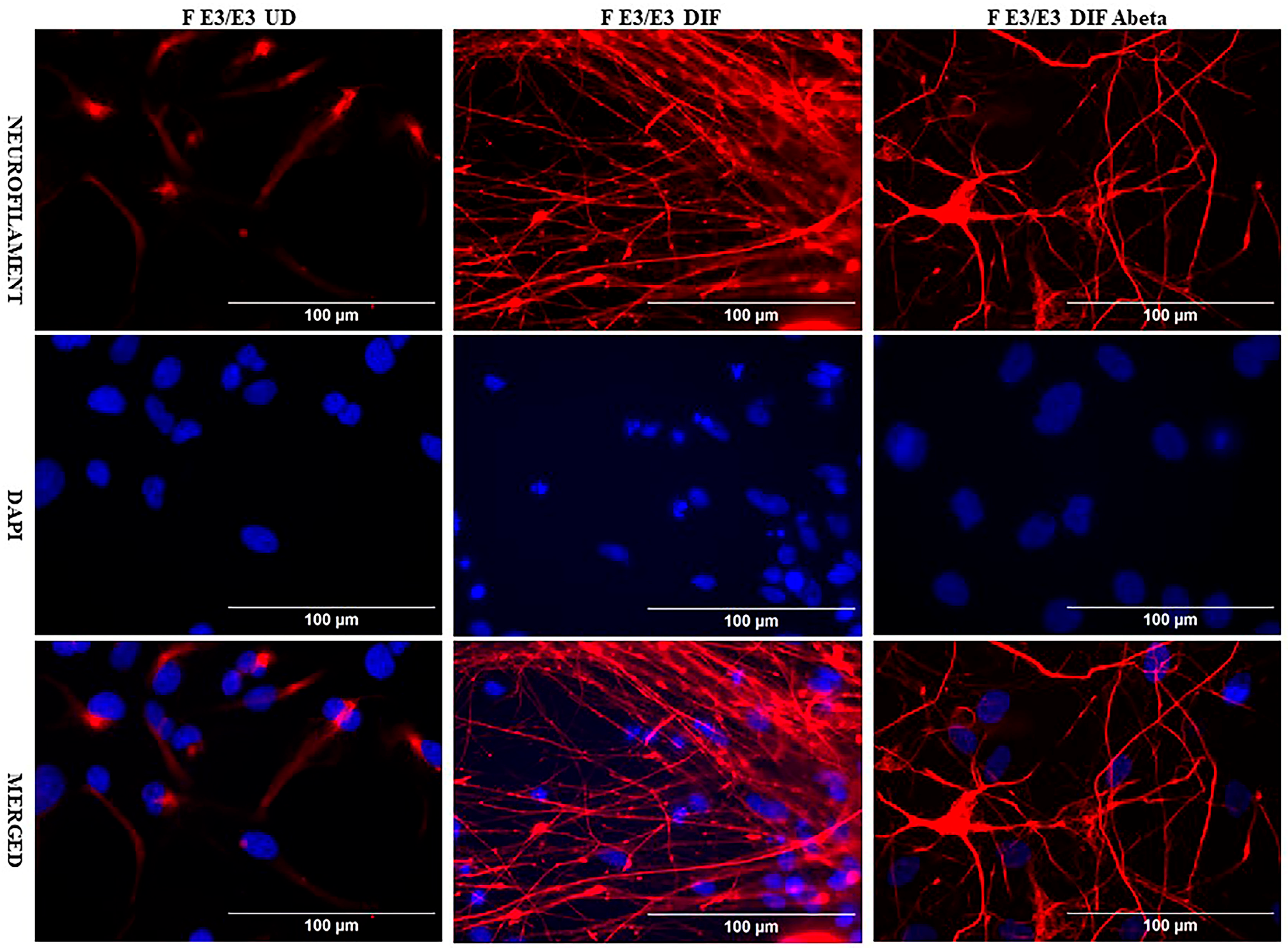
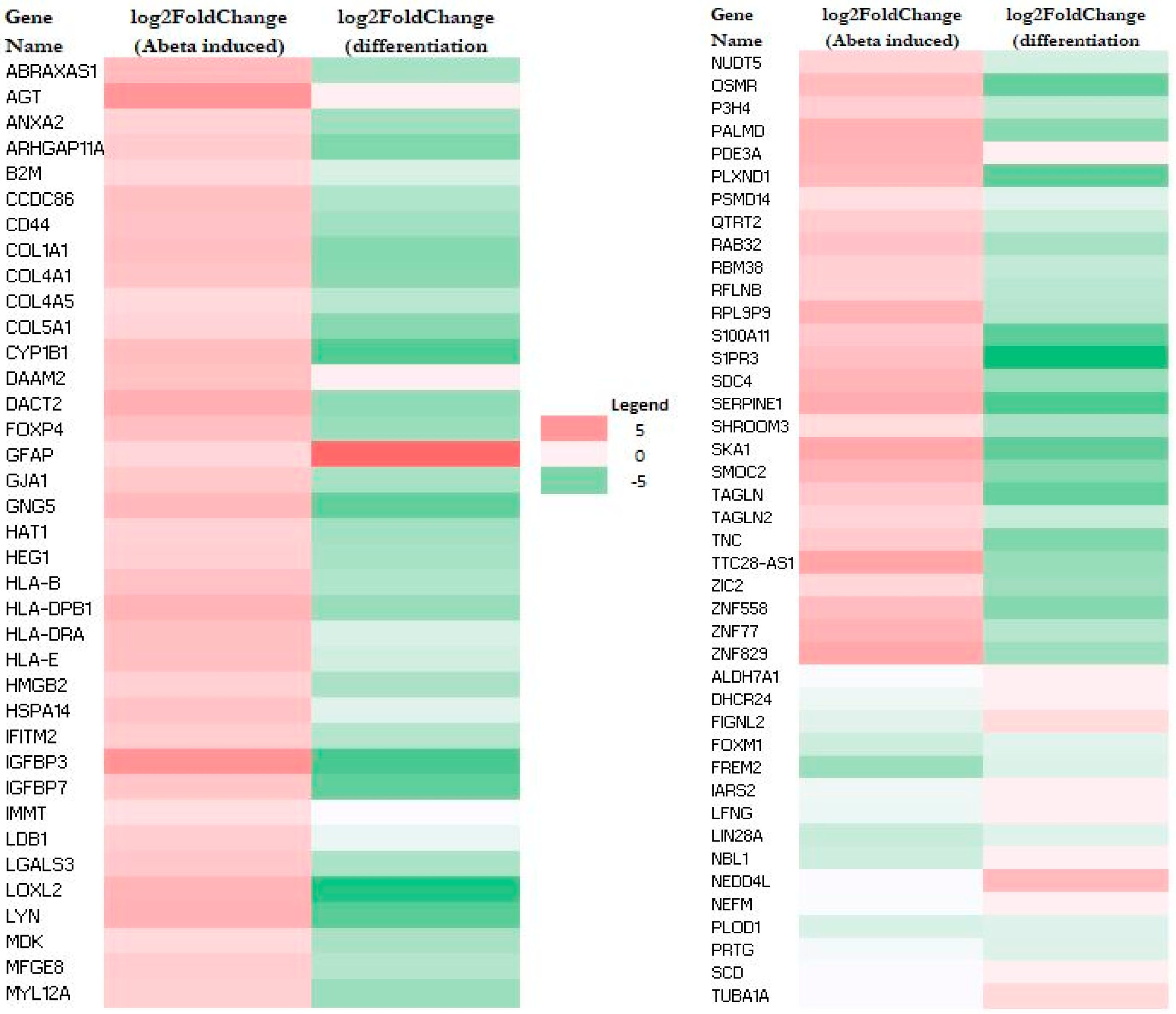
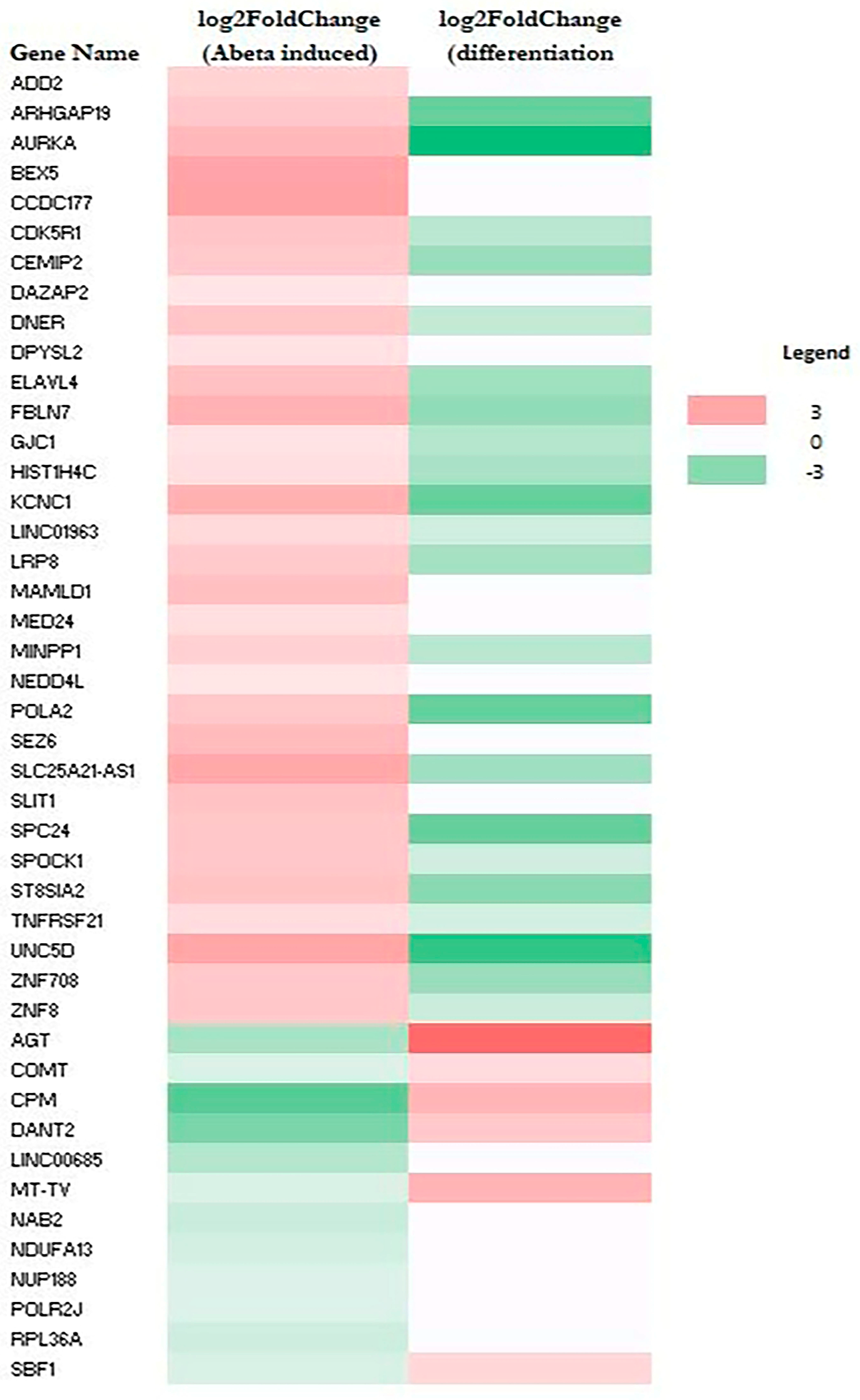

| Name | log2FoldChange | Expected a | Molecule Type | Location |
|---|---|---|---|---|
| TNF pathway | ||||
| AGT | 5.05 | Down | growth factor | Extracellular Space |
| B2M | 1.4 | Up | transmembrane receptor | Plasma Membrane |
| CD44 | 2.51 | Up | other | Plasma Membrane |
| COL1A1 | 2.72 | Up | other | Extracellular Space |
| COL4A1 | 2.4 | Up | other | Extracellular Space |
| COL5A1 | 1.55 | Up | other | Extracellular Space |
| CYP1B1 | 2.82 | Up | enzyme | Cytoplasm |
| GFAP | 1.34 | Up | other | Cytoplasm |
| GJA1 | 2.14 | - - | transporter | Plasma Membrane |
| HLA-B | 2.57 | Up | transmembrane receptor | Plasma Membrane |
| HLA-DRA | 2.62 | Up | transmembrane receptor | Plasma Membrane |
| HLA-E | 2.63 | Up | transmembrane receptor | Plasma Membrane |
| IFITM2 | 1.96 | Up | other | Cytoplasm |
| IGFBP3 | 5.19 | Up | other | Extracellular Space |
| LFNG | −1.35 | Down | enzyme | Cytoplasm |
| LGALS3 | 2.22 | Down | other | Extracellular Space |
| LOXL2 | 3.31 | Up | enzyme | Nucleus |
| LYN | 3.47 | - - | kinase | Cytoplasm |
| NEFM | −0.92 | Up | other | Plasma Membrane |
| OSMR | 2.88 | Up | transmembrane receptor | Plasma Membrane |
| RAB32 | 2.52 | Up | enzyme | Cytoplasm |
| S1PR3 | 2.78 | Up | G-protein coupled receptor | Plasma Membrane |
| SDC4 | 3.3 | Up | other | Plasma Membrane |
| SERPINE1 | 3.82 | Up | other | Extracellular Space |
| TAGLN | 2.19 | Down | other | Cytoplasm |
| TNC | 2.13 | Up | other | Extracellular Space |
| IL1β pathway | ||||
| ALDH7A1 | −0.88 | - - | enzyme | Cytoplasm |
| B2M | 1.4 | Up | transmembrane receptor | Plasma Membrane |
| CD44 | 2.51 | Up | other | Plasma Membrane |
| COL1A1 | 2.72 | Down | other | Extracellular Space |
| CYP1B1 | 2.82 | Up | enzyme | Cytoplasm |
| DAAM2 | 2.6 | Up | other | Cytoplasm |
| GJA1 | 2.14 | Up | transporter | Plasma Membrane |
| HLA-B | 2.57 | Up | transmembrane receptor | Plasma Membrane |
| HLA-DRA | 2.62 | - - | transmembrane receptor | Plasma Membrane |
| HLA-E | 2.63 | Up | transmembrane receptor | Plasma Membrane |
| IFITM2 | 1.96 | Up | other | Cytoplasm |
| IGFBP3 | 5.19 | - - | other | Extracellular Space |
| LYN | 3.47 | Up | kinase | Cytoplasm |
| OSMR | 2.88 | Up | transmembrane receptor | Plasma Membrane |
| S1PR3 | 2.78 | Up | G-protein coupled receptor | Plasma Membrane |
| SDC4 | 3.3 | Up | other | Plasma Membrane |
| SERPINE1 | 3.82 | Up | other | Extracellular Space |
| TGFβ1 pathway | ||||
| ANXA2 | 1.62 | Up | other | Plasma Membrane |
| B2M | 1.4 | - - | transmembrane receptor | Plasma Membrane |
| CD44 | 2.51 | Up | other | Plasma Membrane |
| COL1A1 | 2.72 | Up | other | Extracellular Space |
| COL4A1 | 2.4 | Up | other | Extracellular Space |
| COL5A1 | 1.55 | Up | other | Extracellular Space |
| DHCR24 | −1.37 | Down | enzyme | Cytoplasm |
| FOXM1 | −2.45 | Up | transcription regulator | Nucleus |
| GFAP | 1.34 | Down | other | Cytoplasm |
| GJA1 | 2.14 | Up | transporter | Plasma Membrane |
| HAT1 | 1.65 | - - | enzyme | Nucleus |
| HLA-DRA | 2.62 | Up | transmembrane receptor | Plasma Membrane |
| IGFBP3 | 5.19 | Up | other | Extracellular Space |
| IGFBP7 | 2.28 | Up | transporter | Extracellular Space |
| LDB1 | 1.89 | - - | transcription regulator | Nucleus |
| LGALS3 | 2.22 | Down | other | Extracellular Space |
| LOXL2 | 3.31 | Up | enzyme | Nucleus |
| PLOD1 | −2.01 | Up | enzyme | Cytoplasm |
| RFLNB | 1.71 | Up | other | Unknown |
| S100A11 | 2.18 | Up | other | Cytoplasm |
| S1PR3 | 2.78 | Up | G-protein coupled receptor | Plasma Membrane |
| SCD | −0.86 | Up | enzyme | Cytoplasm |
| SDC4 | 3.3 | - - | other | Plasma Membrane |
| SERPINE1 | 3.82 | Up | other | Extracellular Space |
| SMOC2 | 3.23 | - - | other | Extracellular Space |
| TAGLN | 2.19 | Up | other | Cytoplasm |
| TAGLN2 | 1.51 | Up | other | Cytoplasm |
| TNC | 2.13 | Up | other | Extracellular Space |
| TUBA1A | −0.86 | Up | other | Cytoplasm |
| IL4 | ||||
| ANXA2 | 1.62 | Up | other | Plasma Membrane |
| CCDC86 | 2.67 | Up | other | Nucleus |
| CD44 | 2.51 | Up | other | Plasma Membrane |
| COL1A1 | 2.72 | Up | other | Extracellular Space |
| GJA1 | 2.14 | Down | transporter | Plasma Membrane |
| HLA-DRA | 2.62 | Up | transmembrane receptor | Plasma Membrane |
| HLA-E | 2.63 | Up | transmembrane receptor | Plasma Membrane |
| LFNG | −1.35 | Up | enzyme | Cytoplasm |
| LGALS3 | 2.22 | Up | other | Extracellular Space |
| SERPINE1 | 3.82 | Up | other | Extracellular Space |
| SKA1 | 4.05 | Up | other | Nucleus |
| TNC | 2.13 | Up | other | Extracellular Space |
| log2FoldChange | Expected a | Molecule Type | Location | |
|---|---|---|---|---|
| migration of cells | ||||
| AGT | 5.05 | Up | growth factor | Extracellular Space |
| ANXA2 | 1.62 | Up | other | Plasma Membrane |
| ARHGAP11A | 2.02 | Up | other | Cytoplasm |
| CD44 | 2.51 | Up | other | Plasma Membrane |
| COL1A1 | 2.72 | Up | other | Extracellular Space |
| COL4A1 | 2.4 | Up | other | Extracellular Space |
| COL5A1 | 1.55 | Up | other | Extracellular Space |
| CYP1B1 | 2.82 | Up | enzyme | Cytoplasm |
| DAAM2 | 2.6 | Up | other | Cytoplasm |
| DACT2 | 3.62 | Down | other | Cytoplasm |
| FIGNL2 | −1.77 | Down | other | Plasma Membrane |
| FOXM1 | −2.45 | Up | transcription regulator | Nucleus |
| FOXP4 | 2.72 | Up | transcription regulator | Nucleus |
| GFAP | 1.34 | Up | other | Cytoplasm |
| GJA1 | 2.14 | Up | transporter | Plasma Membrane |
| IGFBP3 | 5.19 | Down | other | Extracellular Space |
| IGFBP7 | 2.28 | Down | transporter | Extracellular Space |
| LDB1 | 1.89 | - - | transcription regulator | Nucleus |
| LFNG | −1.35 | Up | enzyme | Cytoplasm |
| LGALS3 | 2.22 | Up | other | Extracellular Space |
| LIN28A | −2.61 | Up | other | Cytoplasm |
| LOXL2 | 3.31 | Up | enzyme | Nucleus |
| LYN | 3.47 | Up | kinase | Cytoplasm |
| MDK | 1.24 | Up | growth factor | Extracellular Space |
| MFGE8 | 1.96 | Up | other | Extracellular Space |
| NBL1 | −2.4 | Down | other | Extracellular Space |
| NEDD4L | −0.86 | Down | enzyme | Cytoplasm |
| OSMR | 2.88 | Up | transmembrane receptor | Plasma Membrane |
| PLOD1 | −2.01 | Up | enzyme | Cytoplasm |
| PLXND1 | 3.02 | Up | transmembrane receptor | Plasma Membrane |
| S100A11 | 2.18 | Up | other | Cytoplasm |
| S1PR3 | 2.78 | Up | G-protein coupled receptor | Plasma Membrane |
| SCD | −0.86 | Up | enzyme | Cytoplasm |
| SDC4 | 3.3 | Up | other | Plasma Membrane |
| SERPINE1 | 3.82 | Up | other | Extracellular Space |
| SKA1 | 4.05 | Up | other | Nucleus |
| SMOC2 | 3.23 | Up | other | Extracellular Space |
| TAGLN2 | 1.51 | Down | other | Cytoplasm |
| TNC | 2.13 | Up | other | Extracellular Space |
| TUBA1A | −0.86 | - - | other | Cytoplasm |
| ZIC2 | 1.36 | Up | transcription regulator | Nucleus |
| Cell movement | ||||
| AGT | 5.05 | Up | growth factor | Extracellular Space |
| ANXA2 | 1.62 | Up | other | Plasma Membrane |
| ARHGAP11A | 2.02 | Up | other | Cytoplasm |
| CD44 | 2.51 | Up | other | Plasma Membrane |
| COL1A1 | 2.72 | Up | other | Extracellular Space |
| COL4A1 | 2.4 | Up | other | Extracellular Space |
| COL5A1 | 1.55 | Up | other | Extracellular Space |
| CYP1B1 | 2.82 | Up | enzyme | Cytoplasm |
| DAAM2 | 2.6 | Up | other | Cytoplasm |
| DACT2 | 3.62 | Down | other | Cytoplasm |
| FIGNL2 | −1.77 | Down | other | Plasma Membrane |
| FOXM1 | −2.45 | Up | transcription regulator | Nucleus |
| FOXP4 | 2.72 | Up | transcription regulator | Nucleus |
| GFAP | 1.34 | Up | other | Cytoplasm |
| GJA1 | 2.14 | Up | transporter | Plasma Membrane |
| HMGB2 | 1.73 | Up | transcription regulator | Nucleus |
| IGFBP3 | 5.19 | - - | other | Extracellular Space |
| IGFBP7 | 2.28 | Down | transporter | Extracellular Space |
| IMMT | 0.96 | Down | other | Cytoplasm |
| LDB1 | 1.89 | - - | transcription regulator | Nucleus |
| LFNG | −1.35 | Up | enzyme | Cytoplasm |
| LGALS3 | 2.22 | Up | other | Extracellular Space |
| LIN28A | −2.61 | Up | other | Cytoplasm |
| LOXL2 | 3.31 | Up | enzyme | Nucleus |
| LYN | 3.47 | Up | kinase | Cytoplasm |
| MDK | 1.24 | Up | growth factor | Extracellular Space |
| MFGE8 | 1.96 | Up | other | Extracellular Space |
| NBL1 | −2.4 | Down | other | Extracellular Space |
| NEDD4L | −0.86 | Down | enzyme | Cytoplasm |
| OSMR | 2.88 | Up | transmembrane receptor | Plasma Membrane |
| PLOD1 | −2.01 | Up | enzyme | Cytoplasm |
| PLXND1 | 3.02 | Up | transmembrane receptor | Plasma Membrane |
| S100A11 | 2.18 | Up | other | Cytoplasm |
| S1PR3 | 2.78 | Up | G-protein coupled receptor | Plasma Membrane |
| SCD | −0.86 | Up | enzyme | Cytoplasm |
| SDC4 | 3.3 | Up | other | Plasma Membrane |
| SERPINE1 | 3.82 | Up | other | Extracellular Space |
| SKA1 | 4.05 | Up | other | Nucleus |
| SMOC2 | 3.23 | Up | other | Extracellular Space |
| TAGLN2 | 1.51 | Down | other | Cytoplasm |
| TNC | 2.13 | - - | other | Extracellular Space |
| TUBA1A | −0.86 | - - | other | Cytoplasm |
| ZIC2 | 1.36 | Up | transcription regulator | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soladogun, A.S.; Zhang, L. Preexisting Genetic Background Primes the Responses of Human Neurons to Amyloid β. Int. J. Mol. Sci. 2025, 26, 9804. https://doi.org/10.3390/ijms26199804
Soladogun AS, Zhang L. Preexisting Genetic Background Primes the Responses of Human Neurons to Amyloid β. International Journal of Molecular Sciences. 2025; 26(19):9804. https://doi.org/10.3390/ijms26199804
Chicago/Turabian StyleSoladogun, Adedamola Saidi, and Li Zhang. 2025. "Preexisting Genetic Background Primes the Responses of Human Neurons to Amyloid β" International Journal of Molecular Sciences 26, no. 19: 9804. https://doi.org/10.3390/ijms26199804
APA StyleSoladogun, A. S., & Zhang, L. (2025). Preexisting Genetic Background Primes the Responses of Human Neurons to Amyloid β. International Journal of Molecular Sciences, 26(19), 9804. https://doi.org/10.3390/ijms26199804







